An extensive transcriptional regulatory network modulates plant defense chemistry.
Abstract
A key unanswered question in plant biology is how a plant regulates metabolism to maximize performance across an array of biotic and abiotic environmental stresses. In this study, we addressed the potential breadth of transcriptional regulation that can alter accumulation of the defensive glucosinolate metabolites in Arabidopsis (Arabidopsis thaliana). A systematic yeast one-hybrid study was used to identify hundreds of unique potential regulatory interactions with a nearly complete complement of 21 promoters for the aliphatic glucosinolate pathway. Conducting high-throughput phenotypic validation, we showed that >75% of tested transcription factor (TF) mutants significantly altered the accumulation of the defensive glucosinolates. These glucosinolate phenotypes were conditional upon the environment and tissue type, suggesting that these TFs may allow the plant to tune its defenses to the local environment. Furthermore, the pattern of TF/promoter interactions could partially explain mutant phenotypes. This work shows that defense chemistry within Arabidopsis has a highly intricate transcriptional regulatory system that may allow for the optimization of defense metabolite accumulation across a broad array of environments.
An organism’s growth and fitness within its environment are largely determined by its ability to efficiently obtain and utilize energy and elements to create biomass to survive. Central to this process is primary metabolism, which determines the use of energy and chemicals from the environment to produce all of the necessary building blocks for cells and the resulting biomass. To optimize fitness, metabolism must be precisely tuned and coordinated to make the most efficient use of available resources. This basic supposition is central to a wide range of biological fields from the study of organismal growth to the study of interactions with the environment, and has led to strong interest in understanding how metabolism is regulated (Karban and Baldwin, 1997; Smith and Stitt, 2007).
The last decade has seen an upsurge in studies investigating the transcriptional control over metabolism. This has largely been driven by three key areas of focus: regulation of sugar metabolism, amino acid metabolism, and secondary metabolism (Desvergne et al., 2006). In most organisms, Glc levels cause transcriptional reprograming of sugar metabolism and organismal development often by a mitogen-activated protein kinase cascade (Moore et al., 2003; Rolland et al., 2006). Transcriptional control over amino acid metabolism frequently involves General Control Nonderepressible4, whereas other transcriptional activators play a role in coordinating amino acid metabolism (Hope and Struhl, 1986; Neuwald and Landsman, 1997; Li et al., 2013). Within plants, there are also amino acid pathway-specific transcription factors (TFs) known for a couple of plant amino acid biosynthetic pathways (Celenza et al., 2005; Maruyama-Nakashita et al., 2006). In plants, the transcriptional regulation of secondary metabolites has also received significant attention with numerous myeloblastosis (MYB) TFs being identified (Dubos et al., 2010).
The above studies have largely focused on regulation via individual TFs or on a few specific genes within a pathway. Thus, there are a number of unanswered questions surrounding the transcriptional regulation of metabolism. For example, is a pathway a coordinately regulated unit or are there distinctly regulated modules within a pathway? Do converging signals, such as those from defense and growth, converge upstream of a key set of TFs to regulate a pathway or is there any capacity to integrate at the level of the promoters for the regulation of individual genes? Finally, how many TFs have the capacity to interact with a pathway and regulate the level of the metabolite(s) and under which conditions?
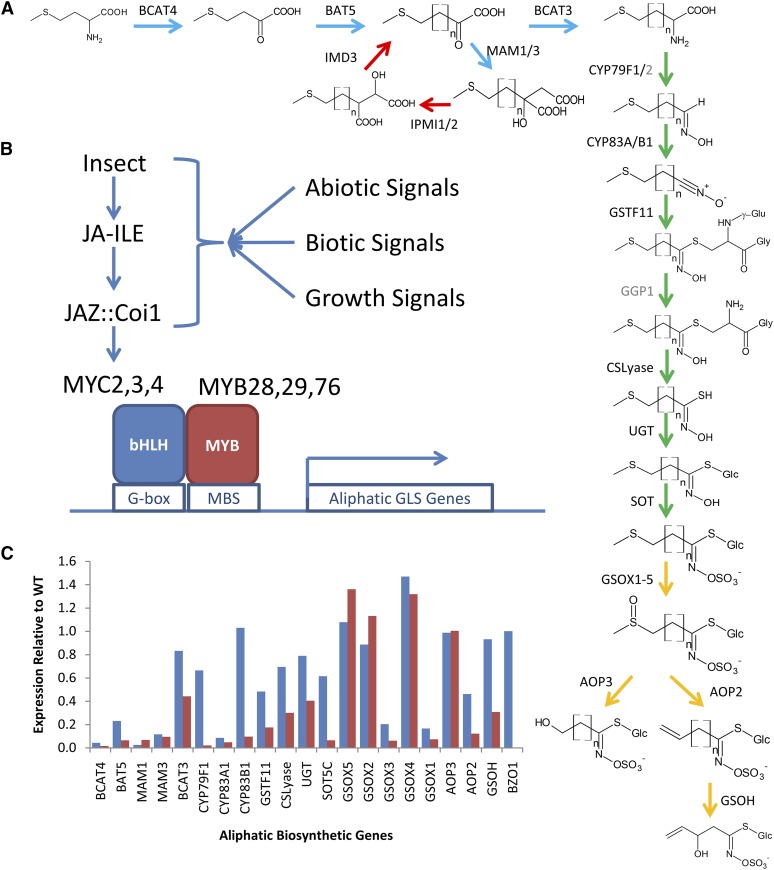
To begin asking these questions, we are using the model metabolic pathway of aliphatic glucosinolates (GLSs) within Arabidopsis (Arabidopsis thaliana; Fig. 1A). This pathway has been used to conduct systems analyses on the function and regulation of metabolism in conjunction with the resulting ecological and evolutionary consequences (Kliebenstein, 2012). Aliphatic GLSs are sulfur-containing secondary metabolites that are the dominant resistance mechanism against chewing insects and a number of bacterial and fungal pathogens (Lambrix et al., 2001; Kliebenstein et al., 2002; Beekwilder et al., 2008; Hansen et al., 2008; Fan et al., 2011; Stotz et al., 2011). The aliphatic GLS pathway also contains genetic variation in enzymatic and regulatory loci that alters fitness at both the local and continental scales (Mauricio, 1998; Kliebenstein et al., 2001a, 2001b, 2001c; Hansen et al., 2007, 2008; Wentzell et al., 2007; Bidart-Bouzat and Kliebenstein, 2008; Chan et al., 2011; Züst et al., 2012). These fitness benefits are constrained by GLS accumulation altering growth and yield, suggesting that the pathway needs fine-tuning to optimize growth and defense (Mauricio, 1998; Lankau and Strauss, 2008; Paul-Victor et al., 2010; Kerwin et al., 2011; Züst et al., 2011). In addition, there is a nearly complete description of the genes and enzymatic steps within the GLS biosynthetic pathways, providing an additional practical advantage to studying these compounds (Fig. 1A; Wittstock and Halkier, 2002; Grubb and Abel, 2006; Halkier and Gershenzon, 2006). Finally, it is possible to cost-effectively, rapidly, and accurately quantify GLSs, further enabling large-scale systems studies (Wentzell et al., 2007; Chan et al., 2010, 2011).
Figure 1.
Proposed aliphatic GLS pathway regulatory model. A, Aliphatic GLS biosynthetic pathway with structures and enzymes. Enzymes in black are ones with promoters included in this study. Those in gray were not cloned. The use of n indicates the number of carbons introduced to the side chain from the elongation cycle and can vary from 1 to 7. The promoters are color coded based on their pathway membership as follows: indolic GLSs are pink, aliphatic regulatory genes are purple, and aliphatic peripheral genes are red. The aliphatic biosynthetic genes are parsed into the core pathway (green), elongation (blue), modifying (yellow), and seed specific (orange) to visualize regulation along the linear model of the pathway. B, A proposed model of aliphatic GLS pathway regulation. In this model, the pathway is regulated by the JA pathway via the bHLH’s MYC2/MYC3/MYC4 interacting with the MYB28/MYB29/MYB76 proteins and binding the respective promoters. C, Transcriptional analysis of the aliphatic GLS pathway in mutants missing the MYB28 and MYB29 TFs (blue) or MYC2/MYC3/MYC4 TFs (red). The value is shown as the relative value of the transcript in the mutant with the wild type set to 1 for each gene. The genes are ordered by their position in the biosynthetic pathway. The transcriptional data presented are from previous publications with full statistical analysis (Sonderby et al., 2010b; Schweizer et al., 2013). WT, Wild type JA-ILE, isoleucyl jasmonic acid; JAZ, jasmonate ZIM domain proteins; Coi1, Coronatine Insensitive1; BCAT4, BRANCHCHAIN AMINOTRANSFERASE4; BAT5, BILE ACID TRANSPORTER5; IMD3, ISOPROPYL MALATE DEHYDROGENASE3; MAM1, METHYLIOALKYLMALATE SYNTHASE1; IPMI1, ISOPROPYL MALATE ISOMERASE1; CYP, CYTOCHROME P450; GGP1, GAMMA GLUTAMYL PEPTIDASE1; GSOH, GLUCOSINOLATE HYDROXYLATION; GSOX1-5, GLUCOSINOLATE OXIDASE1-5; GSTF11, GLUTHATHIONE-S-TRANSFERASE11; CSLyase, CYSTEINE-S-CONJUGATE β-LYASE; UGT, UDP-GLYCOSYLTRANSFERASE; SOT, SULFUR TRANSFERASE; AOP3, ALKENYLHYDROXYPROPYL3; BZO1, BENZOYLOXY1.
The developing body of biosynthetic genetic tools has enabled the identification of TFs that regulate the aliphatic GLS pathway (Gigolashvili et al., 2007a, 2007b, 2008; Hirai et al., 2007; Sønderby et al., 2007, 2008; Schweizer et al., 2013). The current model, which is in general agreement with other defense pathway models, suggests that there are limited sets of TFs that can linearly account for most of the transcriptional regulation in the GLS pathway and that environmental signal integration occurs upstream of these TFs (Fig. 1B; Dombrecht et al., 2007; Beekwilder et al., 2008; Navarro et al., 2008; Hou et al., 2010; Lackman et al., 2011; Wild et al., 2012; Nakata et al., 2013; Xu et al., 2014). Although there are data supporting this model, specifically that removing two MYB (MYB28 and MYB29) and three MYC (MYC2, MYC3, and MYC4) TFs abolishes all aliphatic GLS accumulation, a closer investigation of published transcriptomic data suggests that this MYB/MYC model does not fully explain the biological observations (Beekwilder et al., 2008; Sønderby et al., 2010b; Schweizer et al., 2013). The expression of most genes in the aliphatic GLS pathway is not completely abolished in either of the polygenic mutants as the model suggests (Fig. 1C). Interestingly, it is only the transcripts for genes at the beginning of the pathway that were strongly altered in either polygenic mutant (Fig. 1C). This raises the potential for there to be additional unidentified TFs regulating the pathway.
To begin searching for these additional TFs, we cloned the promoters from 22 of the known enzyme-encoding genes in the aliphatic GLS biosynthetic pathway. We then tested all of these promoters for interactions with 659 TFs in a high-throughput yeast one-hybrid (Y1H) platform (Gaudinier et al., 2011). This analysis found a large number of newly identified potential TFs that could interact with the biosynthetic genes and/or the known regulatory TFs. Among these TFs were ones known to be involved with growth, biotic, and abiotic signaling. Over 75% of the tested mutant TFs found by Y1H altered the accumulation of the aliphatic GLSs similarly to single mutants in the known MYBs/MYCs (Beekwilder et al., 2008; Sønderby et al., 2010b; Schweizer et al., 2013). The phenotypic effects of the TF mutants were highly dependent upon the environment and tissue in which the accumulation was measured. This shows the importance of performing a gene-centered approach coupled with a nonbiased set of TFs to elucidate conditional-dependent regulatory TFs (Arda and Walhout, 2010). We could show that TFs that bound similar aliphatic GLS promoters also had similar phenotypic consequences using a nonlinear matrix analysis. This suggests that, at least for the aliphatic GLSs, the pathway appears to be regulated as a set of overlapping modules. These overlapping modules allow for the integration of multiple signals at the promoter level within the pathway. Future research will be needed to develop a cohesive model involving the dozens of newly identified TFs and the previously known TFs (Fig. 1).
RESULTS
cis-Regulatory Analysis of GLS Pathway Promoters
To develop a detailed graph of the potential transcriptional regulatory network mediating the expression of aliphatic GLS genes, we cloned approximately 2 kb upstream or to the nearest gene if this was less than 2 kb for the 22 known enzyme-encoding genes for this pathway (Figs. 1A and 2; Supplemental Table S1; Sønderby et al., 2010a; Gaudinier et al., 2011). These genes include all of the known genes for the pathway except for two genes (GGP and CYP79F2; Fig. 1A; Geu-Flores et al., 2009; Sønderby et al., 2010a). We also cloned the promoters from the known MYB regulatory factors (MYB28 and MYB29) that are critical for the aliphatic GLS pathway (Fig. 1B; Supplemental Table S1; Sønderby et al., 2007, 2010a, 2010b). In addition, we also included the promoter of MYB76, which is a homolog of MYB28 and MYB29 and also controls aliphatic GLS accumulations (Sønderby et al., 2007, 2010a, 2010b). As a comparison, we also included the promoters from six genes that had been peripherally associated with the GLS pathway either by being shared with the Val biosynthetic pathway or coexpressing with the pathway (IMD3, IPMI2, IPMI1, and PMSR1–PMSR3). Another comparison was provided by including two promoters from the indolic GLS pathway, which is largely discrete from the aliphatic biosynthetic pathway (UGT74B and GSTF9; Sønderby et al., 2010b).
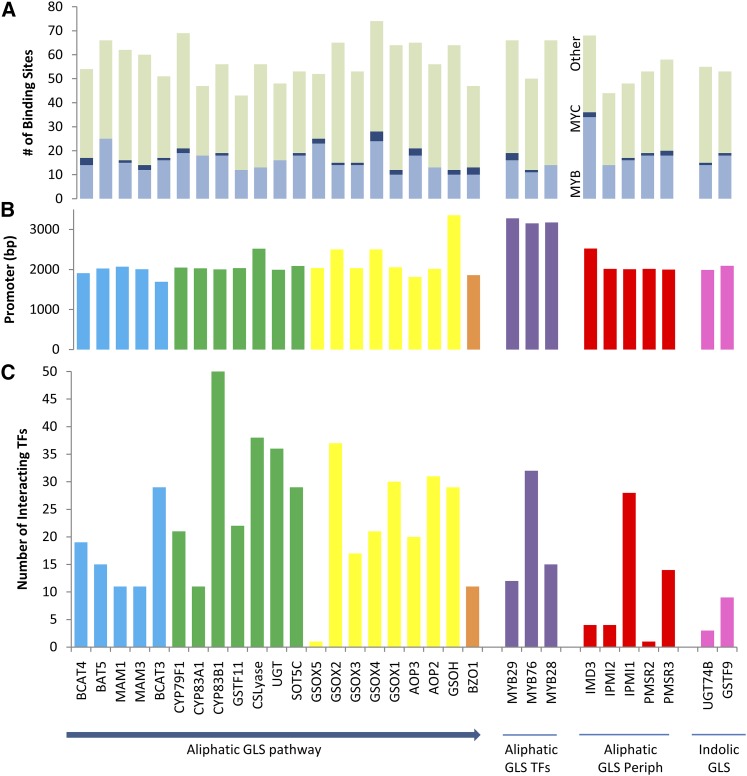
Figure 2.
Y1H identified TF interactions for the biosynthetic pathway promoters. The aliphatic GLS genes are ordered according to the biosynthetic pathway. The promoters are color coded based on their pathway membership as follows: indolic GLSs are pink, aliphatic regulatory genes are purple, and aliphatic peripheral genes are red. The aliphatic biosynthetic genes are parsed into the core pathway (green), elongation (blue), modifying (yellow), and seed specific (orange) to visualize regulation along the linear model of the pathway. A, The number of MYB, MYC, and other TF binding sites per promoter as called by Athena are presented. B, The size of the promoter for each gene that was cloned for Y1H is shown. C, Based on the Y1H analysis, the numbers of TFs found to interact with each promoter are plotted for the different classes of promoters. PMSR3, PROTEIN METHIONINE SULFOXIDE REDUCTASE3.
Using the sequences of the cloned promoters, we queried the TF binding sites that may be enriched within these promoters using the Athena analysis suite (O’Connor et al., 2005). This showed that the aliphatic GLS promoters are enriched in binding sites for the two classes of TFs previously associated with regulating this pathway, the MYBs and basic helix-loop-helix (bHLH) MYCs (Fig. 2; Supplemental Table S2; Gigolashvili et al., 2007a, 2007b, 2008; Hirai et al., 2007; Sønderby et al., 2007, 2010a, 2010b; Schweizer et al., 2013). There was also a strong enrichment in Basic Leucine Zipper Domain (bZIP) and Homeodomain DNA binding elements and a weaker enrichment in binding elements for the MADs, Auxin Response Factor (ARF), and Apetala2 (AP2)/Ethylene-response factor (ERF) TF families (Supplemental Table S2). The Homeodomain, bZIP, MADS, ARFs, and AP2/ERF TF families have not been previously associated with regulating the aliphatic GLS pathway, suggesting that there are potentially a number of unknown TFs that regulate this pathway.
Y1H Survey of Aliphatic Biosynthetic Genes
We then assessed the interaction of each promoter with all 659 TFs of the published stele-expressed TF collection (Fig. 2; Gaudinier et al., 2011). The stele is the predominant tissue in which the aliphatic GLS biosynthetic pathway genes are expressed (Sønderby et al., 2010a; Moussaieff et al., 2013). We also added the MYB28, MYB29, and MYB76 TFs to this collection because they bind the majority of tested GLS promoters in Arabidopsis cell culture assays (Gigolashvili et al., 2007a, 2007b, 2008).
The entire Y1H analysis identified 487 promoter/TF interactions between 140 TFs and the 21 aliphatic biosynthetic gene promoters (Supplemental Table S3). On average, each promoter interacted with 23.3 ± 2.6 TFs (average ± se). By contrast, the majority of TFs interacted with only one or two promoters (Figs. 2 and 3A). To determine an empirical threshold for identifying TFs that likely interact with the aliphatic GLS pathway, we tested the interaction of MYB28, MYB29, and MYB76 with all of the aliphatic GLS pathway promoters in the Y1H system. MYB28, MYB29, and MYB76 have been experimentally validated to bind at least six of the aliphatic GLS pathway promoters using an Arabidopsis tissue culture system (Gigolashvili et al., 2007a, 2007b, 2008). Using our Y1H system, we identified three, zero, and two interactions, respectively, between these TFs and aliphatic GLS biosynthetic promoters. Differences in interaction numbers in tissue cultures compared with Y1H are likely due to false positives and false negatives associated with each system (Yu et al., 2008; Walhout, 2011).
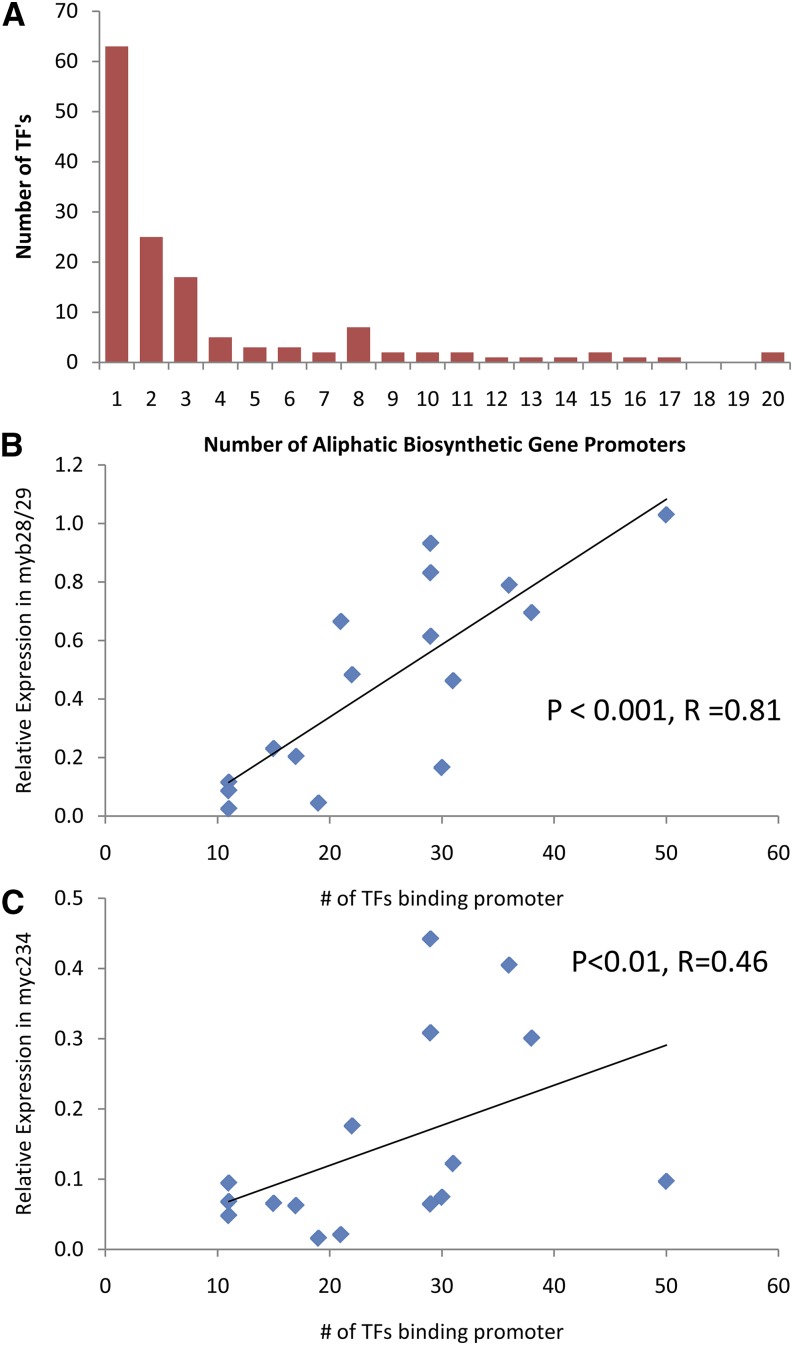
Figure 3.
Relationship between the number of TFs binding a promoter and gene expression. A, Histogram of the number of TFs found to interact with different number of aliphatic biosynthetic gene promoters based on the Y1H analysis. B, Comparison of the relative expression of the aliphatic GLS genes within the myb28/myb29 knockout with the number of TFs found to interact with their promoters based on the Y1H analysis. The predicted linear trendline is shown. C, Comparison of the relative expression of the aliphatic GLS genes within the myc2/myc3/myc4 knockout with the number of TFs found to interact with their promoters based on the Y1H analysis. The predicted linear trendline is shown.
These results using known interacting TFs provide us an ability to develop an empirical threshold for calling TFs that likely interact with the pathway. Thus, as a compromise between false positives and false negatives, we decided to use three interactions as an empirically validated threshold to identify TFs that are likely to be biologically relevant. This threshold requires these TFs to have at least as many interactions as the validated MYB28 TF. Using this threshold, we are likely removing some biologically relevant promoter-TF interactions because the MYB76 TF, which does regulate the pathway but is not a critical regulator of the pathway, had only two interactions and would have been dropped from our analysis. Of the 144 TFs identified in the Y1H analysis, 52 interacted with three or more of the aliphatic biosynthetic gene promoters similar to the validated MYB28, suggesting that we have identified a large collection of TFs potentially regulating aliphatic GLS accumulation (Fig. 3A).
Our main hypothesis is that aliphatic biosynthetic gene expression is regulated partially independently of MYB28/MYB29 and MYC2/MYC3/MYC4. One mechanism by which this independence could occur is if some promoters have a larger suite of regulatory interactions. These other TFs would provide an added level of redundancy, leading to these promoters being less sensitive to knockouts in the MYBs or MYCs. To test whether this model of independence may be present, we compared the number of TFs that interact with a promoter versus the relative expression of that gene within MYB and MYC knockouts. Because the available transcriptomics studies are done in adult foliar tissues, we focused on only the 16 genes expressed in this tissue. This removed four genes that have embryo-specific expression (BZO1, AOP3, GSOX2 GLUCOSINOLATE, and GSOX4; Kliebenstein et al., 2001c, 2007; Hansen et al., 2007; Li et al., 2008). There was a significant positive correlation in the relative expression of the aliphatic biosynthetic genes in the myb28/myb29 double knockout and the number of identified TFs interacting with their promoters (P < 0.001; R = 0.81; n = 16; Sønderby et al., 2010a; Fig. 3B). There was also a significant positive relationship between the aliphatic gene expression within the myc2/myc3/myc4 triple mutant and the number of identified TFs interacting with their promoters (P < 0.01; R = 0.46; n = 16; Schweizer et al., 2013; Fig. 3C). Thus, the expression of 16 foliar aliphatic biosynthesis genes is regulated in a partially MYB- and MYC-independent manner, concordant with the number of TFs identified as binding to their promoters.
Y1H Survey of Aliphatic Regulatory and Peripheral Genes
Using the promoters for the three aliphatic-related MYBs (MYB28, MYB29, and MYB76), we identified 46 TFs that bound one or more of the promoters for these three MYBs, suggesting that they could act as putative upstream regulators (Supplemental Table S3). Thirty-six of these 46 TFs (78%) also interacted with an aliphatic biosynthetic gene promoter with a median of interacting with eight aliphatic biosynthetic gene promoters. This fraction of TFs interacting with both the aliphatic biosynthetic and aliphatic TF gene promoters is higher than expected by random chance when using the distribution of interactions detected using all 659 TF and 34 promoters to generate a random distribution (i.e. a null expectation) to empirically control for false positives and negative errors within this system (χ2 = 17.64; P < 0.05). Thus, over three-quarters of potential TFs that interact with the aliphatic MYB promoters also link to a significant fraction of biosynthetic gene promoters, suggesting a potential for the formation of feed-forward loops (Alon, 2006). Among these potential loops was a feedback loop identified by the interaction of the MYB28 TF with its own promoter. This is in agreement with previously observed nonlinear changes in gene expression in MYB28 transgenic plants (Sønderby et al., 2010b).
Interestingly, we found that 26 of the 44 TFs (59%) that interacted with promoters of genes peripherally related to the aliphatic GLS pathway could also interact with core aliphatic biosynthetic gene promoters. By contrast, less overlap was observed between TFs that interact with indolic GLS biosynthesis genes and those that interact with core aliphatic GLS biosynthetic genes (11 of 21 TFs with a median of 2 interactions). These data are supported by gene expression profiling experiments in which peripheral genes show a higher level of coexpression with genes in the aliphatic biosynthetic pathway, whereas the indolic GLS pathway shows much lower coexpression, suggesting that they are differentially regulated (Gigolashvili et al., 2007a, 2007b, 2008; Sønderby et al., 2007, 2010b; Wentzell et al., 2007).
TF Binding Does Not Reflect Position within the GLS Biosynthetic Pathway
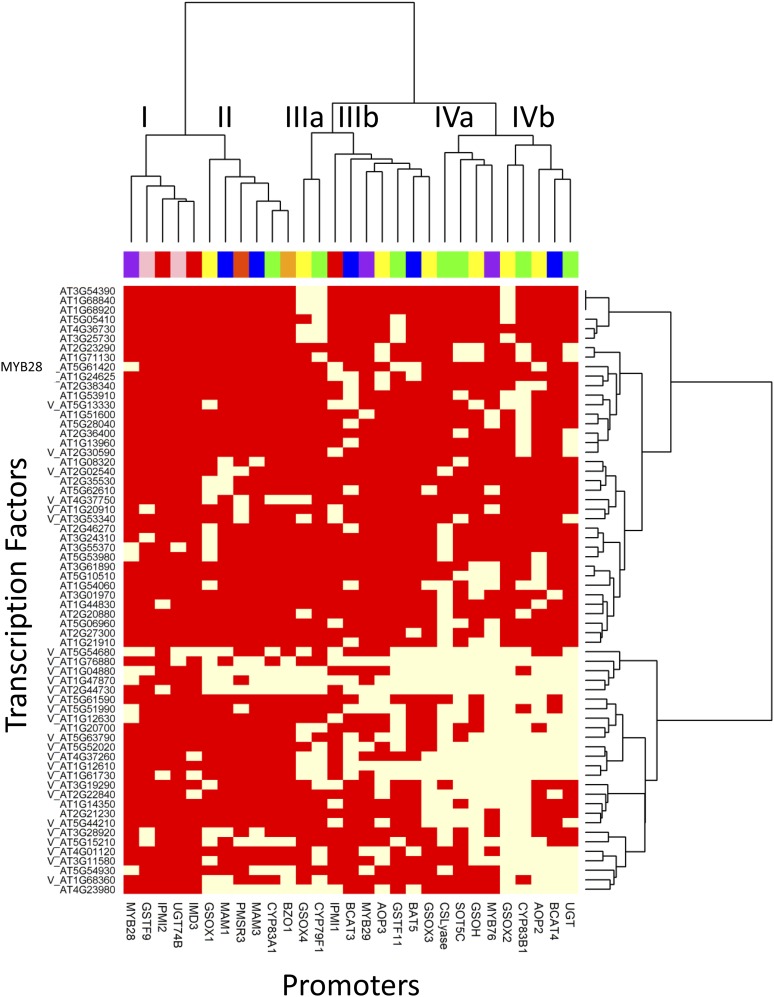
Given the large number of potential candidate TFs identified in this survey, we wanted to query whether there were identifiable TF/promoter modules and whether these modules reflected position within the biosynthetic or regulatory pathways. We first conducted hierarchical clustering using Euclidean distance with the Ward clustering algorithm (Fig. 4). The promoters fit into four to six large groups dependent on the TFs with which they interacted (Fig. 4). Group I is a distinct cluster that contains no aliphatic biosynthetic gene promoters but instead contains MYB28 and peripheral and indolic promoters. This is in contrast with previously published data demonstrating that MYB28 most closely coexpresses with the aliphatic genes and not the indolic genes (Hirai et al., 2007) and indicates that TF binding behavior relative to coexpression is likely quite complex.
Figure 4.
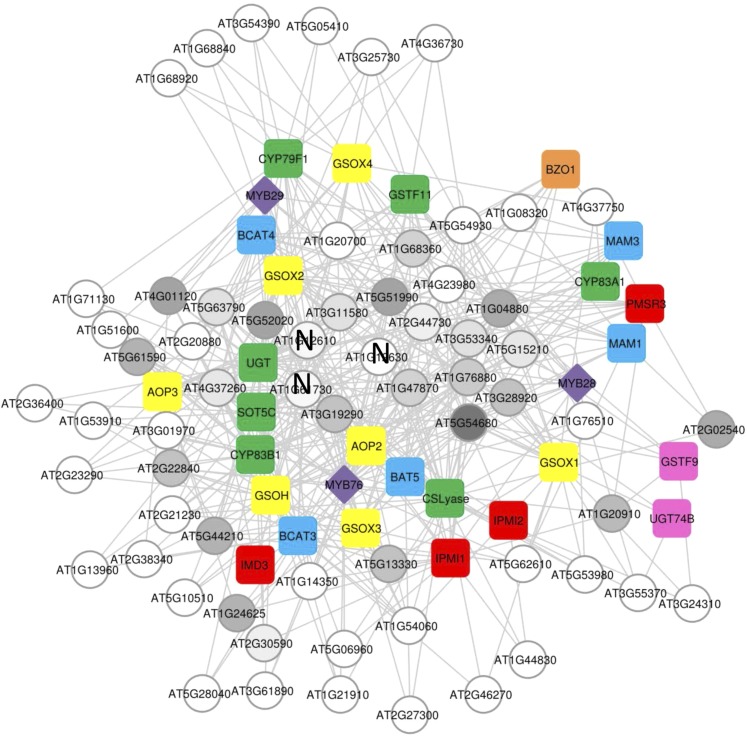
Coclustering of TFs and promoters via their interactions. Hierarchical clustering of promoter to TF interactions. Only promoters or TFs showing three or more interactions were included in the analysis, and the Ward algorithm was used for clustering. A V in front of the TF shows the TFs chosen to validate in mutant analysis. Within the plot, yellow shows a significant Y1H interaction between the promoter and TF, whereas red indicates no interaction. The promoters are color coded as shown in Figure 2 based on their pathway membership as follows: indolic GLSs are pink, aliphatic regulatory genes are purple, and aliphatic peripheral genes are red. The aliphatic biosynthetic genes are parsed into the core pathway (green), elongation (blue), modifying (yellow) and seed specific (orange) to visualize regulation along the linear model of the pathway. The roman numerals show potential promoter groupings.
The other promoter groupings all contained promoters from the aliphatic biosynthetic genes and regulatory genes with no obvious linear relationship to the pathway. For example, cluster IVa included the promoter for MYB29, promoters for the core biosynthetic pathway genes CS-LYASE and SOT2, and the side chain modifying genes GSOH and GSOX3 (Fig. 4). These four genes do not work in a colinear fashion and instead are scattered throughout the pathway (Fig. 1A). This finding suggests that there may be modular TF interactions that do not require the targets to be linear within a biosynthetic pathway. A correlation analysis of TF interactions with respect to gene position within the biosynthetic pathway found no significant correlations between any TF and promoter position within the pathway (data not shown). A spring-embedded clustering approach showed a similar graph in which there was no observable relationship between pathway position and TF-promoter binding behavior for aliphatic GLS biosynthetic pathway genes (Fig. 5). Thus, our analyses demonstrate that TFs are likely to interact with promoters in nonlinear modules of the pathway (Figs. 4 and 5).
Figure 5.
Spring-embedded network linking TFs and promoters via their interactions. A network of promoter to TF interactions as generated using a spring-embedded approach is shown. This approach leads to TFs or promoters with more interactions being present in the center of the diagram and less connected ones being more distal. Only promoters or TFs showing three or more interactions were included in the analysis, and the Ward algorithm was used for clustering. TFs chosen to validate in mutant analysis are shown in dark gray, whereas the other TFs are in light gray. Purple shows TFs that are known and validated to control aliphatic GLS accumulation. Circles show genes present only as the TF, squares are genes present only as promoters, and diamonds are present as both a TF and promoter. The promoters are color coded as shown in Figure 2 based on their pathway membership as follows: indolic GLSs are pink, aliphatic regulatory genes are purple, and aliphatic peripheral genes are red. The aliphatic biosynthetic genes are parsed into the core pathway (green), elongation (blue), modifying (yellow), and seed specific (orange) to visualize regulation along the linear model of the pathway. TFs that validated by mutant analysis are shown in gray with the intensity showing the fraction of aliphatic GLS phenotypes that validated from light gray to dark gray. TFs with a black N are those that were tested but had no significant phenotypic effect on GLS accumulation.
Phenotypic Validation of GLS Regulatory Potential
The Y1H analysis with the whole pathway identified a large number of candidate TFs that may regulate the aliphatic GLS pathway. However, there are currently no high-throughput chromatin immunoprecipitation assays that can rapidly screen through a large number of TFs with tissue-restricted expression. Thus, we decided to focus on the downstream phenotype of GLS metabolite accumulation in TF mutants as an approach to biologically validate these potential interactions. This approach has successfully identified GLS phenotypes in single regulatory TFs even when there are redundant genes providing a similar function (Sønderby et al., 2007, 2010a, 2010b). We focused on validating the potential regulatory activity of 29 TFs. These TFs were present in clusters that bind the most aliphatic GLS promoters or that clustered close to the known GLS TF MYB28 and had available homozygous transfer DNA (T-DNA) insertions (all TFs tested by T-DNAs are marked with a V in Fig. 4). In all possible cases, multiple alleles were identified for each gene (Supplemental Tables S4–S6). Obtaining mutants in both the dense and sparse clusters allow us to test whether overall levels in network connectivity are reflected in the magnitude of perturbations in aliphatic GLS quantities.
Using a randomized block design, we analyzed the accumulation of leaf and seed GLSs in the presence and absence of biotic stress for all TFs. Measuring GLS accumulation across multiple tissues and conditions allows us to sample a broader phenotypic spectrum to test whether TFs function only under specific conditions or developmental stages or more generally. To provide the biotic stress treatment, we utilized two growth chambers that have a similar abiotic environment but contain dramatically different biotic environments, in which one was pest free (Controlled Environment Facility [CEF]) and one had an endogenous pest population (Life Sciences Addition [LSA]). We then utilized a three-way ANOVA with nesting to test whether insertions in a gene affected any aliphatic or indolic GLSs across the experiments. The nesting term directly tested whether any GLS differences were due to individual insertions or if all insertions within a given TF had similar effects. Only results significant for the gene term (effect across all insertions) are discussed and insertion-specific events are ignored (Supplemental Tables S7 and S8).
The statistical analysis showed that all of the 29 TFs affected accumulation of at least one GLS with a median of nine GLSs affected across all TF insertion lines. To minimize the chance for false positive results, we established a baseline of a TF having to affect five GLS phenotypes because a random collection of gene mutations significantly affected levels of 3 ± 1 GLSs (Chan et al., 2011). This left 22 TFs that significantly altered the accumulation of at least five GLSs. Some of these TFs were previously characterized as having a role in abiotic stress responses, including COLD RESPONSIVE BINDING FACTOR (CBF4; for full names and Arabidopsis Genome Initiative codes, see Supplemental Table S5; Haake et al., 2002; Zhou et al., 2011), ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTOR (ABF4; Choi et al., 2005; Xu et al., 2013), NAC DOMAIN-CONTAINING PROTEIN102 (NAC102; Christianson et al., 2009), RELATED TO AP2 (RAP2.6L; Che et al., 2006; Asahina et al., 2011; Krishnaswamy et al., 2011; Liu et al., 2012; Huang et al., 2013), and INDOLE-3-ACETIC ACID-LEUCINE RESISTANT3 (ILR3; Rampey et al., 2006; Long et al., 2010). Another set of TFs had previously been linked to growth and the cell cycle, including E2Fc (del Pozo et al., 2006; Berckmans et al., 2011), G-BOX BINDING FACTOR2 (GBF2; Schindler et al., 1992; Menkens and Cashmore, 1994; Terzaghi et al., 1997), GROWTH REGULATING FACTOR (GRF; (Hewezi et al., 2012; Casadevall et al., 2013), and AINTEGUMENTA (ANT; Elliott et al., 1996; Klucher et al., 1996; Krizek et al., 2000; Liu et al., 2000; Mizukami and Fischer, 2000; Horstman et al., 2014). Of these previously characterized TFs, only ERF9 had been previously associated with biotic stress and the resistance to necrotrophic fungi (Camehl and Oelmüller, 2010; Maruyama et al., 2013). This diversity of TF functions suggests that the aliphatic GLS biosynthetic gene promoters can function as a site of integration for a complex set of signals integrating across growth and defense to both abiotic and biotic stresses with respect to GLS levels.
If these TFs are central to integrating both diverse environmental and developmental inputs, then their effects should be conditional on the environment and developmental tissue in which we measured GLS levels. ANOVA analyses revealed that the majority of the statistically significant validations were specific to the growth conditions or tissue being tested (Table I). For example, ERF9 altered 10 GLSs in a tissue-dependent manner (Table I; Supplemental Tables S7–S9). By contrast, GBF2’s influence on GLS levels had an increased dependency on stress conditions (Table I; Supplemental Tables S7–S9). In total, 22 of the 29 TFs were able to significantly alter the accumulation of multiple GLS in a tissue- or stress-dependent manner arguing that promoters in biosynthetic pathways may allow for specificity to integrate signals.
Table I. Insertion mutant validation of putative TFs.
Data are presented as the number of statistically significant tests for the gene term out of 21 different phenotypes. Gene shows the number of phenotypes that showed as significant difference between the wild type and the insertion mutants in the specific gene listed. This is after controlling for potential variation between the multiple insertion lines using nested ANOVA. KO indicates knockout and shows the number of independent T-DNA insertions tested per TF. Using the same exact model, Chamber:Gene shows the number of phenotypes with a significant interactions between the gene and chamber, and Tissue:Gene shows the number of phenotypes with a significant interaction between tissue and gene. The three-way term shows the number of phenotypes that were controlled by an interaction of chamber, tissue, and gene. The name column shows the published literature name for any gene with an identifiable name, and class shows the class of TF with which a gene is associated.
| Arabidopsis Genome Initiative Code | Name | Class | KO | Total | Gene | Chamber: Gene | Tissue: Gene | TGC |
|---|---|---|---|---|---|---|---|---|
| AT1G12630 | ERF/AP2 | 2 | 1 | 1 | 0 | 1 | 0 | |
| AT1G61730 | Storekeeper | 1 | 2 | 1 | 0 | 1 | 0 | |
| AT1G12610 | DDF1 | ERF/AP2 | 2 | 2 | 0 | 1 | 0 | 2 |
| AT2G30590 | WRKY21 | WRKY | 2 | 3 | 2 | 0 | 2 | 0 |
| AT5G15210 | HB3 | HB | 1 | 4 | 1 | 3 | 0 | 2 |
| AT4G37260 | MYB73 | MYB | 2 | 4 | 3 | 1 | 3 | 0 |
| AT2G44730 | MYB-Like | 1 | 3 | 3 | 2 | 2 | 1 | |
| AT3G53340 | NF-YB1 | NF-Y | 1 | 5 | 2 | 4 | 2 | 1 |
| AT1G68360 | C2H2 ZF | 1 | 7 | 4 | 2 | 4 | 1 | |
| AT3G11580 | AP2/B3 | 1 | 5 | 4 | 2 | 3 | 2 | |
| AT1G76880 | MYB-Like | 2 | 8 | 6 | 0 | 6 | 0 | |
| AT3G19290 | ABF4 | bZIP | 2 | 9 | 4 | 0 | 8 | 0 |
| AT1G47870 | E2FC | E2F | 2 | 7 | 5 | 2 | 4 | 2 |
| AT5G44210 | ERF9 | ERF/AP2 | 3 | 11 | 3 | 0 | 10 | 0 |
| AT5G63790 | NAC102 | NAC | 1 | 5 | 2 | 5 | 2 | 4 |
| AT3G28920 | HB34 | HB | 3 | 9 | 6 | 5 | 0 | 3 |
| AT5G13330 | RAP2.6L | ERF/AP2 | 2 | 9 | 2 | 4 | 6 | 3 |
| AT1G20910 | Arid/Bright | 2 | 10 | 10 | 0 | 7 | 0 | |
| AT2G22840 | GRF1 | C3H ZF | 1 | 9 | 5 | 5 | 5 | 2 |
| AT2G02540 | HB21 | HB | 1 | 12 | 8 | 2 | 2 | 8 |
| AT1G24625 | ZFP7 | C2H2 ZF | 1 | 11 | 5 | 4 | 10 | 2 |
| AT1G04880 | Arid/Bright | 2 | 12 | 10 | 4 | 6 | 2 | |
| AT4G01120 | GBF2 | bZIP | 1 | 13 | 2 | 12 | 1 | 8 |
| AT5G51990 | CBF4 | ERF/AP2 | 1 | 13 | 3 | 5 | 6 | 9 |
| AT5G52020 | ERF/AP2 | 1 | 13 | 4 | 8 | 3 | 9 | |
| AT5G61590 | ERF/AP2 | 1 | 11 | 4 | 7 | 6 | 8 | |
| AT1G66140 | ZFP4 | C2H2 ZF | 2 | 17 | 8 | 11 | 9 | 10 |
| AT5G54680 | ILR3 | bHLH | 2 | 19 | 16 | 6 | 8 | 6 |
| AT4G37750 | ANT | AP2 | 1 | 21 | 15 | 16 | 18 | 9 |
Connectivity versus Phenotypic Validation
Systems biology studies like to focus validation efforts on hub genes, genes that show the highest level of connectivity, with the assumption that these genes are more likely to have phenotypic consequences. For instance, it was previously shown in Arabidopsis that genes bound by a relatively large number of TFs are significantly positively correlated with conferring a morphological phenotype (Brady et al., 2011). Thus, we proceeded to test this assumption using the phenotypes of our TF mutants compared with their connectivity. Within our data, there was no correlation between the number of promoters that a TF interacted with and the number of GLSs affected by in the corresponding mutant (n = 29; R2=0.06; P = 0.185). Thus, filtering Y1H data sets simply by the number of interactions will not provide an increase in the successful validation of new genes controlling a phenotype. Similarly, there was no pattern in the level of connectivity within the spring-embedded network diagram. Finally, the three TFs whose insertional mutants had no phenotype were among the most connected at the center of the diagram (Fig. 5). This has also been observed in yeast (Saccharomyces cerevisiae) pseudohyphal growth and in a Caenorhabditis elegans neuronal gene regulatory network, suggesting that it is a broader phenomenon than plant secondary metabolism (Borneman et al., 2006; Vermeirssen et al., 2007). In addition, this finding suggests that transcriptional regulation of secondary plant metabolism may be distinct from that of cell-type specification and differentiation and organ morphogenesis (Taylor-Teeples et al., 2014).
ANT and ILR3 Are Strong Regulators of GLS Biosynthesis
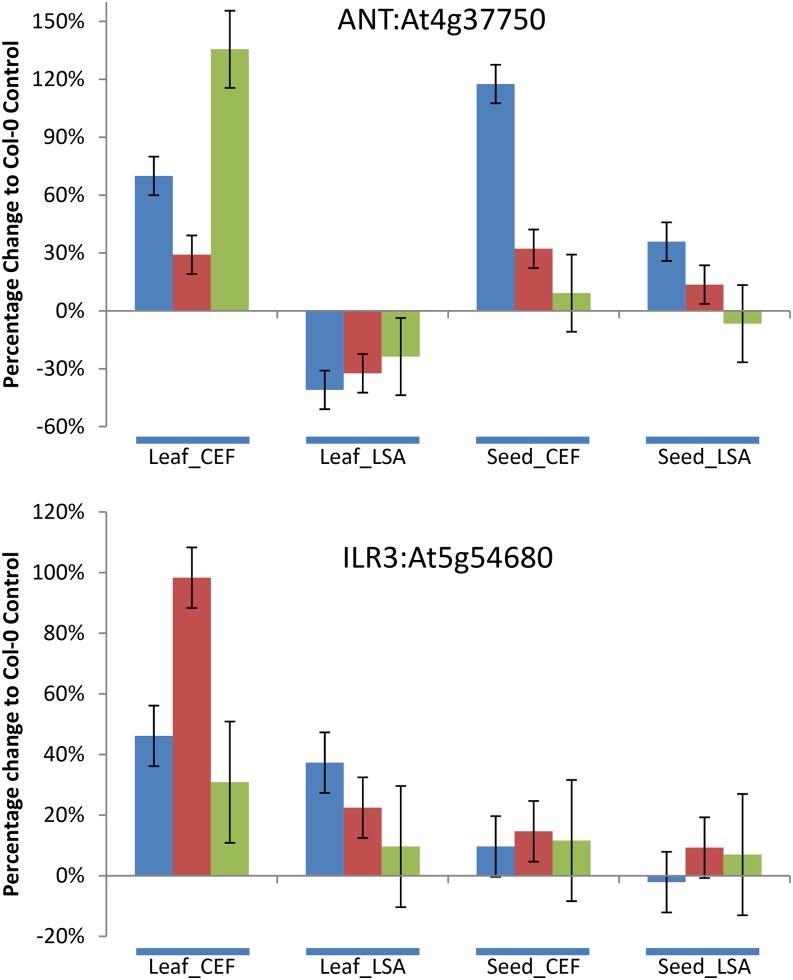
ANT, a TF important for controlling plant organ size, growth, and cell number, was the TF with the strongest effects both in terms of effect size and number of GLSs affected (Table I; Supplemental Table S9; Klucher et al., 1996; Mizukami and Fischer, 2000). However, we could only identify interactions of ANT with promoters of four aliphatic biosynthetic genes (Fig. 5). Although ANT affected aliphatic GLSs in all conditions, the direction and magnitude changed dependent on the condition (Fig. 6). ANT also influences leaf indole GLS accumulation even though we identified no promoter interactions with any indolic GLS-associated promoter, suggesting additional feedback between aliphatic and indolic GLS biosynthesis (Fig. 6). In contrast with ANT, loss of ILR3, a bHLH TF that has been connected to auxin conjugation and response to iron deprivation, had consistent effects on aliphatic GLS accumulation in the leaves but no effect in the seed (Rampey et al., 2006; Long et al., 2010; Fig. 6). This suggests that ILR3 functions as a tissue-specific repressor of aliphatic GLS biosynthesis (Fig. 6).
Figure 6.
Effects and networks for ANT and ILR3. The average effect of insertions in ANT and ILR3 on short-chain (blue), long-chain (red), and indolic (green) GLS accumulation in the different chambers and tissues. The 95% confidence limits are presented. If the bars do not cross 0, then there is a statistically significant difference from ecotype Columbia-0 of Arabidopsis (Col-0) for that mutant in that tissue and condition. For ANT, Col-0 and ANT are represented by six independent samples per chamber per tissue. For ILR3, Col-0 and ILR3 are represented by 12 independent samples per chamber per tissue.
Phenotypic Network Analysis
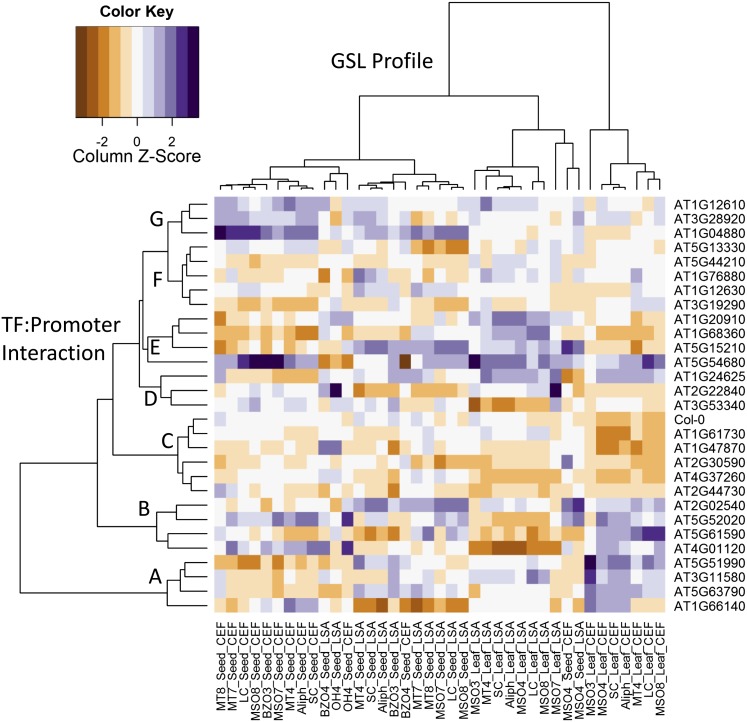
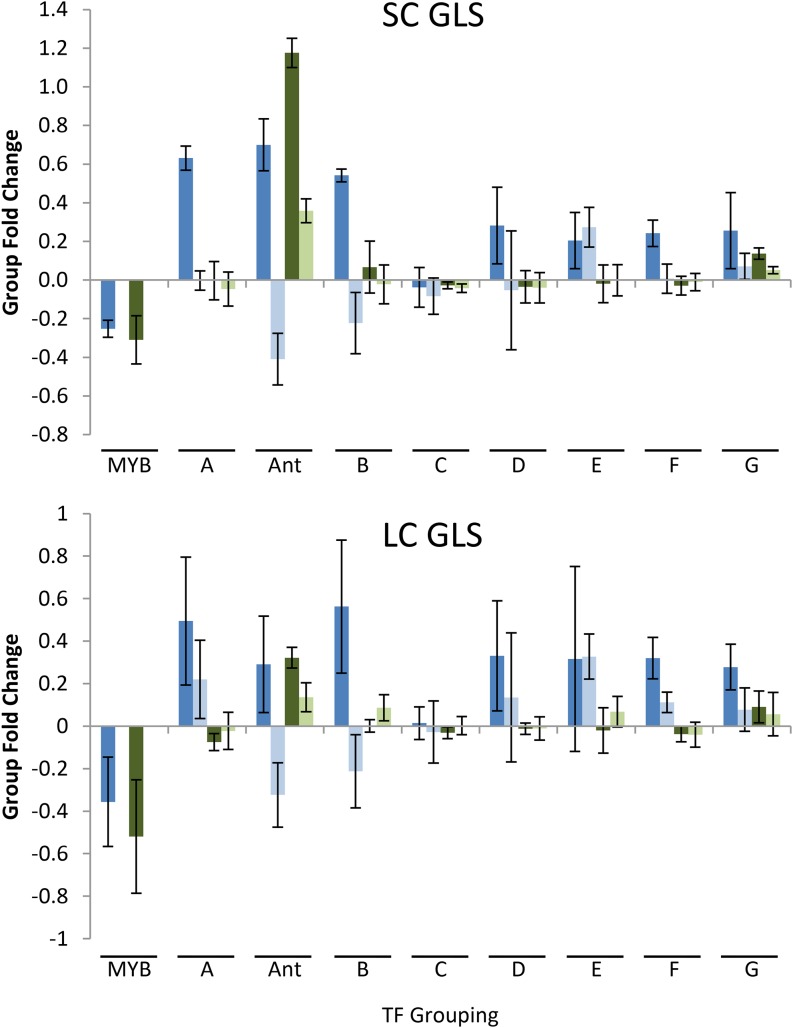
To relate the phenotypic effects of the TF insertional mutations to the pattern of Y1H interactions we clustered the genes by their effect on aliphatic GLS accumulation compared with Col-0. We found that the TFs clustered into eight groupings (Fig. 7). Group C closely clustered with Col-0 and contained all of the TFs with the fewest significant phenotypic changes in either long-chain or short-chain aliphatic GLS accumulation (Figs. 7 and 8). The rest of the clusters all showed a significant effect on short-chain or long-chain aliphatic GLS accumulation of a magnitude similar to that seen in the single knockouts of the validated MYB28/MYB29 and MYB76 (Fig. 8). To our knowledge, although the MYB insertions always lead to lower GLS accumulation, most TF mutants in this study led to higher GLS in the absence of biotic stresses (Fig. 8, CEF chamber). However, this increase disappeared in the presence of pests (Fig. 8, LSA chamber), suggesting that they are TFs that repress aliphatic GLS accumulation in the absence of biotic or abiotic stress. Whether negative GLS regulators are more numerous than positive regulators, or the assay is more prone toward identifying them, is yet to be determined. However, these stele-expressed TFs appear to act as general repressors of aliphatic GLS accumulation rather than activators.
Figure 7.
Clustering TFs based upon their aliphatic GLS phenotype. Hierarchical clustering of the aliphatic GLS phenotype across the TF mutants. For each TF, the fold change compared with Col-0 was utilized to standardize across experiments. The fold changes were then Z-scaled within a phenotype to balance the clustering and the Ward algorithm used for clustering. The color key in the top left shows the effect scale of the mutants upon the aliphatic GLS phenotypes in a Z-scale. Col-0 was included in the analysis with a value of 0-fold change for each phenotype for comparison purposes. For this analysis, only phenotypes describing the accumulation of aliphatic GLS were utilized. The letters on the left show the described clustering of TFs. ANT was not included in the analysis because of its strong single gene effects that biased the clustering. The dendrograms are labeled based on what is being clustered.
Figure 8.
Average GLS change in TF groups. The average change in short-chain and long-chain aliphatic GLSs of the TF groups A to G as defined by the phenotypic clustering are shown with 95% confidence intervals for the group change. The corresponding changes in ANT are also shown. For comparison, the average changes in the leaf and seed phenotypes in single insertion mutants in MYB28, MYB29, and MYB76 are shown. For the MYBs, the se of the phenotype across published results in these same chambers is shown. Blue bars are values from leaves, with dark blue showing the CEF chamber and light blue showing the LSA chamber. Green bars are from seeds, with dark green showing the CEF chamber, and light green is from seeds in the LSA chamber. To estimate the average group changes for the TF clusters shown in Figure 7, we took the average change of each TF mutant in relation to wild-type Col-0. We then grouped the TFs into the appropriate groups based on Figure 7. We then estimated the average percent change and 95% confidence interval for each group of TFs. This was done for the total level of short-chain and long-chain aliphatic GLS. LC, Long chain; SC, short chain.
In contrast with this general pattern, mutants in TFs within cluster B (containing HOMEOBOX PROTEIN21 [HB21], GBF1, and ERF/AP2s At5g52020 and At5g61590) were most similar to the effects seen in ANT, showing opposing effects with higher aliphatic GLS in the pathogen-free conditions and lower aliphatic GLS in response to pathogens (Fig. 8). Cluster B, however, did not display the seed effects of ANT, suggesting that these TFs function with different tissue specificity than ANT. TF group E (containing ILR3, HB30, C2H2 Zinc Finger At1g68360, and ARID/Bright At1g20910) showed the most consistent effects across conditions (Fig. 8). Thus, we can use Y1H to identify TFs that affect aliphatic GLS accumulation in a range of conditions with a high rate of validation.
TF/Promoter Interactions Link to Metabolic Consequence in TF Mutants
The above analysis found that T-DNA insertion mutants in 22 of the 29 tested TFs had significant effects on GLS accumulation. This shows that we can utilize the Y1H approach to identify TFs for the aliphatic GLS pathway with a success rate of 75% (Fig. 5). However, no clear pattern was observed with respect to TF-promoter interaction and specific mutant effects on GLS accumulation for the TFs (Figs. 4 and 7). Thus, we used a variety of linear modeling approaches (multiple regression and principal component) to test for a correlation between the TF-promoter interactions and their mutants’ associated metabolic changes. No such correlation was observed, suggesting nonlinear control of TF regulation of GLS biosynthesis (data not shown).
The above approaches to link mutant phenotype and promoter binding of specific TFs are limited because they function by focusing on linear analysis of the connections. To avoid the limitations of linear approaches, we tested whether it was possible to identify connections in the entire data set using a Mantel test (Mantel, 1967). The Mantel test allows us to conduct a correlation of all TF-promoter interactions against the GLS phenotypes of all of the TF mutants in a single nonlinear test. The Mantel test conducts a comparison of matrices. As such, we used the TF-promoter interactions and the relative GLS phenotypes of the TF mutants to generate two separate distance matrices. One distance matrix describes the similarity of how the TFs interact with the tested promoters, whereas the other distance matrix describes the similarity of the TFs GLS (either aliphatic or indole) phenotypes relative to the wild-type control depending on tissue or stress treatment (Supplemental Tables S2 and S3).
We first tested for a correlation between the TF-promoter and TF-GLS phenotype matrices in each of the four tissue/chamber combinations specifically for TFs that bound to aliphatic GLS-related promoters. For both seed data sets (stress-LSA and nonstress-CEF), there was a significant correlation between the aliphatic GLS phenotypes of TF mutants and the aliphatic GLS-related promoter interactions of these same promoters (CEF: r = 0.29; P = 0.042; and LSA: r = 0.28; P = 0.020). There was also a significant correlation between TF-promoter interactions and indolic GLS phenotypes in the seeds independent of stress treatment (CEF, r = 0.2607; P = 0.014; and LSA, R = 0.36; P = 0.001). This significant influence of the TFs on indolic GLS phenotypes is in contrast with the fact that we found these TFs based on their binding to aliphatic GLS-related promoters, and further supports our conclusion that regulation of aliphatic GLS biosynthesis is interconnected with indolic GLS biosynthesis, likely in an indirect manner. Using the leaf GLS data, we only found a single significant relationship between the TF-promoter matrix under nonstress conditions with indolic GLS (R = 0.26; P = 0.0480). No significant interactions were found for aliphatic GLS under either treatment, or for indolic GLS under stress treatment.
Thus, the Mantel tests show that there is a significant similarity between the pattern of how the TFs interact with the aliphatic GLS promoters and the resulting GLS phenotype of the TF mutants in seeds. This relationship is strongest within the seeds potentially because the seeds contain GLS transported from the maternal plant and thus represent an integrative measurement of GLS regulation across the life span of the maternal plant (Nour-Eldin et al., 2012; Andersen et al., 2013). By contrast, the leaves represent a more temporal measurement specific to that time and condition and may require more spatial or temporal information to identify the linkage between promoter interaction and metabolic phenotype for the different TFs. In addition, the TF library is generated from stele-expressed TFs, which may indicate that this collection of TFs provides more information about loading of GLS into seeds than it does about foliar GLS accumulation (Gaudinier et al., 2011). It will require the use of a complete collection of Arabidopsis TFs to test whether we can generate a similarly significant relationship between TF-promoter interactions and foliar GLS accumulation.
cis-Element Motif Enrichment Largely Reflects TF Binding
To test whether there was a link between the cis-elements within a promoter and the Y1H identification of associated TF families, we assessed if specific TF families were enriched in their Y1H identification across the aliphatic GLS pathway promoters (Supplemental Table S4). This showed that the bZIP, AP2/ERF, ABSCISIC ACID INSENSITIVE3, GLABROUS ENHANCER BINDING PROTEIN, and Trihelix TF families interacted more often with the aliphatic GLS pathway gene promoters than expected by chance given their occurrence in the Y1H library. This agreed well with the enrichment of bZIP, AP2/ERF, and Trihelix binding motifs in these promoters (Supplemental Table S2). Our finding that nearly all of the TFs in these families that we tested affect aliphatic GLS accumulation suggests a previously unrecognized regulatory role for these TFs in Arabidopsis GLS biosynthesis.
By contrast, the enrichment in MYB, Homeodomain, and bHLH binding motifs in these promoters did not coincide with an increased identification of the corresponding TFs (Supplemental Tables S2 and S4). Given the high level of validation of phenotypes for TFs in these families, this could suggest that these TF families have a higher level of secondary sequence specificity such that each MYB binds a sufficiently different sequence from the predicted core motif that it is not possible to accurately predict enrichment (Weirauch et al., 2014). Alternatively, these TF families may have a higher requirement for dimerization partners to bind DNA and thus may generate a higher false-negative rate in Y1H for these families. This second hypothesis agrees with the low frequency of identifying known MYB28, MYB29, and MYB76 interactions with these promoters because these TFs are thought to interact with the MYCs to bind their promoters (Supplemental Table S3; Zhao et al., 2008; Schweizer et al., 2013).
DISCUSSION
Conducting large-scale Y1H with 22 promoters from the aliphatic GLS genes representing nearly the entire biosynthetic pathway and known regulators identified 52 TFs that interacted with at least the same number of promoters as MYB28, which has been validated to interact with these promoters (Figs. 4 and 5). This was conducted using a library that only had approximately one-third of the known TFs in Arabidopsis, albeit a collection that was focused on TFs functioning in the major biosynthetic cell type for aliphatic GLSs, the vasculature (Sønderby et al., 2010a, 2010b; Gaudinier et al., 2011). None of these TFs had been previously linked to determining the function of the aliphatic GLS pathway. Using metabolic profiling to test the role of 29 of these TFs in controlling this pathway showed that we could validate that 75% of the identified TFs had the capacity to alter the accumulation of aliphatic GLSs (Table I). Obtaining this level of validation required measuring the aliphatic GLSs in multiple tissues and environments in accordance with the concept that these TFs may be playing a role in coordinating the aliphatic GLS defense pathway across a complex environment (Fig. 6). Thus, it is possible to use Y1H assays to identify new TFs that can influence a metabolic pathway with a high success rate using metabolic profiling for validation.
Signal Integration Can Occur at the TF and Promoter Level
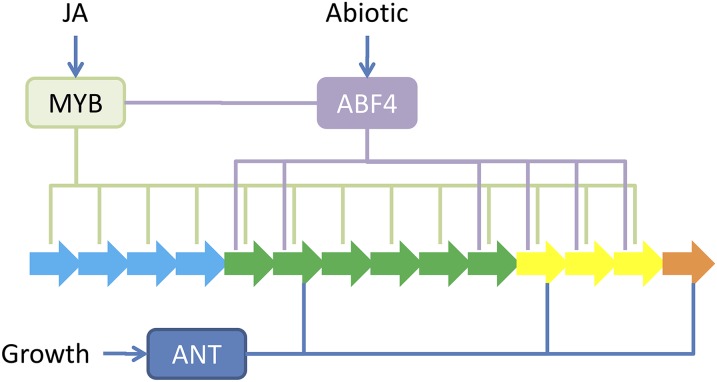
The TFs identified as aliphatic GLS regulators in this study include a number of known TFs controlling the responses to a variety of abiotic and biotic stresses as well as internal growth and development switches (Table I). This suggests that these TFs provide the ability to integrate a complex mixture of signals to regulate the pathway (Fig. 9). Furthermore, the fact that the TFs caused independent alterations in the amount and profile of aliphatic GLSs suggests that this integration can occur by interaction of the TFs with nonlinear components of the pathway. For example, comparing the predicted binding capacity of ABF4 and ANT shows that there is both overlap and independence in their promoter binding and the chemical profile of mutants in these genes (Figs. 7 and 9). This modularity involved some TFs such as ABF4 having the capacity to interact with the known MYB promoters allowing for feed-forward loops to regulate the pathway (Figs. 4 and 9). By contrast, other TFs such as ANT did not show any interaction with the MYBs but instead only connected directly to the biosynthetic gene promoters (Figs. 4 and 9). Thus, it is likely that this metabolic pathway has a mixture of direct regulatory processes that bypass the major MYC/MYB regulators as well as feed-forward loops that incorporate the MYC/MYB regulators.
Figure 9.
Multiple sites of integration and noncoordinate pathway regulation. Shown is a simplified model containing the interactions of the known MYBs to the aliphatic GLS pathway genes (blue arrows) using green lines. Putative inputs of abiotic and growth signals via predicted ANT and ABF4 interactions are shown in blue and purple lines, respectively. Potential connections to the MYB promoters are also shown. JA, Jasmonic acid.
This diverse set of independent and overlapping regulatory links between TFs and the aliphatic GLS pathway is potentially a way to optimize the ability of the aliphatic GLS to protect the plant at minimal cost in a highly diverse biotic environment. For example, a number of the TF mutants lead to a significant change in the blend of aliphatic GLS within the plant. This ability to modulate the blend may be a response to the fact that each aliphatic GLS is most effective as a defense against a specific set of biotic attackers (Lambrix et al., 2001; Kliebenstein et al., 2002; Kroymann and Mitchell-Olds, 2005; Hansen et al., 2008; Stotz et al., 2011; Züst et al., 2012). Thus, altering the blend may be a way to optimize the defense against a specific set of biotic pests and away from another (Wentzell and Kliebenstein, 2008; Burow et al., 2009). By having a mixture of TFs that directly bypass and TFs that create feed-forward loops with the MYBs, this creates a system in which the plant may be able to precisely fine-tune the regulation of the biosynthetic genes to optimize the defense of the system using a myriad of inputs including biotic, abiotic, and developmental signals.
Master Regulators in Development versus Defense Metabolism
Studies routinely classify genes as master or key regulators controlling a process. In developmental biology, this term reflects the ability of a single TF to be both necessary and sufficient to drive a developmental program or cell identity. MYB28 and MYB29 were proposed to act as a master regulator of this process because their absence results in a complete absence of GLS biosynthesis and because their transcript levels are strongly correlated with GLS abundance. However, the master regulator terminology may be a misleading interpretation because it assumes that the biological pathway associated with the trait being measured has a largely hierarchical regulatory architecture. In contrast with this assumption, it has long been known that development and defense metabolism should be differently regulated (Waddington, 1942). Developmental characters such as floral tissue specification must be tightly coordinated and likely involve irreversible regulatory switches. An organism gains no benefit from having stamens and pistils rapidly interconverting because their functions are defined by their end states. In plants, these irreversible regulatory switches comprised the homeotic floral regulators.
By contrast, metabolism is characterized by rapid interconversion between compounds that is likely critical to allow for specific fine-tuning of metabolism (Samal and Jain, 2008; Chandrasekaran and Price, 2010; Watson and Walhout, 2014). Thus, metabolism likely has very few if any irreversible regulatory switches and a greater potential for fine-tuning. Hence, a myb28/myb29 double mutant that abolishes a compound without abolishing the majority of pathway transcripts does not fit the concept of master regulator as defined by classical studies of developmental processes. The idea that metabolic and developmental pathways may have different regulatory architectures is supported by systems studies, which show that that the molecular architecture of regulatory networks is highly dependent on the phenotype or output that is being controlled (Milo et al., 2002, 2004; Sullivan et al., 2014)
Molecular Validation
We used high-throughput metabolic profiling to validate the potential of the identified TFs to alter metabolite accumulation. The goal of this approach was to identify a small subset of candidates upon which to conduct further molecular validation studies. Instead, however, we were able to find an effect of 75% of the tested TFs mutants upon aliphatic GLS accumulation. Although it is tempting to assume that these are indirect effects, the 75% rate of validation is dramatically higher than a random null sample of genes, and these genes were identified via a potential direct in yeasto interaction with promoters for aliphatic GLS biosynthetic genes (Fig. 4; Chan et al., 2011). Current methods to validate direct regulatory interactions would require performing transgenic plant generation to independently develop lines containing tagged versions of each putative TF and then conducting a chromatin immuno-precipitation assay in each line coupled with high-throughput expression profiling. Furthermore, this would have to be conducted in multiple tissues and environments given the fact that the TFs were highly conditional in their effect. This illuminates a critical need to develop new methodologies to conduct high-throughput in vivo validation under a variety of abiotic and biotic stress treatments of potential TF-promoter interactions or other molecular interactions to fully develop the complete model for how this pathway is regulated.
One potential method to provide further evidence of direct regulatory interactions would be to use the regulatory network model to identify putative double mutants that would epistatically interact and create significantly larger or even opposing effects upon the pathway. For example, our model predicts that ANT directly regulates the aliphatic GLS pathway as a repressor, whereas MYB28/MYB29 activates the pathway. Because these two processes are independent, our model would predict that a triple myb28/myb29/ant mutant would potentially rescue aliphatic GLS accumulation. Similar work in yeast has suggested that double mutants between regulatory modules have epistatic effects in contrast with those within modules (Segrè et al., 2005). Thus, it may be possible to further refine the direct regulatory module by working to predict the phenotype of double or higher order mutants.
CONCLUSION
By utilizing greater than 90% of the promoters for aliphatic biosynthetic genes with a large, focused Y1H library, we were able to develop a better image of the potential scale of GLS pathway transcriptional regulation. Incorporating metabolic profiling, we were able to show that there are at least dozens of TFs that have the capacity to significantly affect this pathway’s function. Furthermore, the effects of these TFs effects are highly conditional, indicating that the aliphatic GLS pathway is regulated to optimize growth and defense in a multifaceted environment. Future work is required to fully map out the molecular connections in this pathway and to expand this analysis to the full Arabidopsis complement of TFs. This will then allow the development of a regulatory model that is more inclusive of multiple environmental and growth inputs then have currently been proposed.
MATERIALS AND METHODS
Promoter Cloning
PCR primers were designed to amplify about 2,000 bp of each of the aliphatic GLS promoters upstream of the predicted translational l start codon or the promoter sequence to the start of the next gene. The promoters were amplified by Phusion High-Fidelity DNA Polymerase (M0530S; New England Biolabs) and cloned into pENTR 5′ TOPO TA Cloning vector (K591-20; Invitrogen). The promoters were then transferred into the pMW2 and pMW3 destination vectors by LR reaction (11791-019; Invitrogen) and confirmed by DNA sequencing (Gaudinier et al., 2011).
MYB TF Cloning
To include the known aliphatic GLS MYB TFs MYB28, MYB29, and MYB76 into the existing Y1H library, we cloned their open reading frames from Col-0 complementary DNA of leaf material by Phusion High-Fidelity DNA Polymerase (M0530S), and the PCR products were cloned into pENTR Directional TOPO Cloning vector (K2400-20; Invitrogen). Finally, the three MYB TFs were transferred into the pDESTAD-2μ destination vector and confirmed by DNA sequencing (Gaudinier et al., 2011).
Y1H
The root vascular-expressed TF collection used for the Y1H screening is as previously described (Gaudinier et al., 2011). The MYB28, MYB29, and MYB76 proteins were added to this collection. All of the GLS bait promoters were screened against the stele-expressed TF collection using the Y1H protocol as previously described (Gaudinier et al., 2011; Taylor-Teeples et al., 2011).
Plant Material and Experimental Conditions
For the GLS analysis of the T-DNA insertion lines, Arabidopsis (Arabidopsis thaliana) plants were grown in two different controlled-environment chambers with 16-h light at 100- to 120-mE light intensity with two complete independent replicates of the whole experiment. One growth chamber (CEF) is routinely cleaned and sterilized to maintain it in a pest-free format and used exclusively for Arabidopsis study. The other walk-in growth chamber (LSA) is used for multiple plant species, including Arabidopsis, tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum), and has an endogenous pest population composed of a mix of aphids and fungus gnats. This chamber is a model of a stressful pest and pathogen environment. In each growth chamber, each line was planted in 6-fold replication with four other mutants and wild-type controls within a flat using a randomized complete block design. Wild-type Col-0 controls were included in each flat to minimize any spatial aspects of the growth chamber. This experiment was replicated in both growth chambers providing 12 biological repeats in total for most of the mutant genotype.
T-DNA Lines
T-DNA lines for the selected TFs were ordered from the Arabidopsis Biological Resource Center (Sussman et al., 2000; Alonso et al., 2003), and lines with homozygous insertions were identified by PCR-based genotyping (Supplemental Tables S5 and S6). Multiple independent insertions within each gene were obtained and phenotyped where possible (Table I).
GLS Analysis
Four weeks after sowing, we harvested a leaf from each plant for GLS analysis. One leaf from the first fully mature leaf pair of each plant was removed and stored in 90% (v/v) methanol to inhibit enzymatic breakdown of chemical compounds and begin the extraction. The plants were then allowed to continue to develop and produce seeds. We collected seeds from each plant individually when survived and extracted GLSs from 40 seeds. GLSs from both tissues were extracted and analyzed by HPLC according to previously described methods (Kliebenstein et al., 2001b).
Statistical Analysis
To test the effect of each TF upon GLS accumulation, all GLSs were analyzed via ANOVA using a general linear model within R software (R Development Core Team, 2014). This was done using a nested model to account for potential differences between independent insertions when comparing the wild type with the mutants. The following model was used to test for differences in GLS accumulation in the mutants within each specific gene to the respective wild-type plants grown concurrently: yragt = µ+ Gg +Aa(Gg) + Tt + Cc + GgxTt + Aa(Gg)xTt + GgxCC + Aa(Gg)xCc + GgxTtxCC + Aa(Gg)xTt xCc + εragt, where εrgt represents the error and is assumed to be normally distributed with mean 0 and variance σε2. In this model, yrgc denotes the GLS accumulation in each plant with T-DNA allele a (insertion line 1…3), with genotype (Gg) representing the presence or absence of a T-DNA (the wild type versus mutant), from tissue (Tt; leaf or seed), from chamber (Cc). This nested architecture allows us to directly test whether the wild-type differs from the mutants independent of any difference between independent insertions. Means and se for each genotype class were obtained using R software (R Development Core Team, 2014). All presented P values are false discovery rates adjusted to 0.2 and least square means are presented (Supplemental Tables S7 and S8). The model was run independently for each TF.
Matrix Comparisons
To test for a link between the matrix of interactions between TFs and GLS promoters and the matrix of GLS phenotypes in the TF knockout mutants, we utilized the vegan R package for running Mantel and partial Mantel tests (Dixon, 2003; Oksanen et al., 2013; R Development Core Team, 2014). Mantel and partial Mantel tests were implemented using the Pearson’s coefficient with 1,000 permutations to test significance. The Mantel test calculates the correlation between two distance matrices. The TF-promoter interaction matrix and the relative mutant TF-GLS phenotype matrix (Supplemental Table S2) were transformed into distances matrices in R. To generate a TF/promoter interaction matrix, the TF-promoter interactions were recorded in two states (0 = no interaction observed; 1 = interaction observed), and the distance matrix was computed using binary correlation. Binary correlation was utilized because the data are in a binary format. Only TFs tested by TF-T-DNA lines were included in creating the matrix. To generate the TF-T-DNA/metabolite distance matrices, we correlated the relative change of each GLS in each TF-T-DNA with respect to the wild type across all of the TFs using a Euclidean distance within R. We independently created a distance matrix TFs using the metabolites for each treatment and tissue types. This was done independently for the aliphatic and indolic GLSs, creating eight total TF/metabolite matrices.
We also generated a third set of distance matrices to describe the TF relationships using the gene expression levels of the same TFs across stress and developmental conditions. Average gene expression values for the TFs were obtained from the Arabidopsis Expression Browser (Bio-Analytic Resource, http://bar.utoronto.ca), specifically the tissue and biostress series within this database (Patel et al., 2012). The average expression of the TFs across the data sets were then used to create coexpression matrices with Euclidean distance (TF/expression matrices). We then tested whether these TF/expression matrices altered the relationship between the TF/promoter and TF/metabolite matrices using partial Mantel tests. This was conducted separately using either the tissue series or the biostress series as the control matrix.
Group Changes
To estimate the average group changes for the TF clusters shown in Figure 7, we took the obtained the average change of each TF mutant in relation to wild-type Col-0. We then grouped the TFs into the appropriate groups based on Figure 7 and also included the previously published MYB28 and MYB29 single-mutant data. We then estimated the average percent change and 95% confidence interval for each group of TFs.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Promoters cloned for Y1H analysis.
Supplemental Table S2. Promoter motif enrichment in the tested promoters.
Supplemental Table S3. List of all promoter TF interactions identified.
Supplemental Table S4. TF family enrichment in interactions with aliphatic glucosinolate promoters.
Supplemental Table S5. T-DNAs used to validate specific transcription factor capacity to affect aliphatic glucosinolate accumulation.
Supplemental Table S6. Primers used for validating T-DNAs and cloning MYB promoters.
Supplemental Table S7. P values of significance for models testing the effect of TF insertions on glucosinolate.
Supplemental Table S8. Type III sums-of-squares for models testing the effect of TF insertions on glucosinolate.
Supplemental Table S9. Least square means and standard error for glucosinolate accumulation in the TF insertions and the comparator Col-0 samples.
Supplementary Material
Glossary
- TF
transcription factor
- GLS
glucosinolate
- Y1H
yeast one-hybrid
- T-DNA
transfer DNA
- Col-0
ecotype Columbia-0 of Arabidopsis
Footnotes
This work was supported by the National Science Foundation (grant nos. DBI 0820580 to D.J.K. and MCB 1330337 to S.M.B. and D.J.K.), the Hellman Fellows Fund (fellowship to S.M.B.), the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch project no. CA–D–PLS–7033–H to D.J.K.), and the Danish National Research Foundation (grant no. DNRF99 to D.J.K.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alon U. (2006) An Introduction to Systems Biology: Design Principles of Biological Circuits, CRC Press, Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersen TG, Nour-Eldin HH, Fuller VL, Olsen CE, Burow M, Halkier BA (2013) Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 25: 3133–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Walhout AJM (2010) Gene-centered regulatory networks. Brief Funct Genomics 9: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, et al. (2008) The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3: e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Lammens T, Van Den Daele H, Magyar Z, Bögre L, De Veylder L (2011) Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol 157: 1440–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart-Bouzat MG, Kliebenstein DJ (2008) Differential levels of insect herbivory in the field associated with genotypic variation in glucosinolates in Arabidopsis thaliana. J Chem Ecol 34: 1026–1037 [DOI] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M (2006) Target hub proteins serve as master regulators of development in yeast. Genes Dev 20: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow M, Losansky A, Müller R, Plock A, Kliebenstein DJ, Wittstock U (2009) The genetic basis of constitutive and herbivore-induced ESP-independent nitrile formation in Arabidopsis. Plant Physiol 149: 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camehl I, Oelmüller R (2010) Do ethylene response factorS9 and -14 repress PR gene expression in the interaction between Piriformospora indica and Arabidopsis? Plant Signal Behav 5: 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P (2013) Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 25: 3570–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Hansen BG, Kliebenstein DJ (2010) The complex genetic architecture of the metabolome. PLoS Genet 6: e1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Price ND (2010) Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci USA 107: 17845–17850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY (2005) Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol 139: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W (2006) Transcriptional regulation of metabolism. Physiol Rev 86: 465–514 [DOI] [PubMed] [Google Scholar]
- Dixon P. (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930 [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C (2011) Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331: 1185–1188 [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-Paz JL, Kim D, Kay SA, et al. (2011) Enhanced Y1H assays for Arabidopsis. Nat Methods 8: 1053–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Nielsen MT, Nafisi M, Møldrup ME, Olsen CE, Motawia MS, Halkier BA (2009) Glucosinolate engineering identifies a gamma-glutamyl peptidase. Nat Chem Biol 5: 575–577 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI (2007a) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50: 886–901 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Müller C, Flügge UI (2008) HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol 177: 627–642 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI (2007b) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51: 247–261 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11: 89–100 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J, Halkier BA, Kliebenstein DJ (2008) A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol 148: 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA (2007) Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J 50: 902–910 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ (2012) The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol 159: 321–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Struhl K (1986) Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46: 885–894 [DOI] [PubMed] [Google Scholar]
- Horstman A, Willemsen V, Boutilier K, Heidstra R (2014) AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci 19: 146–157 [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Huang Z, Zhao P, Medina J, Meilan R, Woeste K (2013) Roles of JnRAP2.6-like from the transition zone of black walnut in hormone signaling. PLoS ONE 8: e75857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (1997) Induced Responses to Herbivory, University of Chicago Press, Chicago [Google Scholar]
- Kerwin RE, Jiménez-Gomez JM, Fulop D, Harmer SL, Maloof JN, Kliebenstein DJ (2011) Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2012) Plant defense compounds: systems approaches to metabolic analysis. Annu Rev Phytopathol 50: 155–173 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, D’Auria JC, Behere AS, Kim JH, Gunderson KL, Breen JN, Lee G, Gershenzon J, Last RL, Jander G (2007) Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J 51: 1062–1076 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2001a) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001b) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001c) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Verma S, Rahman MH, Kav NN (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75: 107–127 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Mitchell-Olds T (2005) Epistasis and balanced polymorphism influencing complex trait variation. Nature 435: 95–98 [DOI] [PubMed] [Google Scholar]
- Lackman P, González-Guzmán M, Tilleman S, Carqueijeiro I, Pérez AC, Moses T, Seo M, Kanno Y, Häkkinen ST, Van Montagu MCE, et al. (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci USA 108: 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13: 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankau RA, Strauss SY (2008) Community complexity drives patterns of natural selection on a chemical defense of Brassica nigra. Am Nat 171: 150–161 [DOI] [PubMed] [Google Scholar]
- Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA (2008) Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol 148: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MW, AuYeung WK, Lam HM (2013) The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biol (Stuttg) 15: 13–18 [DOI] [PubMed] [Google Scholar]
- Liu P, Sun F, Gao R, Dong H (2012) RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol Biol 79: 609–622 [DOI] [PubMed] [Google Scholar]
- Liu Z, Franks RG, Klink VP (2000) Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220 [PubMed] [Google Scholar]
- Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K, Sato T, Yamaguchi J (2013) The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci 213: 79–87 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R. (1998) Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat 151: 20–28 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Cashmore AR (1994) Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA 91: 2522–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Itzkovitz S, Kashtan N, Levitt R, Shen-Orr S, Ayzenshtat I, Sheffer M, Alon U (2004) Superfamilies of evolved and designed networks. Science 303: 1538–1542 [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, Yativ M, Brady SM, Benfey PN, Aharoni A (2013) High-resolution metabolic mapping of cell types in plant roots. Proc Natl Acad Sci USA 110: E1232–E1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Landsman D (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci 22: 154–155 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Oksanen F, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) vegan: Community Ecology Package, R package version 2.0-10, R Project for Statistical Computing, Vienna [Google Scholar]
- Patel RV, Nahal HK, Breit R, Provart NJ (2012) BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J 71: 1038–1050 [DOI] [PubMed] [Google Scholar]
- Paul-Victor C, Züst T, Rees M, Kliebenstein DJ, Turnbull LA (2010) A new method for measuring relative growth rate can uncover the costs of defensive compounds in Arabidopsis thaliana. New Phytol 187: 1102–1111 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2014) R: A Language and Environment for Statistical Computing, R Project for Statistical Computing, Vienna [Google Scholar]
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B (2006) An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Samal A, Jain S (2008) The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response. BMC Syst Biol 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR (1992) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J 11: 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P (2013) Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrè D, Deluna A, Church GM, Kishony R (2005) Modular epistasis in yeast metabolism. Nat Genet 37: 77–83 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA (2010a) A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol 153: 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA (2010b) Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Sawada Y, Shimada Y, Hirai MY, Sasaki E, Krischke M, Brown PD, Saito K, Kamiya Y (2011) Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J 67: 81–93 [DOI] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M, Gaudinier A, Zhang LF, Reece-Hoyes J, Ahnert S, Walhoug M, Ware D, Brady SM (2011) Mapping spatiotemporal gene regulatory networks in the Arabidopsis root stele. Dev Biol 356: 110 [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Doherty C, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, et al. (2014) Environmental, developmental and genotype-dependent regulation of xylem cell specification and secondary cell wall biosynthesis in Arabidopsis thaliana. Nature (in press) [Google Scholar]
- Terzaghi WB, Bertekap RL Jr, Cashmore AR (1997) Intracellular localization of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultured Arabidopsis and soybean cells. Plant J 11: 967–982 [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo CA, Babon JAB, Sequerra R, Doucette-Stamm L, Barabási AL, Walhout AJM (2007) Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res 17: 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Walhout AJM. (2011) What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Walhout AJM (2014) Caenorhabditis elegans metabolic gene regulatory networks govern the cellular economy. Trends Endocrinol Metab 25: 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Kliebenstein DJ (2008) Genotype, age, tissue, and environment regulate the structural outcome of glucosinolate activation. Plant Physiol 147: 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière J-M, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon DY, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, et al. (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zhou MQ, Shen C, Wu LH, Tang KX, Lin J (2011) CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit Rev Biotechnol 31: 186–192 [DOI] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA (2012) Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]
- Züst T, Joseph B, Shimizu KK, Kliebenstein DJ, Turnbull LA (2011) Using knockout mutants to reveal the growth costs of defensive traits. Proc Biol Sci 278: 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.