The nicotine transporter NUP1 is required for jasmonate-mediated expression of a transcription factor that affects enzyme expression for the biosynthesis of tobacco alkaloids.
Abstract
The down-regulation of a tobacco (Nicotiana tabacum) plasma membrane-localized nicotine uptake permease, NUP1, was previously reported to reduce total alkaloid levels in tobacco plants. However, it was unclear how this nicotine transporter affected the biosynthesis of the alkaloid nicotine. When NUP1 expression was suppressed in cultured tobacco cells treated with jasmonate, which induces nicotine biosynthesis, the NICOTINE2-locus transcription factor gene ETHYLENE RESPONSE FACTOR189 (ERF189) and its target structural genes, which function in nicotine biosynthesis and transport, were strongly suppressed, resulting in decreased total alkaloid levels. Conversely, NUP1 overexpression had the opposite effect. In these experiments, the expression levels of the MYC2 transcription factor gene and its jasmonate-inducible target gene were not altered. Inhibiting tobacco alkaloid biosynthesis by suppressing the expression of genes encoding enzymes in the nicotine pathway did not affect the expression of ERF189 and other nicotine pathway genes, indicating that ERF189 is not regulated by cellular alkaloid levels. Suppressing the expression of jasmonate signaling components in cultured tobacco cells showed that NUP1 acts downstream of the CORONATINE INSENSITIVE1 receptor and MYC2, but upstream of ERF189. These results suggest that although jasmonate-activated expression of MYC2 induces the expression of both NUP1 and ERF189, expression of ERF189 may actually be mediated by NUP1. Furthermore, NUP1 overexpression in tobacco plants inhibited the long-range transport of nicotine from the roots to the aerial parts. Thus, NUP1 not only mediates the uptake of tobacco alkaloids into root cells, but also positively controls the expression of ERF189, a key gene in the biosynthesis of these alkaloids.
Nicotine and structurally related pyridine alkaloids accumulate throughout the plant in Nicotiana spp., and contribute to chemical defense against insect herbivory (Steppuhn et al., 2004). Nicotine is synthesized from the amino acids Asp and Orn (Shoji and Hashimoto, 2011a; Dewey and Xie, 2013). Early in the NAD biosynthetic pathway, Asp is metabolized to nicotinate mononucleotide, which is incorporated into the pyridine ring of nicotine by unknown enzyme reactions. Quinolinate phosphoribosyltransferase (QPT) catalyzes the last step of this pathway to yield nicotinate mononucleotide (Katoh et al., 2006), and one of the duplicated QPT genes functions in nicotine formation, rather than in NAD biosynthesis (Shoji and Hashimoto, 2011b). The pyrrolidine ring of nicotine is synthesized from Orn by three enzymes: Orn decarboxylase, putrescine N-methyltransferase (PMT; Hibi et al., 1994), and N-methylputrescine oxidase. A nicotinate derivative from the Asp pathway and the N-methyl-Δ1-pyrrolinium cation from the Orn pathway are then condensed into nicotine, but these reactions are poorly understood. Berberine bridge enzyme-like enzyme (BBL) is required for the late biosynthetic pathway of nicotine (Kajikawa et al., 2011).
Insect herbivory on tobacco (Nicotiana tabacum) leaves initiates the jasmonate signaling cascade, which ultimately activates nicotine biosynthesis genes in the roots (Shoji and Hashimoto, 2013). Tobacco uses canonical jasmonate signaling components, such as the jasmonate receptor CORONATINE INSENSITIVE1 (COI1) and the JASMONATE ZIM domain transcriptional repressors (JAZs), to activate structural genes involved in nicotine biosynthesis (Paschold et al., 2007; Shoji et al., 2008). The basic helix-loop-helix Leu zipper-type transcription factor MYC2 is released from JAZ-mediated repression upon jasmonate perception by COI1, and activates nicotine biosynthesis genes in Nicotiana spp. (Todd et al., 2010; De Boer et al., 2011; Shoji and Hashimoto, 2011c; Zhang et al., 2012). The evolutionarily conserved MYC2 transcription factor induces the expression of the master regulator genes of nicotine biosynthesis, which encode the NICOTINE2 (NIC2)-locus ethylene response factor (ERF)-type transcription factors (Shoji and Hashimoto, 2011c). At least seven ERF genes encoding a subclade of the tobacco group IXa subfamily are clustered at the NIC2 locus, also known as the B locus (Legg and Collins, 1971), and all of these genes are deleted in the low-nicotine tobacco nic2 mutant (Shoji et al., 2010). The NIC2-locus ERF genes are expressed in the root and up-regulated by jasmonate with kinetics distinct among the locus members (Shoji et al., 2010). The NIC2-locus ERFs activate the transcription of nicotine biosynthetic genes by directly binding to GCC box-like elements in their promoter regions (Shoji and Hashimoto, 2011b, 2012; Shoji et al., 2010, 2013). Although these ERF homologs are likely somewhat functionally redundant, ERF189 and its closest homolog ERF199 are thought to be the major regulators of the nicotine biosynthetic pathway; overexpression of ERF189 upregulates the expression of structural genes of this pathway to a greater extent than does that of the other ERF members examined (Shoji et al., 2010), and the expression patterns of ERF189 and ERF199 overlap with those of genes in the nicotine biosynthetic pathway (Shoji and Hashimoto, 2014; Shoji et al., 2010). In addition to containing ERF-binding sites, the promoters of nicotine biosynthesis genes contain functional binding sites for MYC2, which are required for their jasmonate-inducible expression (De Boer et al., 2011; Shoji and Hashimoto, 2011c). Thus, tobacco MYC2 has dual roles in nicotine biosynthesis; it positively regulates the expression of NIC2-locus ERFs and, together with these ERFs, directly binds to and activates the promoters of structural genes involved in nicotine biosynthesis (Shoji and Hashimoto, 2011c).
Several nicotine transporters have been reported in tobacco. Two highly homologous MULTIDRUG AND TOXIC COMPOUND EXTRUSION-family transporters (MATE1 and MATE2) are localized to the vacuolar membrane and transport tobacco alkaloids from the cytosol into the vacuole in alkaloid-synthesizing root cells (Shoji et al., 2009). MATE1 and MATE2 genes are coordinately regulated with nicotine biosynthesis genes with respect to spatial expression patterns in the root and NIC2-locus ERF-mediated expression (Shoji et al., 2009, 2010). Reducing the expression of MATE1/MATE2 in tobacco plants renders root growth more sensitive to exogenously applied nicotine, indicating that these transporters ameliorate nicotine toxicity in root cells during the active production of nicotine (Shoji et al., 2009). Another tonoplast-localized MATE-type transporter, tobacco JASMONATE-INDUCIBLE ALKALOID TRANSPORTER1, also transports nicotine, and is preferentially expressed in leaves, suggesting that the transporter is involved in nicotine sequestration in the leaf vacuole (Morita et al., 2009).
NICOTINE UPTAKE PERMEASE1 (NUP1) is a plasma membrane-localized tobacco transporter belonging to the purine uptake permease family (Hildreth et al., 2011). When expressed in yeast (Saccharomyces cerevisiae) cells or in cultured tobacco cells, NUP1 facilitates the import of tobacco alkaloids (nicotine and anatabine) from the culture medium into the cells (Hildreth et al., 2011; Kato et al., 2014). Although NUP1 preferentially transports pyridine alkaloids, it also transports pyridoxine and pyridoxamine (vitamin B6; Kato et al., 2014). Unexpectedly, when NUP1 expression was suppressed in tobacco plants, total nicotine levels were reduced and seedling root growth was enhanced (Hildreth et al., 2011). Jasmonate-treated transgenic tobacco hairy root lines in which NUP1 expression was suppressed (Hildreth et al., 2011) did not show statistically significant differences in the expression levels of five nicotine biosynthesis genes tested, including PMT and QPT. However, in the absence of exogenous jasmonate, some of these root lines exhibited reduced expression levels of nicotine biosynthesis genes encoding Orn decarboxylase, PMT, and quinolinate synthase. These results suggest the existence of a homeostatic control mechanism that maintains apoplastic nicotine levels in tobacco roots and indicate that NUP1 is unlikely to control nicotine biosynthesis directly.
In this study, we explored the enigmatic functions of NUP1 using transgenic tobacco lines, including cultured cells, hairy roots, and plants, in which NUP1 was either efficiently suppressed or overexpressed. Transcriptome analysis of tobacco genes involved in jasmonate-inducible nicotine biosynthesis identified the NIC2-locus ERF transcription factors as the molecular targets of NUP1. Furthermore, we demonstrated that NUP1 affects the long-range transport of nicotine from the roots to the aerial parts of tobacco plants.
RESULTS
Expression Profiles of NUP1
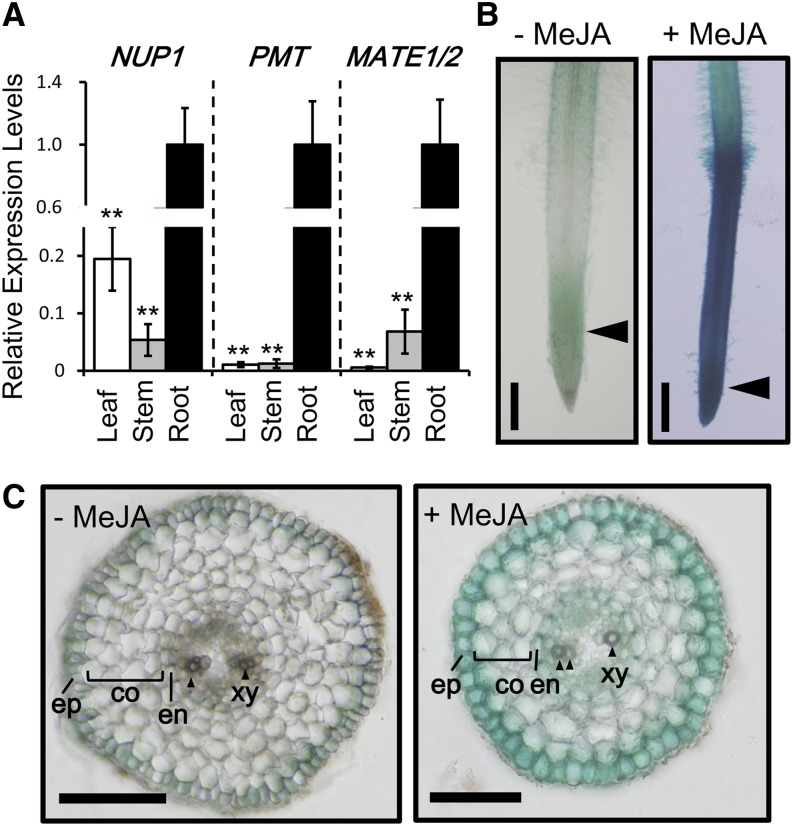
NUP1 transcripts were reported to be more abundant in the roots, especially the root tips, than in the leaves of 2-week-old hydroponically grown tobacco ‘Xanthi’ plants (Hildreth et al., 2011). Furthermore, NUP1 transcript levels in 4-week-old tobacco ‘Burley 21’ plants were higher in the roots than in the leaves and stems, although the transcript levels in the aerial parts of tobacco plants were considerably higher than those of genes encoding enzymes (e.g. PMT) and transporters (e.g. MATE1/MATE2) involved in nicotine biosynthesis and storage (Fig. 1A).
Figure 1.
Expression patterns of NUP1. A, Quantitative RT-PCR analysis of NUP1, PMT, and MATE1/MATE2 expression in the third leaf, stem (approximately 5 cm above the soil) and root of 8-week-old tobacco ‘NC95’ plants. The data are the mean values (±sd) for three independent plants. Significant differences between root values and other sample values were determined using Dunnett’s test and are indicated by asterisks (double asterisks for P < 0.01). B and C, Histochemical GUS staining of tobacco hairy roots expressing ProNUP1::GUS grown in the absence (−) or presence (+) of 100 μm MeJA for 24 h. B, Hairy roots at the tip region. C, Cross sections of ProNUP1::GUS roots. Sections were made at the approximate positions indicated by arrowheads in B. co, Cortex; en, endodermis; ep, epidermis; xy, xylem. Arrowheads indicate xylem. Bars = 1 mm in B and 200 μm in C. [See online article for color version of this figure.]
To examine the cell- and tissue-specific expression pattern of NUP1, we fused a 1.4-kb 5′-upstream regulatory region of NUP1 to the GUS gene, and stably expressed this reporter construct (ProNUP1::GUS) in tobacco hairy roots. The GUS reporter was expressed throughout the root, but expression was greatest in the root tip region, including the root meristem and elongation zone (Fig. 1B). A 24-h exposure to exogenously applied methyljasmonate (MeJA; 100 μm) greatly enhanced GUS staining, consistent with the MeJA-induced expression of NUP1 previously reported in tobacco hairy roots (Hildreth et al., 2011) and cultured cells (Kato et al., 2014). In cross sections of MeJA-treated roots prepared from the root tip region, GUS staining was strongest in the epidermal cells (Fig. 1C). GUS was expressed in root hairs, in the epidermal layer of the root cap (Supplemental Fig. S1), and in parenchyma cells in the stele.
Phenotypes of Tobacco Plants with Reduced or Increased NUP1 Expression
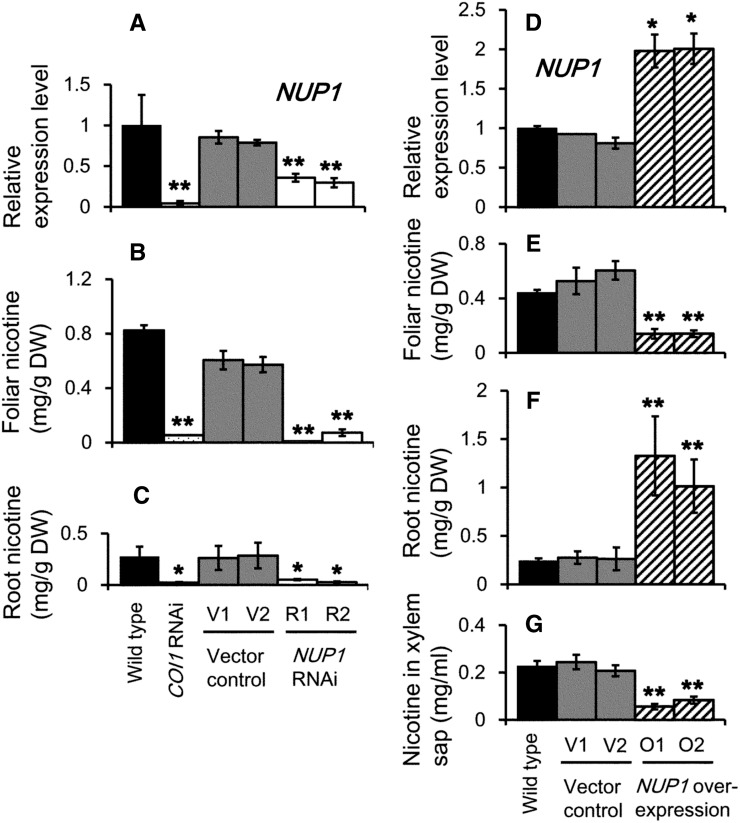
Next, we examined whether down-regulation of NUP1 decreased nicotine accumulation in tobacco plants, as reported for NUP1-RNA interference (RNAi) plants regenerated from tobacco hairy roots (Hildreth et al., 2011). Using Agrobacterium tumefaciens-mediated transformation of leaf discs, we generated two independent transgenic tobacco ‘SR1’ lines in which NUP1 expression in the root was down-regulated to less than one-half of that in the wild type and two independent empty vector-transformed controls, collectively referred to as the control plants (Fig. 2A). The nicotine content was severely reduced in the leaves and roots of these RNAi lines (R1 and R2), to less than 20% of that in the control plants (Fig. 2, B and C), and was comparable in level to that reported in tobacco COI1-RNAi plants in which jasmonate perception was blocked (Shoji et al., 2008).
Figure 2.
Down-regulation or overexpression of NUP1 affects the accumulation and distribution of nicotine in tobacco plants. Down-regulation data are shown in A to C, whereas overexpression data are presented in D to G. A to C, Quantitative RT-PCR analysis of NUP1 transcript levels in the roots (A) and foliar (B) and root (C) nicotine contents of 10-week-old wild-type, COI1 RNAi, vector control (V1 and V2), and NUP1 RNAi (R1 and R2) tobacco lines. D to G, Quantitative RT-PCR analysis of NUP1 expression in the roots (D), and nicotine content of the leaves (E), roots (F), and xylem sap (G) of 10-week-old wild-type, vector control (V1 and V2), and NUP1-overexpressing (O1 and O2) tobacco lines. The data are the mean values (±sd) of three independent plants. Significant differences between the wild type and test samples were determined using Dunnett’s test and are indicated by asterisks (single asterisk for P < 0.05 and double asterisks for P < 0.01). DW, Dry weight.
Because nicotine biosynthesis is largely blocked in the roots of NUP1-RNAi plants, it is challenging to examine the root-to-shoot translocation of nicotine in these plants. Therefore, we next generated two independent transgenic tobacco lines (referred to as O1 and O2) in which NUP1 was overexpressed under the strong constitutive Cauliflower mosaic virus 35S promoter. The level of NUP1 transcript in the roots of the NUP1-overexpressing plants was approximately 2-fold that of the control plants (Fig. 2D). Interestingly, the nicotine content in the roots of O1 and O2 was approximately 4-fold that of the control roots (Fig. 2F), whereas the foliar nicotine content was reduced to approximately 30% that of the control plants (Fig. 2E). These results indicate that NUP1 overexpression inhibits the long-range transport of nicotine from the root to the aerial parts of tobacco plants. To test this possibility, we measured the alkaloid content in xylem sap collected from the base of tobacco stems of the various lines (Fig. 2G). The nicotine content in the xylem sap of NUP1-overexpressing plants was approximately 30% that of the control plants, suggesting that NUP1 overexpression inhibits the xylem loading of nicotine in the root.
NUP1 Affects Nicotine Accumulation by Modulating the Expression of ERF189 and Nicotine Biosynthesis Genes
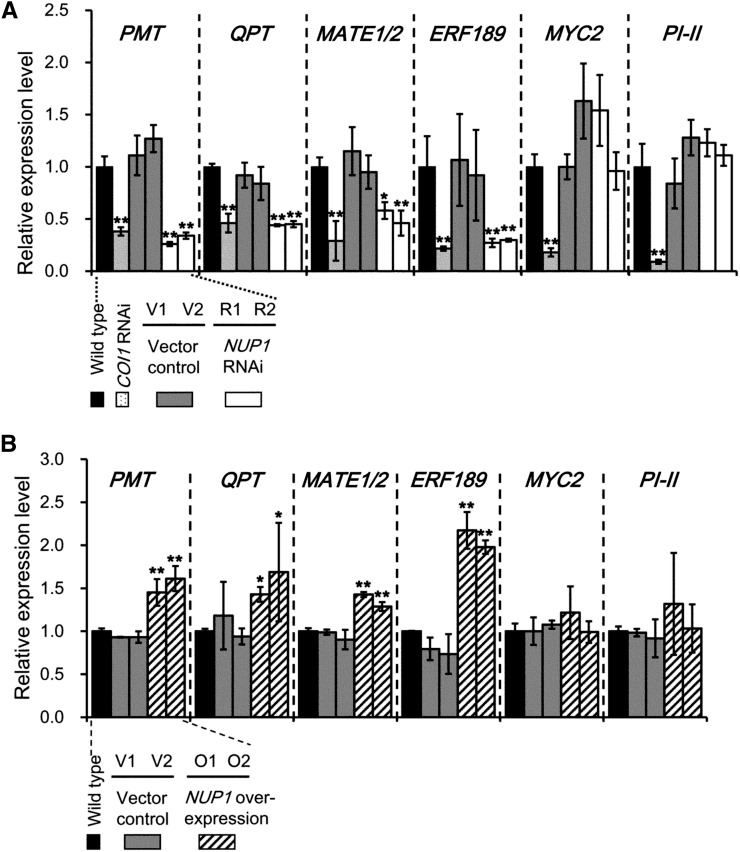
To examine the molecular mechanisms by which NUP1 affects nicotine accumulation, we studied the expression of structural genes involved in nicotine biosynthesis (PMT and QPT) and transport (MATE1/MATE2) and their upstream regulatory genes in the jasmonate signaling pathway (ERF189 and MYC2). In the roots of two NUP1-RNAi lines (R1 and R2), the transcript levels of PMT, QPT, and MATE1/MATE2 were reduced to around one-half or less than one-half of those in the wild-type and vector control plants (Fig. 3A). The transcript levels of ERF189 were similarly reduced to less than 40% of control plants in the RNAi roots, whereas those of MYC2 were not significantly affected. Because MYC2 is regulated not only transcriptionally but also posttranslationally, by its physical interaction with JAZ repressors (Chini et al., 2007), we analyzed the expression of the MYC2-regulated PROTEINASE INHIBITOR II (PI-II; Balandin et al., 1995; Shoji and Hashimoto, 2011c) to establish whether the transcriptional activator activity of MYC2 was affected in the silenced lines. PI-II expression was similar in the NUP1-RNAi and control lines, indicating that NUP1 regulates ERF189 but not MYC2. By contrast, the expression of both MYC2 and ERF189, and thus the expression of their downstream structural genes, was suppressed altogether in COI1-RNAi plants. COI1-dependent MeJA-induced up-regulation of MYC2 has been reported in Arabidopsis (Arabidopsis thaliana; Lorenzo et al., 2004). We tested whether ERF189 and its target genes would be up-regulated in NUP1-overexpressing plant lines (O1 and O2) subjected to MeJA treatment (Fig. 3B). The transcript levels of PMT, QPT, and MATE1/MATE2 in NUP1-overexpressing roots were significantly higher than those in wild-type and control roots. The level of ERF189 transcript was strikingly higher in the roots of NUP1-overexpressing plants than in those of the control plants, whereas the transcript levels of MYC2 and PI-II did not differ significantly between the NUP1-overexpressing and control plants.
Figure 3.
Down-regulation or overexpression of NUP1 affects the expression of genes involved in the nicotine biosynthetic pathway in tobacco plants. A, Quantitative RT-PCR analysis of PMT, QPT, MATE1/MATE2, ERF189, MYC2, and PI-II in the roots of 10-week-old wild-type, COI1 RNAi, vector control (V1 and V2), and NUP1 RNAi (R1 and R2) tobacco lines. B, Quantitative RT-PCR analysis of PMT, QPT, MATE1/MATE2, ERF189, MYC2, and PI-II expression in the roots of 10-week-old wild-type, vector control (V1 and V2), and NUP1 overexpressing (O1 and O2) tobacco lines. The data are the mean values (± sd) of three independent plants. Significant differences between the wild type and test samples were determined by Dunnett’s test and are indicated by asterisks (single asterisk for P < 0.05 and double asterisks for P < 0.01).
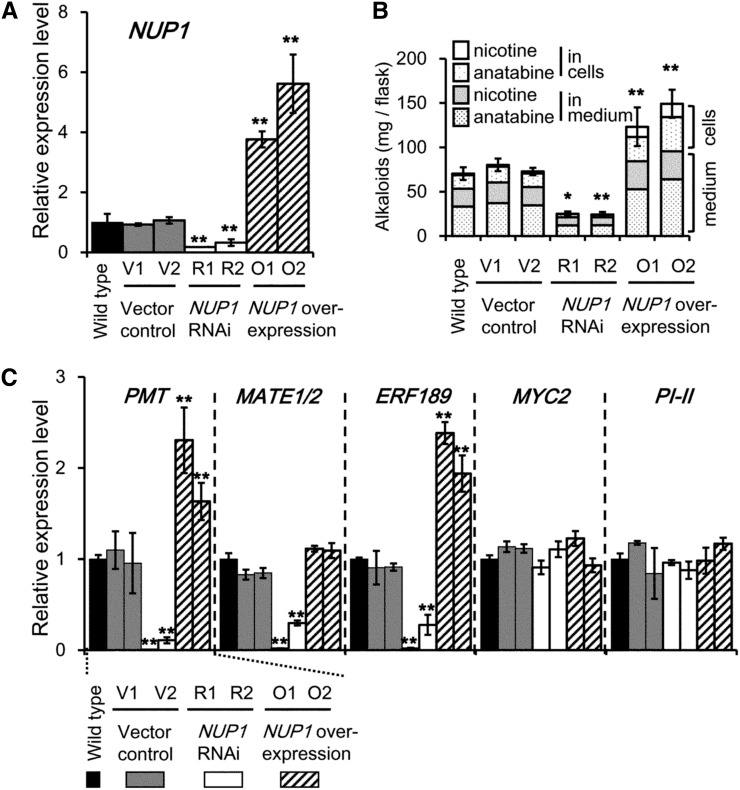
To substantiate these results obtained in transgenic tobacco plants, we generated tobacco cultured BY-2 cell lines in which NUP1 expression was either down- or up-regulated. Although BY-2 cells do not biosynthesize alkaloids when cultured in growth medium containing auxin, they produce the alkaloids anatabine and, to a lesser extent, nicotine, when grown on auxin-free medium supplemented with MeJA (Shoji and Hashimoto, 2008). When grown under culture conditions that promote alkaloid biosynthesis, two independent RNAi lines (R3 and R4) exhibited NUP1 transcript levels that were less than 30% those of control plants, whereas two independent overexpression lines (O3 and O4) had NUP1 transcript levels that were more than 4-fold those of the control lines (Fig. 4A). The total amount of tobacco alkaloids in the culture (i.e. the sum of anatabine and nicotine in the cells and culture medium) was severely decreased in the NUP1 down-regulated lines and was considerably increased in the NUP1-overexpressing lines, compared with those in the control lines (Fig. 4B). We calculated the relative ratios of tobacco alkaloids in the cells and in the medium, and found that the proportion of alkaloids in the medium was significantly increased in the RNAi lines, but was significantly decreased in the overexpression lines (Supplemental Fig. S2). Therefore, NUP1 not only promotes the uptake of tobacco alkaloids from the medium into the cells, but also positively regulates alkaloid biosynthesis. The expression level of an alkaloid biosynthesis gene (PMT), nicotine transporters (MATE1/MATE2), and their direct upstream regulator (ERF189) was highly decreased in the NUP1 RNAi lines, but increased in the NUP1-overexpressing lines, although the MATE1/MATE2 transcript level in the overexpression lines was only slightly increased and was not statistically different from that in the controls (Fig. 4C). The expression levels of MYC2 and its downstream target gene PI-II were not affected by the down-regulation or overexpression of NUP1.
Figure 4.
Down-regulation or overexpression of NUP1 in cultured tobacco BY-2 cells affects the expression of genes involved in the nicotine biosynthetic pathway and the accumulation of alkaloids. A to C, Quantitative RT-PCR analysis of NUP1 (A) and PMT, MATE1/MATE2, ERF189, MYC2, and PI-II (C) expression, and the amount of alkaloid in a culture flask (B) of wild-type, vector control (V1 and V2), NUP1 RNAi (R1 and R2), and NUP1 overexpressing (O1 and O2) BY-2 lines. Tobacco cells were treated with 100 μm MeJA for 24 h for RNA extraction and for 72 h for alkaloid analysis. The data are the mean values (±sd) of three biological replicates. Significant differences between the wild type and test samples were determined by Dunnett’s test and are indicated by asterisks (single asterisk for P < 0.05 and double asterisks for P < 0.01).
These results in transgenic plants and in cultured cells suggest that the NIC2-locus ERF transcription factor genes, but not MYC2, are regulated by NUP1. The changes in tobacco alkaloid levels and in the expression of genes involved in alkaloid biosynthesis in lines in which NUP1 had either been down- or up-regulated are likely caused by changes in the transcript levels of ERF189, and possibly other NIC2-locus ERF genes.
Reducing Alkaloid Biosynthesis by Metabolic Blocks Does Not Affect ERF189 Expression
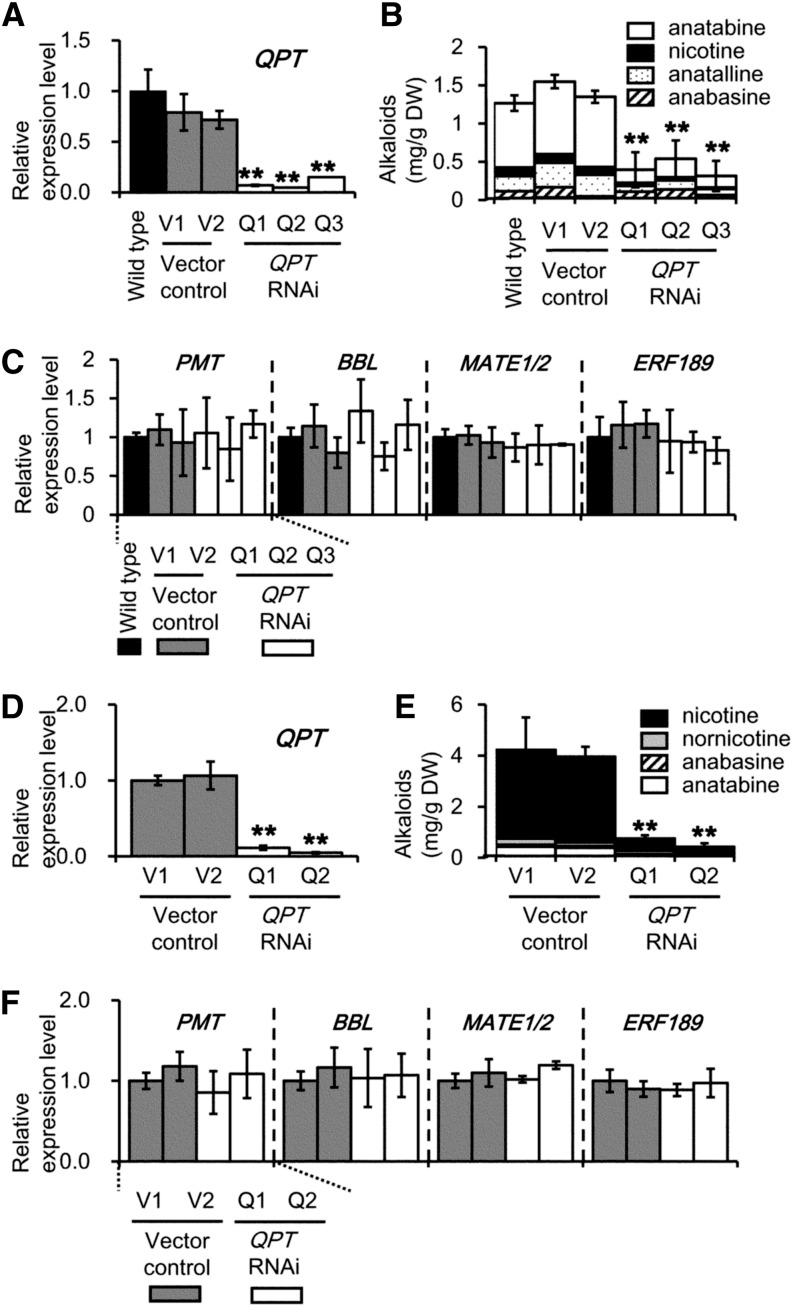
NUP1 suppression reduces the uptake of tobacco alkaloids from the extracellular space, and is expected to lower their intracellular levels. To examine the possibility that a certain threshold level of cellular alkaloids is required to maintain the expression of the NIC2-locus ERF genes, we inhibited alkaloid biosynthesis by RNAi-mediated suppression of QPT (Xie et al., 2004). Suppressing QPT expression (Fig. 5A) in jasmonate-treated cultured tobacco BY-2 cells (Q1, Q2, and Q3) resulted in significant decreases in nicotine, anatabine, and anatalline (to below 30% of control values; Fig. 5B). Likewise, in QPT-suppressed tobacco hairy root lines (Q1 and Q2; Fig. 5D), accumulation of nicotine, nornicotine, and anatabine was highly reduced (Fig. 5E). In these alkaloid-deficient cell and root lines, the transcript levels of ERF189 and of its target structural genes involved in nicotine biosynthesis and transport (PMT, BBL, and MATE1/MATE2) were not significantly different from those of the control plants (Fig. 5, C and F). Similar results were obtained in jasmonate-treated tobacco cells and tobacco hairy roots, in which expression of another nicotine biosynthesis gene (BBL; Kajikawa et al., 2011) was suppressed (Supplemental Fig. S3). We therefore conclude that the cellular levels of tobacco alkaloids do not affect the expression of their master transcription factor genes (e.g. ERF189) and the structural genes involved in nicotine biosynthesis, and that the transcriptional control of ERF189 by NUP1 is not influenced by the cellular levels of tobacco alkaloids.
Figure 5.
Down-regulation of QPT does not affect the expression of genes involved in alkaloid biosynthesis and transport. Data from cultured tobacco BY-2 cells are shown in A to C, whereas data from tobacco hairy roots are shown in D to F. Quantitative RT-PCR analysis of QPT (A and D), PMT, BBL, MATE1/MATE2, and ERF189 (C and F) expression, and the accumulation of tobacco alkaloids (B and E) in wild-type, vector control (V1 and V2), and QPT RNAi (Q1, Q2, and Q3 in A–C, and Q1 and Q2 in D–F) transgenic lines. The cultured tobacco cells were treated with 100 μm MeJA for 24 h for RNA extraction and for 72 h for alkaloid analysis. The data are the mean values (±sd) of three biological replicates. Significant differences between the wild type and test samples were determined by Dunnett’s test and are indicated by asterisks (double asterisks for P < 0.01).
Epistasis Analysis of NUP1 and Genes Involved in Jasmonate-Induced Nicotine Biosynthesis
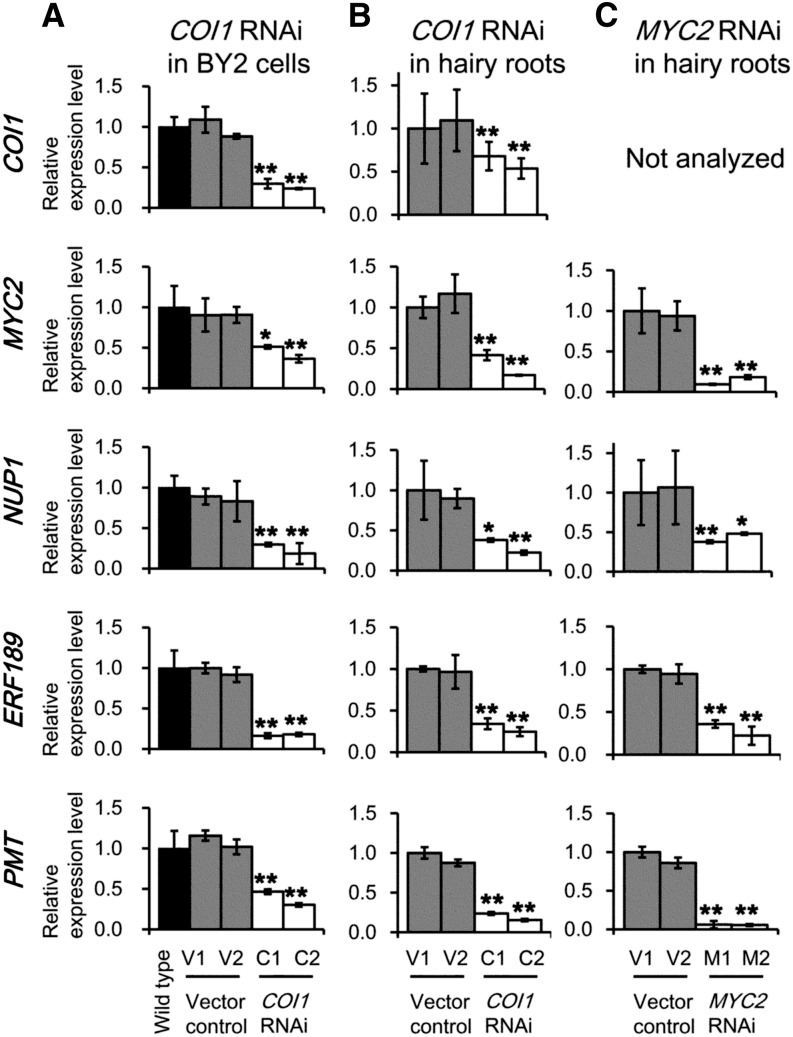
To gain insight into the regulatory hierarchy of NUP1, ERF189, and jasmonate signaling components, we suppressed the expression of COI1 or MYC2 in tobacco BY-2 cells or in tobacco hairy roots and examined their expression levels after alkaloid-inducing MeJA treatment. When COI1 was suppressed in cultured cells (Fig. 6A) and in hairy roots (Fig. 6B) in two independent transgenic lines (C1 and C2, and C3 and C4, respectively), the expression levels of NUP1, PMT, and MATE1/MATE2 were all significantly down-regulated compared with the control levels. NUP1 was also down-regulated in the roots of transgenic tobacco plants in which COI1 was suppressed (Fig. 3A). In hairy root lines in which MYC2 expression was suppressed (M1 and M2), the expression levels of NUP1, PMT, and MATE1/MATE2 were also significantly down-regulated (Fig. 6C).
Figure 6.
NUP1 is regulated by the canonical COI1-MYC2 jasmonate signaling pathway. A to C, Quantitative RT-PCR analysis of COI1, MYC2, NUP1, ERF189, and PMT expression in COI1 RNAi lines (C1 and C2, and C3 and C4, respectively) of cultured tobacco BY-2 cells (A) or tobacco hairy roots (B) and in MYC2 RNAi lines (M1 and M2) of tobacco hairy roots (C). Tobacco cells or roots were treated with 100 μm MeJA for 24 h. The data are the mean values (±sd) of three biological replicates. Significant differences between the wild type (A) or vector controls (B and C) and the test samples were determined by Dunnett’s test and are indicated by asterisks (single asterisk for P < 0.05 and double asterisks for P < 0.01).
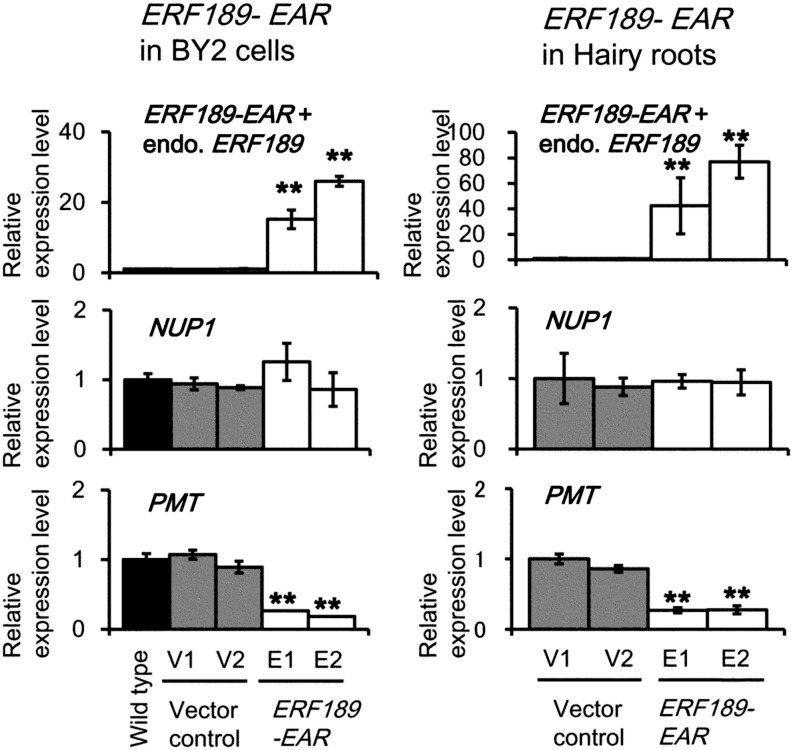
To suppress the function of partially redundant NIC2-locus ERF genes, we constitutively expressed a dominant-negative form of ERF189 (ERF189-EAR; Shoji et al., 2010) in two independent transgenic lines (E1 and E2, and E3 and E4, respectively) of MeJA-treated tobacco BY-2 cells (Fig. 7A) or tobacco hairy roots (Fig. 7B). In both culture systems, the expression of ERF189-EAR effectively down-regulated PMT, as reported previously (Shoji et al., 2010), but did not affect the expression of NUP1. These results suggest that MeJA-induced expression of NUP1 is mediated by the canonical jasmonate signaling components, including COI1 and MYC2, and that NUP1 acts upstream of the NIC2-locus ERF genes and their target structural genes involved in nicotine biosynthesis and transport.
Figure 7.
NUP1 is not regulated by ERF189. Quantitative RT-PCR analysis of ERF189, NUP1, and PMT expression in cultured tobacco BY-2 cells or tobacco hairy roots of wild-type, vector control (V3 and V4, and V5 and V6, respectively), and ERF189-EAR overexpressing (E1 and E2, and E3 and E4, respectively) lines. An expression assay for ERF189 detected transcripts of both the ERF189-EAR transgene and endogenous ERF189. Tobacco cells or roots were treated with 100 μm MeJA for 24 h. The data are the mean values (±sd) of three biological replicates. Significant differences between the wild type (A) or vector controls (B) and the test samples were determined by Dunnett’s test and are indicated by asterisks (double asterisks for P < 0.01).
DISCUSSION
NUP1 Is Expressed in Root Epidermal Cells
The NUP1 promoter is primarily expressed in the epidermal cells of the root tip and in the outermost cell layer of the lateral root cap, with weaker expression in the parenchyma cells of the stele. Low but substantial expression of NUP1 was also observed in the leaf and stem of tobacco plants, although NUP1-expressing cell types were not identified in the aerial organs in this study. These spatial expression patterns of NUP1 in tobacco roots are distinct from those of known enzyme genes involved in nicotine biosynthesis, which are strongly expressed in the root cortex and not in the aerial parts (Shoji et al., 2000, 2002; Kajikawa et al., 2011; Shoji and Hashimoto, 2011b). Tobacco MATE1/MATE2 genes, whose products sequester nicotine into the vacuole during active alkaloid biosynthesis, are also expressed in the root cortex, in a similar pattern as nicotine enzyme genes (Shoji et al., 2009). The spatially coordinated expression of these tobacco genes is likely controlled by key transcription factors for nicotine biosynthesis and accumulation, such as ERF189 (Shoji et al., 2010). The preferential expression of NUP1 in the epidermis indicates that its transcriptional control does not depend on ERF189. Indeed, when ERF189 and its related transcription factors were functionally suppressed, the expression of nicotine biosynthesis genes, but not of NUP1, was highly reduced (Shoji et al., 2010; Fig. 7). Thus, although NUP1 transports nicotine, its transcriptional regulation does not constitute an ERF189-mediated nicotine regulon.
The physiological functions of NUP1-mediated tobacco alkaloid uptake are not well understood. Epidermis-localized NUP1 may prevent apoplastic nicotine, which is synthesized in the cortex, from being secreted into the rhizosphere at the time of active nicotine synthesis or may even retrieve secreted nicotine back into the root tissues. NUP1 overexpression in tobacco roots would be expected to decrease nicotine concentrations in the root apoplast, and may affect the root-to-shoot transport of nicotine via xylem. Indeed, in NUP1-overexpressing plants, the concentration of nicotine in the xylem sap was significantly decreased, resulting in decreased foliar nicotine contents and increased root nicotine contents (Fig. 2). These results indicate that NUP1 controls the apoplastic distribution of tobacco alkaloids in the root, and potentially influences the long-range shoot-bound transport of root alkaloids. Thus, it is noteworthy that Nicotiana alata, a wild Nicotiana sp. containing very low levels of foliar alkaloids, does not efficiently transport root-synthesized alkaloids to the aerial parts, and that this nontransporting phenotype (i.e. the retention of alkaloids in the root) is genetically dominant over the transporting phenotype (Pakdeechanuan et al., 2012). It would be interesting to determine whether NUP1 or a functionally similar nicotine transporter is overexpressed in N. alata roots.
NUP1 Regulates Nicotine Biosynthesis via ERF189
The suppression of NUP1 expression in tobacco hairy roots and the resulting regenerated plants unexpectedly reduced the total nicotine contents by unknown molecular mechanisms (Hildreth et al., 2011). Down-regulation of NUP1 in cultured tobacco cells significantly decreased the total amounts of tobacco alkaloids in the culture, whereas NUP1 overexpression increased these amounts (Fig. 4B), indicating that NUP1 either promotes alkaloid biosynthesis or inhibits alkaloid degradation.
Expression analysis of PMT and QPT revealed that these enzyme-encoding genes involved in nicotine biosynthesis are positively regulated by NUP1 in plant roots and cultured cells (Figs. 3 and 4). Moreover, the vacuolar-localized nicotine transporter genes MATE1/MATE2 are similarly regulated by NUP1. Because these structural genes involved in nicotine biosynthesis and transport are the direct targets of the NIC2-locus ERF genes (Shoji et al., 2010), we analyzed the expression of a representative NIC2-locus gene, ERF189, which was found to be up- and down-regulated by the up- and down-regulation of NUP1 expression, respectively. Although ERF189 expression is regulated by MYC2, modulating NUP1 expression levels did not affect the expression of MYC2 (Figs. 3 and 4). The expression levels of PI-II, a gene that functions downstream of MYC2, did not change upon modulation of NUP1 expression, indicating that MYC2 was not activated at the transcriptional or posttranscriptional levels under these conditions. Therefore, the regulation of ERF189 by NUP1 is not mediated by MYC2. The functional suppression of ERF189 and the related ERF subclade members in the ERF189-EAR overexpressing lines confirmed that NUP1 acts upstream of ERF189, but not vice versa (Fig. 7).
Hildreth et al. (2011) did not find reduced transcript levels of nicotine enzyme genes encoding Orn decarboxylase, quinoline synthase, QPT, PMT, and N-methylputrescine oxidase in the jasmonate-treated hairy root lines induced from NUP1-RNAi tobacco plants. Upon exogenous jasmonate treatment, NUP1 transcript levels in their NUP1-RNAi roots increased severalfold from the untreated values, although the increased levels were still lower than the levels in wild-type tobacco roots (Hildreth et al., 2011). The elevated levels of NUP1 transcript might have obscured the impact of NUP1 suppression in their experimental conditions. It should be noted that, in the absence of jasmonate, the expression levels of ORN DECARBOXYLASE, QUINOLINE SYNTASE, and PMT were significantly suppressed in several NUP1-RNAi hairy root lines (Hildreth et al., 2011).
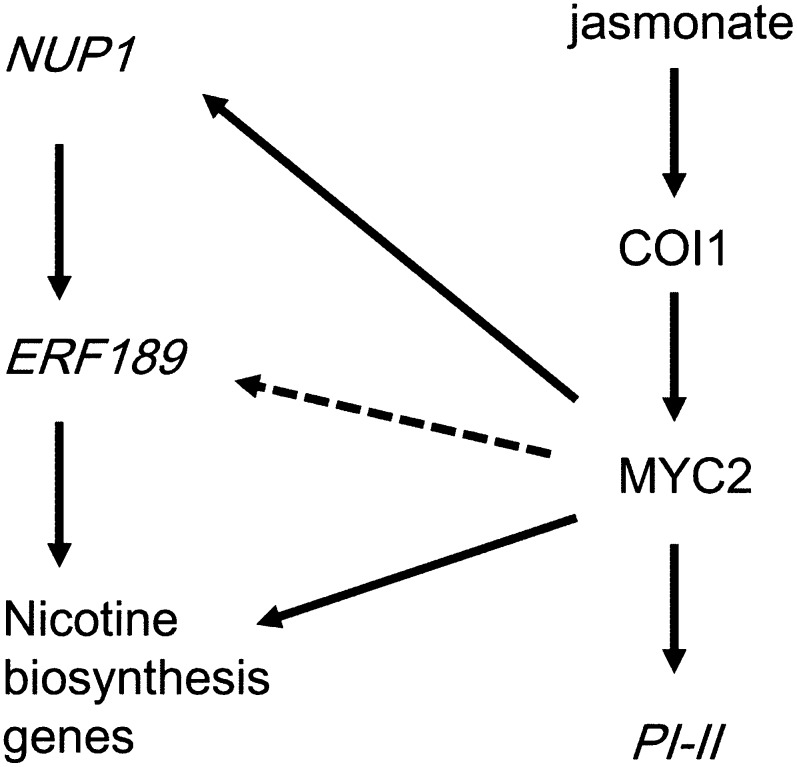
NUP1 is known to be up-regulated in hairy root and other cultures by the exogenous application of jasmonates (Hildreth et al., 2011; Kato et al., 2014). In this study, we found that the jasmonate-mediated induction of NUP1 relies on COI1 and MYC2 (Fig. 6). These results are summarized in Figure 8. After jasmonate perception by COI1, activated MYC2 induces the expression of the NIC2-locus ERF genes (e.g. ERF189), as well as of NUP1 and general jasmonate-responsive genes (e.g. PI-II). However, the jasmonate-elicited induction of ERF189 is modulated by the expression levels of NUP1. Successfully induced ERF189 and MYC2 then bind to and activate structural genes involved in nicotine biosynthesis.
Figure 8.
Model of NUP1-mediated regulation of tobacco alkaloid biosynthesis. Bioactive jasmonate is perceived by COI1 and activates MYC2, which induces the expression of general wound-inducible genes (e.g. PI-II) and nicotine biosynthesis genes. MYC2 and the NIC2-locus transcription factors (e.g. ERF189) specifically regulate nicotine biosynthesis. This study demonstrates that jasmonate-induced expression of ERF189 requires the MYC2-mediated expression of NUP1. Previously reported MYC2-dependent ERF189 expression (dashed arrow; Shoji and Hashimoto, 2011c) may be mediated by NUP1.
It is unknown how a nicotine transporter (i.e. NUP1) affects the expression of a transcription factor gene ERF189. Inhibiting the biosynthesis and accumulation of tobacco alkaloids by suppressing the expression of nicotine biosynthesis enzyme genes (i.e. QPT and BBL) did not affect ERF189 expression (Fig. 5), indicating that the cellular levels of tobacco alkaloids are unlikely to mediate the transcriptional activation function of NUP1. Because the uptake substrates of NUP1 are not restricted to tobacco alkaloids (Kato et al., 2014), unidentified bioactive metabolites that are secreted from MeJA-treated tobacco cells or are present in the culture medium might be transported by NUP1. Alternatively, NUP1 might possess a unique biochemical function distinct from, and in addition to, its transporter activity. In this scenario, a NUP1 mutant in which the transporter activity is abolished may activate ERF189. It would be interesting to explore whether Arabidopsis NUP1-related transporters (i.e. PURINE PERMEASE1 [PUP1], PUP2, and PUP3) have biological functions that are not readily explained from their known transporter activities.
MATERIALS AND METHODS
Plant Materials and Transformation
Nicotiana tabacum ‘Burley 21’ or ‘Petit Havana’ line SR1 plants were grown in the greenhouse. The tobacco BY-2 cell suspension was cultured as described (Nagata et al., 1992). To induce alkaloid biosynthesis, 4-d-old BY-2 cells were first rinsed extensively to remove 2,4-dichlorophenoxyacetic acid and then transferred to fresh auxin-free medium at an inoculum density of a 10-mL packed cell volume per 90 mL of culture medium. After incubation for 12 h, MeJA was added to a final concentration of 100 μm. All samples were frozen in liquid nitrogen immediately after harvest and were kept at −80°C until use.
Transgenic tobacco plants (SR1) were produced using Agrobacterium tumefaciens strain EHA105 and a leaf disc transformation protocol (Horsch et al., 1985). Transgenic plants of the T1 generation were analyzed and nontransgenic T1 progeny were excluded from the analysis. Tobacco BY-2 cells were transformed as described (An, 1985) using A. tumefaciens strain EHA105. Tobacco hairy roots were induced from tobacco ‘SR1’ leaf discs by Agrobacterium rhizogenes strain ATCC15834 and cultured in liquid Gamborg B5 medium at 25°C, as described (Shoji et al., 2010).
Plasmid Construction
The 5′-upstream regulatory region (1.4 kb) of NUP1 was obtained by PCR using genomic DNA extracted from tobacco ‘Burley 21.’ PCR primers (Supplemental Table S1) were designed based on the genome fragment sequence C32375 in the tobacco Methylation Filtered Genome TGI:V.1 (http://solgenomics.net). After the identity of the cloned NUP1 promoter region was confirmed by sequencing, it was flanked by the HindIII and BamHI sites, and then cloned into pBI121 to generate ProNUP1::GUS.
To generate the NUP1-RNAi vector, two complementary DNA (cDNA) fragments of NUP1 (+750 to +1,129, relative to the translation initiation ATG) were placed in front of a pyruvate dehydrogenase kinase intron and just after it in the reverse orientation in the pHANNEBAL vector (Wesley et al., 2001), and the resulting RNAi expression cassette was inserted into pBI121 at the BamHI and SacI sites. The QPT1-RNAi vector was constructed in a similar way, using the QPT1 cDNA fragment (+1 to +510). The sequences of the PCR primers used for cloning are listed in Supplemental Table S1.
Transformation vectors used to overexpress NUP1 (Kato et al., 2014), to down-regulate COI1 (Shoji et al., 2008), MYC2 (Shoji and Hashimoto, 2011c), or BBL (Kajikawa et al., 2011), or to dominantly suppress the expression of ERF189-related genes (Shoji et al., 2010) have been reported.
Histochemical GUS Assay
GUS activity was detected histochemically and cross sections of roots were prepared as described previously (Shoji et al., 2000). GUS-stained cultured roots were rinsed with 20 mm phosphate buffer, pH 7.0, embedded in 5% (w/v) agar containing 1 mm dithiothreitol and 20 mm phosphate buffer (pH 7.0), and sliced into 75- to 100-μm-thick cross sections using a microslicer (DTK-1500; Dohan EM). For the sections in Supplemental Figure S1, roots were embedded in Technovit 7100 (Heraeus Kulzer) and sliced to give 10-μm-thick cross and longitudinal sections using a MICROM HM335E Rotary Microtome (Thermo Fisher Scientific). The GUS-stained sections were then counterstained with neutral red. Images of the stained tissues and sections were captured with an SZX12 (Olympus) or an Eclipse E-1000 (Nikon) microscope, equipped with a DP-70 digital camera (Olympus).
Quantitative Reverse Transcription PCR
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen) from samples that had been ground in liquid nitrogen. cDNA was synthesized from 1 μg of total RNA using Super Script II Reverse Transcriptase (Invitrogen) and an oligo(dT) primer. The cDNA templates were amplified using a LightCycler 480 (Roche) with SYBR Premix Ex Taq (Takara) under the following conditions: 95°C for 10 min, followed by 55 cycles of 94°C for 10 s, 55°C for 10 s, and 72°C for 15 s. EF1a was used as a reference gene. The PCR primers used are listed in Supplemental Table S2. The QPT primers amplified both QPT1 and QPT2 (Shoji and Hashimoto, 2011b), while all four BBL members, BBLa, BBLb, BBLc, and BBLd, were collectively detected by the BBL primers (Kajikawa et al., 2011).
Alkaloid Analysis
The alkaloids in freeze-dried plant samples were purified as described (Shoji et al., 2009). When BY-2 cell cultures and hairy root cultures were harvested, the culture medium was separated from the cultured samples after filtering through a paper filter (0.26-mm thickness × 55 ø; Advantech) and centrifuged at 17,400g for 10 min. The supernatant was then applied directly to an Extrelut-1 column (Merck), eluted with chloroform, and dried as described (Shoji et al., 2009). Purified tobacco alkaloids were analyzed by gas-liquid chromatography (Shoji et al., 2009). The xylem sap was collected from the base of 8-week-old tobacco plants as described (Pakdeechanuan et al., 2012) and was analyzed for alkaloids in the same manner as described above for alkaloids in the culture medium.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU174267 (NUP1), AB433899 (COI1), HM466975 (MYC2), AB827951 (ERF189), AB004323 (PMT), AJ748263 (QPT1), AJ748263 (QPT2), AB604219 (BBLa), AM851017 (BBLb), AB604220 (BBLc), AB604221 (BBLd), AB286961 (MATE1), AB286962 (MATE2), Z29537 (PI-II), and D63396 (EF1α).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Histochemical GUS staining of ProNUP1::GUS tobacco hairy roots.
Supplemental Figure S2. NUP1 expression affects the distribution of tobacco alkaloids in cells and the culture medium.
Supplemental Figure S3. Down-regulation of BBL does not affect the expression of genes involved in alkaloid biosynthesis and transport.
Supplemental Table S1. PCR primers used for vector construction.
Supplemental Table S2. Quantitative reverse transcription-PCR primers used in this study.
Supplementary Material
Glossary
- QPT
quinolinate phosphoribosyltransferase
- PMT
putrescine N-methyltransferase
- ERF
ethylene response factor
- MeJA
methyljasmonate
- RNAi
RNA interference
- cDNA
complementary DNA
- RT
reverse transcription
Footnotes
This work was supported in part by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research no. 26440144 to T.S.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- An G. (1985) High efficiency transformation of cultured tobacco cells. Plant Physiol 79: 568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandin T, van der Does C, Albert JMB, Bol JF, Linthorst HJ (1995) Structure and induction pattern of a novel proteinase inhibitor class II gene of tobacco. Plant Mol Biol 27: 1197–1204 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A (2011) APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J 66: 1053–1065 [DOI] [PubMed] [Google Scholar]
- Dewey RE, Xie J (2013) Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum. Phytochemistry 94: 10–27 [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth SB, Gehman EA, Yang H, Lu RH, Ritesh KC, Harich KC, Yu S, Lin J, Sandoe JL, Okumoto S, et al. (2011) Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc Natl Acad Sci USA 108: 18179–18184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Shoji T, Kato A, Hashimoto T (2011) Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol 155: 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Shitan N, Shoji T, Hashimoto T (June 16, 2014) Tobacco NUP1 transports both tobacco alkaloids and vitamin B6. Phytochemistry http://dx.doi.org/10.1016/j.phytochem.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Katoh A, Uenohara K, Akita M, Hashimoto T (2006) Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg PD, Collins GB (1971) Inheritance of percent total alkaloids in Nicotiana tabacum L.: II. genetic effects of two loci in Burley21 x LA Burley21 populations. Can J Genet Cytol 13: 287–291 [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K, Van Montagu MC, Inzé D, Rischer H, Goossens A, Oksman-Caldentey KM, Moriyama Y, Yazaki K (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 106: 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Pakdeechanuan P, Shoji T, Hashimoto T (2012) Root-to-shoot translocation of alkaloids is dominantly suppressed in Nicotiana alata. Plant Cell Physiol 53: 1247–1254 [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT (2007) Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51: 79–91 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2008) Why does anatabine, but not nicotine, accumulate in jasmonate-elicited cultured tobacco BY-2 cells? Plant Cell Physiol 49: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2011a) Nicotine biosynthesis. InAshihara H, Crozier A, Komamine A, eds, Plant Metabolism and Biotechnology, Wiley & Sons, New York, pp 191–216 [Google Scholar]
- Shoji T, Hashimoto T (2011b) Recruitment of a duplicated primary metabolism gene into the nicotine biosynthesis regulon in tobacco. Plant J 67: 949–959 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2011c) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol 52: 1117–1130 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2012) DNA-binding and transcriptional activation properties of tobacco NIC2-locus ERF189 and related transcription factors. Plant Biotechnol 29: 35–42 [Google Scholar]
- Shoji T, Hashimoto T (2013) Biosynthesis and regulation of tobacco alkaloids. InWallner F, ed, Herbaceous Plants: Cultivation Methods, Grazing and Environmental Impacts, Nova Biomedical, New York, pp 37–67 [Google Scholar]
- Shoji T, Hashimoto T (June 16, 2014) Stress-induced expression of NICOTINE2-locus genes and their homologs encoding Ethylene Response Factor transcription factors in tobacco. Phytochemistry http://dx.doi.org/10.1016/j.phytochem.2014.05.017 [DOI] [PubMed] [Google Scholar]
- Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, et al. (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22: 3390–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Mishima M, Hashimoto T (2013) Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiol 162: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Ogawa T, Hashimoto T (2008) Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol 49: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Shoji T, Winz R, Iwase T, Nakajima K, Yamada Y, Hashimoto T (2002) Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol Biol 50: 427–440 [DOI] [PubMed] [Google Scholar]
- Shoji T, Yamada Y, Hashimoto T (2000) Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol 41: 831–839 [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT (2004) Nicotine’s defensive function in nature. PLoS Biol 2: E217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AT, Liu E, Polvi SL, Pammett RT, Page JE (2010) A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J 62: 589–600 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Xie JH, Song W, Maksymowicz W, Jin W, Cheah K, Chen WX, Carnes C, Ke J, Conkling MA (2004) Biotechnology: a tool for reduced risk tobacco products – the nicotine experience from test tube to cigarette pack. Rev Adv Tob Sci 30: 17–37 [Google Scholar]
- Zhang HB, Bokowiec MT, Rushton PJ, Han S-C, Timko MP (2012) Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol Plant 5: 73–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.