A β-amylase that works during the day and under osmotic stress is more active at high pH and more thermostable than a mesophyll cell β-amylase that works at night and under cold stress.
Abstract
Starch degradation in chloroplasts requires β-amylase (BAM) activity, which is encoded by a multigene family. Of nine Arabidopsis (Arabidopsis thaliana) BAM genes, six encode plastidic enzymes, but only four of these are catalytically active. In vegetative plants, BAM1 acts during the day in guard cells, whereas BAM3 is the dominant activity in mesophyll cells at night. Plastidic BAMs have been difficult to assay in leaf extracts, in part because of a cytosolic activity encoded by BAM5. We generated a series of double mutants lacking BAM5 and each of the active plastidic enzymes (BAM1, BAM2, BAM3, and BAM6) and found that most of the plastidic activity in 5-week-old plants was encoded by BAM1 and BAM3. Both of these activities were relatively constant during the day and the night. Analysis of leaf extracts from double mutants and purified BAM1 and BAM3 proteins revealed that these proteins have distinct properties. Using soluble starch as the substrate, BAM1 and BAM3 had optimum activity at pH 6.0 to 6.5, but at high pH, BAM1 was more active than BAM3, consistent with its known daytime role in the guard cell stroma. The optimum temperature for BAM1, which is transcriptionally induced by heat stress, was about 10°C higher than that of BAM3, which is transcriptionally induced by cold stress. The amino acid composition of BAM1 and BAM3 orthologs reflected differences that are consistent with known adaptations of proteins from heat- and cold-adapted organisms, suggesting that these day- and night-active enzymes have undergone thermal adaptation.
Starch is an important energy and carbon storage compound in most plants. In leaves, starch accumulates in chloroplasts during the day and is broken down at night, when photosynthesis is inactive, to supply sugars for local use and export (Zeeman et al., 2010). During the day, about one-half of the reduced carbon is exported from plastids as triose phosphate, and the other one-half is converted to starch (Smith and Stitt, 2007). Degradation of starch at night begins with phosphorylation of the outer glucan chains by glucan/water dikinase and phosphoglucan/water dikinase (Ritte et al., 2002; Kötting et al., 2005). These enzymes phosphorylate the C6 and C3 positions of individual glucosyl residues, and the consequent ionic repulsion leads to hydration of the outer glucan chains (Hejazi et al., 2008). The chains are then simultaneously dephosphorylated by phosphoglucan phosphatases (Kötting et al., 2009; Comparot-Moss et al., 2010), linearized by debranching enzymes (Streb et al., 2012), and hydrolyzed, primarily by β-amylases (BAMs) that generate maltose (Scheidig et al., 2002; Lu and Sharkey, 2004; Weise et al., 2004; Zeeman et al., 2004; Kaplan and Guy, 2005). Although some Glc is also produced and exported from chloroplasts (Cho et al., 2011), the major product of starch degradation is maltose, which is exported to the cytosol for additional metabolism (Niittylä et al., 2004).
In Arabidopsis (Arabidopsis thaliana), there are nine genes encoding BAM-like proteins. The first to be isolated, BAM5 (also called βAMYLASE1 [BMY1] or REDUCED β-AMYLASE1 [RAM1] but for simplicity, the BAM names are used here and refer to the Arabidopsis genes), encodes a catalytically active cytosolic enzyme found primarily in phloem tissue (Monroe and Preiss, 1990; Monroe et al., 1991; Wang et al., 1995). Mutants lacking BAM5 have no obvious phenotype (Laby et al., 2001). BAM5 is transcriptionally induced by sugars (Mita et al., 1995) and overexpressed in several different starchless mutants that accumulate sugar (Caspar et al., 1989; Monroe and Preiss, 1990). BAM5 activity also changes developmentally (Doyle et al., 2007), and under some conditions (e.g. high light), BAM5 can account for up to 80% of total leaf BAM activity (Lin et al., 1988; Caspar et al., 1989), but its function is unknown.
BAM3 encodes a catalytically active, plastid-localized enzyme expressed in mesophyll cells that plays an important role in leaf starch degradation at night (Lao et al., 1999; Kaplan and Guy, 2005; Fulton et al., 2008). The potato (Solanum tuberosum) ortholog of BAM3 has a similar function (Scheidig et al., 2002). BAM1 also encodes a catalytically active, plastid-localized enzyme (Sparla et al., 2006; Fulton et al., 2008), but in young leaves, its expression is restricted to guard cells, where it functions during the day (Valerio et al., 2011), likely to provide carbon skeletons for malate and Suc accumulation accompanying stomatal opening (Outlaw and Manchester, 1979). After plants begin flowering, BAM1 is also expressed in mesophyll cells (Valerio et al., 2011). Unlike BAM3, BAM1 contains an internal disulfide bond that is reduced by thioredoxin (Sparla et al., 2006). The enzyme is only active in its reduced state, but the extent of BAM1 reduction in vivo is not known. Both BAM1 and BAM3 were expressed in Escherichia coli, and no differences in their catalytic properties were reported (Fulton et al., 2008; Li et al., 2009). BAM2 also encodes a plastid-localized enzyme, but the specific activity of the expressed form was reported to be far lower than that of BAM1 or BAM3, and no mutant phenotypes were described (Fulton et al., 2008; Li et al., 2009). Li et al. (2009) found that BAM2 bound starch only weakly, perhaps because of a four-amino acid insertion in a surface loop very near the active site. Little is known about BAM6, but it apparently evolved from a relatively recent duplication of a chromosome segment containing BAM5 (Fulton et al., 2008). Like BAM5, BAM6 is catalytically active (C. Torres and J. Monroe, unpublished data), but unlike BAM5, it is located in plastids (S. Zeeman, personal communication).
BAM4 encodes a plastid-localized protein, but it lacks some of the conserved residues important for catalysis and has no observable catalytic activity (Fulton et al., 2008; Li et al., 2009). However, mutants lacking BAM4 have a starch excess phenotype, and therefore, it was hypothesized to play a regulatory role in starch metabolism (Fulton et al., 2008). BAM9 is similar to BAM4 in that it also lacks conserved residues important for catalytic activity (Fulton et al., 2008), has no observable catalytic activity (K. Fedkenheuer and J. Monroe, unpublished data), and is located in plastids (S. Zeeman, personal communication). It is possible that BAM4 and BAM9 catalyze reactions with as yet unidentified substrates. BAM7 and BAM8 are the most unusual members of the family, in that they encode proteins with N-terminal BRASSINAZOLE RESISTANT1-like domains, are targeted to nuclei, have little or no catalytic activity, and function as DNA-binding transcription factors (Reinhold et al., 2011). Their BAM domains were recently shown to be involved in their function as transcription factors (Soyk et al., 2014), but the ligands that they bind have not been identified.
Very little is known about the in vivo catalytic rates and properties of the plastid-localized BAM enzymes, partly because leaf extracts may contain multiple isoforms and partly because of the masking effect of cytosolic BAM5. In addition, because BAM5 activity is influenced by developmental and environmental parameters, it is difficult to relate induced changes in activity to a particular BAM. Gene expression studies have also been ambiguous. Microarray data indicate that BAM1 and BAM3 transcripts are present throughout the day and the night, but they both reach peaks at dawn (Smith et al., 2004) when starch is nearly exhausted. This could indicate a period of increased demand for BAM protein; however, there is often little correlation between the levels of transcripts and the proteins that they encode (Gibon et al., 2004).
Before the genomic era, there were reports of abiotic stress causing elevated BAM activity in plants (Kaplan et al., 2006), but it was not known which genes were affected. More recently, measurements of mRNA levels in Arabidopsis indicated that BAM1 expression is induced by heat, dehydration, and osmotic stress (Kaplan and Guy, 2004; Maruyama et al., 2009; Valerio et al., 2011), whereas BAM3 expression is induced by cold stress (Kreps et al., 2002; Kaplan and Guy, 2004, 2005; Maruyama et al., 2009; Sicher, 2011). Moreover, Maruyama et al. (2009) found that dehydration and cold stress lead to increased expression of BAM1 and BAM3, respectively, and a reciprocal decreased expression of BAM3 and BAM1, respectively. Stress-induced accumulation of maltose provided correlative support for the postulated secondary roles for these enzymes in contributing to the accumulation of osmoprotectants and cryoprotectants (Kaplan and Guy, 2005; Kaplan et al., 2006; Kempa et al., 2008; Sicher, 2011). Strong evidence for the role of BAM3 in cold stress-induced maltose accumulation was found using an RNA interference knockdown experiment (Kaplan and Guy, 2005). However, there have been no direct measurements of BAM1 and BAM3 activities in abiotically stressed plants.
Here, we report on the activities of two chloroplast-localized BAM enzymes using double mutants that lack BAM5 and each of the plastid-localized BAMs as well as purified recombinant proteins. Most of the plastidic activity in leaves of vegetative Arabidopsis plants is encoded by BAM1 and BAM3, and both of these activities are relatively constant over the course of a day. Interestingly, we observed differences in the pH and temperature sensitivities of these two enzymes, which may suggest how their activities are modulated in situ. Differences in their amino acid composition suggest that they have undergone thermal adaptation.
RESULTS
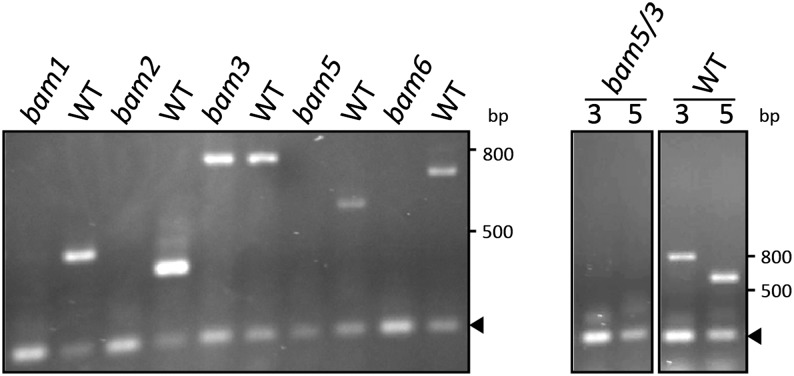
Arabidopsis contains nine BAM-like genes, but three (BAM5, BAM7, and BAM8) encode proteins that are not located in plastids (Wang et al., 1995; Reinhold et al., 2011) and two more (BAM4 and BAM9) encode proteins that are apparently not catalytically active (Fulton et al., 2008; Li et al., 2009; S. Zeeman, personal communication; K. Fedkenheuer and J. Monroe, unpublished data). The remaining four genes (BAM1, BAM2, BAM3, and BAM6) encode plastidic proteins with putative BAM activity, although BAM2 may not contribute much activity (Fulton et al., 2008; Li et al., 2009). To determine which of these genes contributes to leaf plastidic BAM activity that acts on starch, we reasoned that transfer DNA (T-DNA) mutants lacking an active plastidic BAM should, under some conditions, accumulate leaf starch and have lower BAM activity in crude leaf extracts. T-DNA mutants used for these investigations were obtained from the SALK collection (Alonso et al., 2003), and PCR was used to verify that they were homozygous at the appropriate locus (data not shown; Supplemental Table S1). We then used reverse transcription (RT) -PCR and gene-specific primers (Supplemental Table S1) to determine that each single mutant lacked the corresponding transcript (Fig. 1). The only mutant that contained mRNA for the putatively mutated BAM gene was bam3 (SALK_041214), which was previously found to contain some of the expressed mRNA (Kaplan and Guy, 2005; Fulton et al., 2008). This is most likely because the insertion is in the promoter region of BAM3. However, we observed a starch accumulation phenotype in this bam3 line (see below) that was similar to that of the BAM3 mutant used by Fulton et al. (2008), which contains a premature stop codon in the fourth exon. Interestingly, the bam5 bam3 (bam5/3) double mutant that we constructed appeared to contain no observable BAM3 transcript (Fig. 1). We cannot explain this discrepancy, but the lack of BAM3 transcript in bam5/3 leaves agrees with our biochemical data (described below), indicating that the bam5/3 T-DNA line used here contains very little, if any, BAM3 activity.
Figure 1.
RT-PCR screening of T-DNA insertion mutants. Total RNA was extracted from bam mutant Arabidopsis leaves and assayed for transcripts by RT-PCR as described in “Materials and Methods.” Wild-type (WT) RNA was used as a control. The double mutant bam5/3 was tested for the presence of both BAM3 and BAM5. Arrowheads indicate the PP2A internal control. Standard sizes are given in base pairs on the side. Expected amplicon sizes: BAM1, 484 bp; BAM2, 422 bp; BAM3, 807 bp; BAM5, 600 bp; BAM6, 716 bp; and PP2A, 232 bp.
A common method for assessing the contribution of a gene to plastidic starch degradation (whether it is catalytic or regulatory) is to observe starch levels at the end of the night using an iodine stain (Caspar et al., 1991). In wild-type plants, almost all of the leaf starch is exhausted by dawn, regardless of the length of the night (Gibon et al., 2004; Smith and Stitt, 2007). Fulton et al. (2008) used this approach along with catalytic activity of proteins expressed in E. coli to establish that both BAM3 and BAM1 contribute to leaf starch degradation at night in 5-week-old (preflowering) plants, and they found that BAM3 was the dominant enzyme. Plants lacking BAM1 only accumulated starch if BAM3 was also lacking; no starch accumulated in bam1 alone, because BAM3 activity was apparently sufficient to completely degrade all of the starch. Fulton et al. (2008) also found that no starch accumulated in bam2 or the double mutant bam3/2, suggesting that BAM2 does not contribute significantly to leaf starch degradation at this developmental stage. We used this approach to test the roles of all active plastidic BAMs in leaf starch degradation in 5- and 8-week-old plants before and after the onset of flowering.
Among the single mutants tested (bam1, bam2, bam3, and bam6), only bam3 accumulated leaf starch in 5-week-old plants, but in 8-week-old plants that had begun flowering, bam1, bam2, and bam6 also accumulated some starch (Fig. 2). Leaf-to-leaf variation in iodine staining was noticeably higher at this stage of development. As observed by Fulton et al. (2008), we also found that, before flowering, our bam3/2 double mutant and bam3 accumulated a similar level of starch. However, after flowering, bam3/2 accumulated more starch than bam3 (Fig. 2). We also observed similar results with bam3/6. From these results and those in Fulton et al., 2008, we concluded that, before flowering, starch degradation is carried out primarily by BAM3 and to a lesser extent, by BAM1, but that after flowering, all of the plastid-localized active BAMs probably play roles in starch degradation, at least under the growing conditions used here. In this report, we focused our studies on leaf BAM activity in plants harvested at 5 to 6 weeks of age, which was before flowering, and therefore, BAM1 and BAM3 were expected to be the primary contributors.
Figure 2.
Starch content in leaves of 5- and 8-week-old wild-type (WT) and mutant Arabidopsis plants. Plants were grown under a 12-h-light/12-h-dark photoperiod, harvested at the start of the light period, decolorized with hot 80% ethanol, and stained with iodine. Representative leaves are shown.
BAM1 and BAM3 Both Contribute to Leaf Plastidic BAM Activity in Vegetative Plants
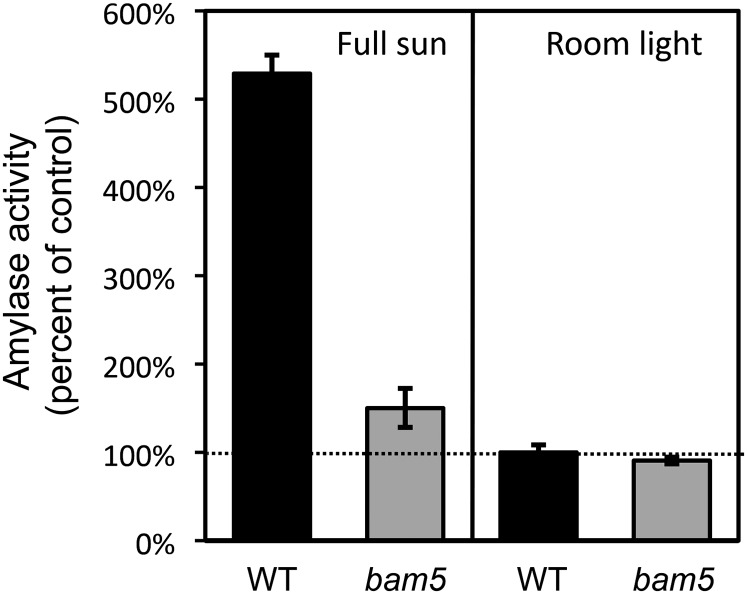
To assess the effect of deleting BAM5 on total extractable leaf BAM activity, we first measured the activity in wild-type and bam5 plants at pH 6.0 using soluble starch as the substrate and the Somogyi-Nelson method to detect the reducing sugars produced (Nelson, 1944). To minimize the contribution of α-amylase in these assays, we included 5 mm EDTA in the extraction buffer and did not include Ca+2 in the assay buffer. Ca+2 is a requirement for α-amylase but not BAM activity, and treatment of pea leaf extracts with 5 mm EDTA strongly inhibited α-amylase activity (Swain and Dekker, 1966; Ziegler, 1988). Comparing extracts from wild-type and bam5 leaves, BAM5 activity represented over 70% of the total activity in greenhouse-grown plants exposed to high light under full sun, but in a growth room under lower light levels, BAM5 comprised less than 10% of total activity (Fig. 3). Activity in bam5 leaves grown under full sun was about 50% higher than activity in low light-grown bam5 leaves, and therefore, one or more of the plastidic BAM activities was probably elevated in the full sun-grown plants. For consistency, all subsequent experiments were performed with plants that were grown under growth room or growth chamber conditions.
Figure 3.
Effect of light conditions under which wild-type (WT) and bam5 mutant plants were grown on total leaf BAM activity. Assays were conducted at 37°C with soluble starch as the substrate. Leaves were harvested at 6 weeks of age. Values are means ± sd (n = 3).
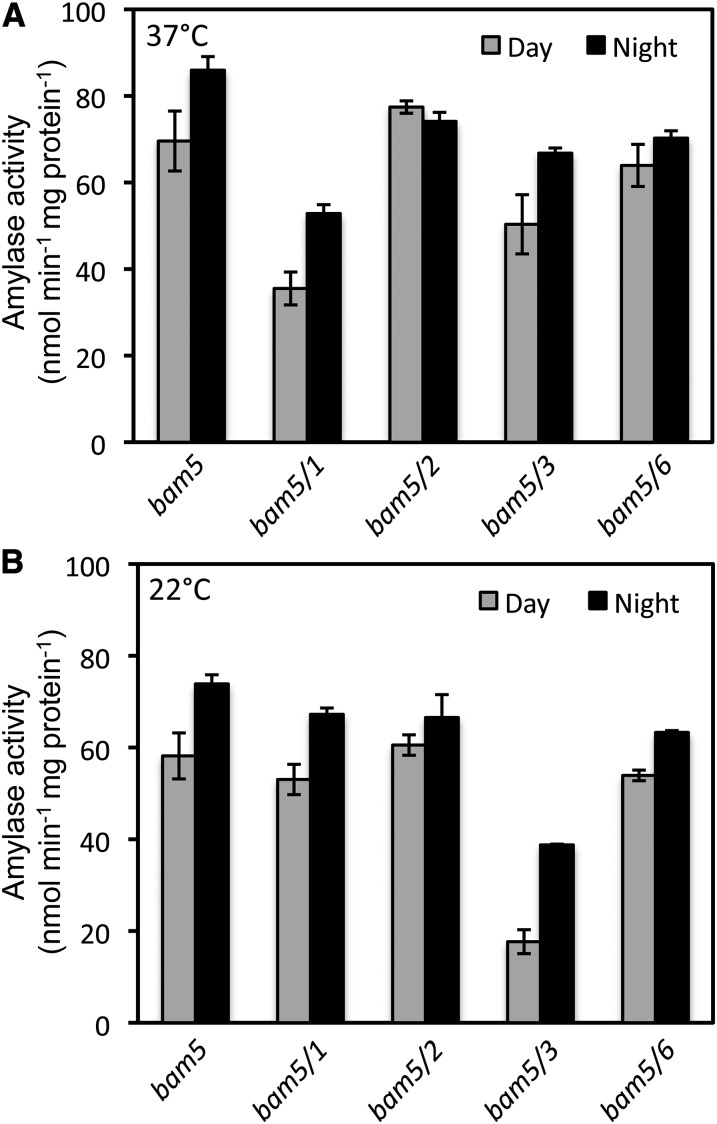
To assess the relative contributions of the BAM genes to plastidic BAM activity, we compared the activity in crude extracts from leaves harvested 4 h into the day and 4 h into the night from 5- to 6-week-old bam5, bam5/1, bam5/2, bam5/3, and bam5/6 plants. Because BAM5 encodes the only active BAM with high specific activity that is not located in plastids (Wang et al., 1995; Reinhold et al., 2011), we reasoned that the activity in mutants lacking BAM5 must be plastidic. Initially, we conducted these assays at 37°C to be consistent with previous studies. Compared with bam5, extracts from bam5/1 and bam5/3 leaves both had lower activity in day- and night-harvested plants (Fig. 4A). Activity in extracts from bam5/2 and bam5/6 leaves differed only slightly from that from bam5 leaves, indicating that BAM2 and BAM6 did not contribute much to leaf BAM activity at this developmental stage; thus, most of the activity in bam5 was probably encoded by BAM1 and BAM3. Similarly, Fulton et al. (2008) observed lower activity in leaf extracts from mutants lacking BAM1 and BAM3 but not the mutant lacking BAM2 compared with the wild type, but their differences were smaller than those that we observed, perhaps because all of their extracts contained BAM5 activity.
Figure 4.
Total BAM activity in crude leaf extracts from 5-week-old single and double mutants of Arabidopsis grown under a 12-h-light/12-h-dark photoperiod. Leaves were harvested 4 h into the day (gray bars) or 4 h into the night (black bars). Extracts were assayed at two different temperatures using soluble starch as the substrate. A, Assays conducted at 37°C. B, Assays conducted at 22°C. Each replicate included all of the leaves from three plants. Values are means ± sd (n = 3).
BAM1 is only active when in a reduced state (Sparla et al., 2006) but including dithiothreitol (DTT) in the assay buffer results in a high level of background color in the Somogyi-Nelson assay that we used for measuring reducing sugars. In this case, the activity being detected is the activity of all reduced BAM1 and not necessarily the total activity of all BAM1 protein present in the extract. Preincubation of a bam5/3 extract, which should contain mostly BAM1 activity, with 20 mm DTT for 60 min at 37°C resulted in a 16% increase in subsequent activity, and therefore, a small portion of the BAM1 protein in our extracts was probably in its oxidized, inactive state, despite the inclusion of 2 mm DTT in the extraction buffer. Low levels of DTT are typically included in extraction buffers to prevent nonspecific oxidation of proteins. We observed 35% less activity in a bam5/3 extract and 41% less activity in a bam5/1 extract when 2 mm DTT was omitted from our extraction buffer (data not shown). There is presently no evidence that BAM3 is specifically regulated by its redox state, and therefore, these results indicate that the inclusion of DTT in the extraction buffer is necessary to prevent nonspecific oxidation leading to enzyme inactivation.
Given the important role of BAM3 in leaf starch degradation, it was surprising that there was only a 20% to 30% decrease in activity from bam5 to bam5/3 leaf extracts assayed at 37°C (Fig. 4A). Because BAM3 is thought to be active at night (Fulton et al., 2008) and is induced by cold stress (Kaplan and Guy, 2005), we wondered if the assay temperature might have been too high to accurately measure BAM3 activity. To test this hypothesis, the same extracts reported in Figure 4A were assayed at 22°C, and it was found that the relative contributions of BAM1 and BAM3 to the activity in bam5 were reversed (Fig. 4B). Activity in bam5/1 was only slightly lower than that of bam5, but activity in bam5/3 was much lower, suggesting that BAM3 encodes the majority of the activity in the bam5 extract when assayed at 22°C. Therefore, when measured at 37°C, BAM3 activity in the bam5/1 leaf extract was probably declining over the course of the assay. Unless noted, all subsequent assays were conducted at 22°C. When measured at 22°C, the rates of activity in bam5/1 and bam5/3 extracts were constant for 2 h (data not shown). Importantly, these results indicated that BAM1 and BAM3 might have different thermal properties.
Whether the assays were conducted at 37°C or 22°C, activities in extracts from bam5/2 and bam5/6 differed little from the activity in bam5 (Fig. 4), suggesting that BAM2 and BAM6 probably contributed little to the activity in bam5 extracts, but their presence in the extracts from bam5, bam5/1, and bam5/3 cannot be ruled out. Also, differences in the activity of extracts harvested during the night and the day were small, indicating that BAM1 and BAM3 were both present during the night and the day (Fig. 4). Nighttime activities were slightly higher than daytime activities in most of the genotypes tested.
BAM1 and BAM3 Have Distinct Properties
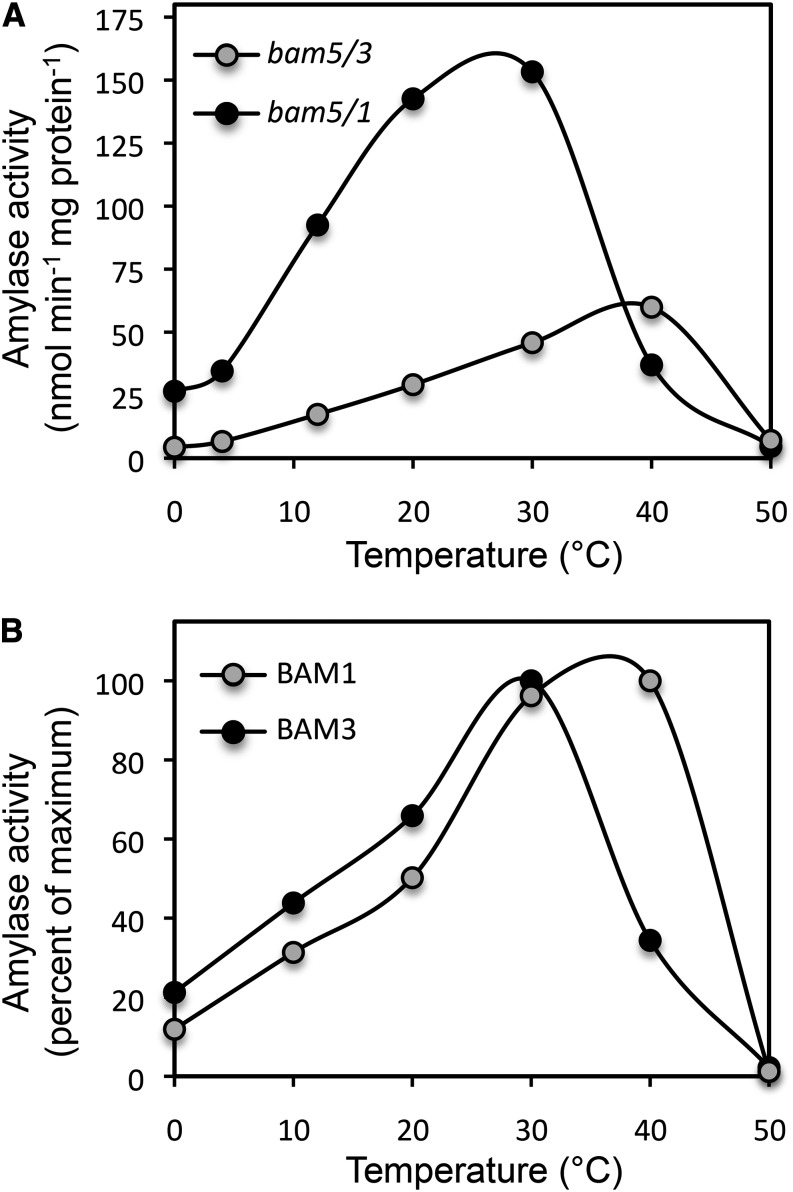
The thermal properties of the bam5/3 and bam5/1 extracts were measured at a range of temperatures. The optimum temperature for activity in bam5/1, which we suspect contains mostly BAM3 activity, was about 10°C lower than the optimum temperature for activity in bam5/3, which we suspect contains mostly BAM1 activity (Fig. 5A). At temperatures typical of cold stress (0°C–10°C), the extract from bam5/1 was about five times more active than that of bam5/3 on a per-milligram total protein basis, whereas at temperatures typical of heat stress (35°C–45°C), bam5/3 activity was up to two times higher than that of bam5/1. To determine whether the thermal properties of the activities in extracts from bam5/3 and bam5/1 leaves could be attributed to BAM1 and BAM3, respectively, we purified recombinant BAM1 and BAM3 expressed in E. coli and conducted similar assays. Treatment of purified recombinant BAM1 with 20 mm reduced DTT for 60 min at 37°C before assaying the enzyme at 22°C resulted in only a small increase in activity compared with untreated enzyme, and therefore, most of the enzyme was in its reduced, active state before initiating the assays (data not shown). A similar treatment of purified BAM1 with 20 mm oxidized DTT caused a sharp decline in activity, revealing that the BAM1 protein used in our experiments was redox sensitive, which was reported by Sparla et al. (2006). Like the leaf extract enzymes, the rates of activity of purified BAM1 and BAM3 were linear for 2 h at 22°C. We conducted assays of the purified proteins at a range of temperatures and expressed the results as percentages of the maximum activity. The optimum temperatures for activity of the recombinant proteins were similar to those of the corresponding leaf extracts, with BAM1 having a higher optimum temperature than BAM3 (Fig. 5B).
Figure 5.
Effect of temperature on BAM1 and BAM3 activity. A, Activity in leaf extracts from two double mutants bam5/3 (gray circles) and bam5/1 (black circles) expressed on a per-milligram protein basis. B, Activity of purified recombinant BAM1 (gray circles) and BAM3 (black circles) expressed as a percentage of the maximum value. All assays were conducted at pH 6 with soluble starch as the substrate.
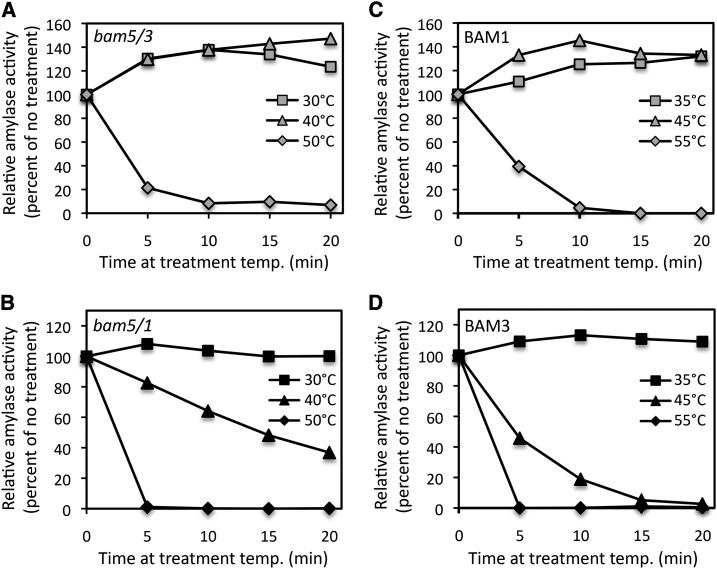
Temperature can affect both catalytic rate constants and the overall thermostability of enzymes (Hochachka and Somero, 2002). To determine if the different temperature profiles for the two activities were caused by a difference in thermostability of BAM1 and BAM3, we held extracts from bam5/1 and bam5/3 at 30°C, 40°C, and 50°C for up to 20 min and then assayed each extract at 22°C. If the decrease in activity for bam5/1 extracts above 30°C shown in Figure 5 was caused by denaturation of BAM3, then we expected to see no restoration of activity when extracts were subsequently assayed at a lower temperature. Treatment at 30°C did not cause any decline in activity of either extract (Fig. 6, A and B). Treatment at 40°C did not cause a loss of activity from bam5/3, but activity of bam5/1 steadily declined with increasing incubation time up to 20 min. At 50°C, both extracts lost activity rapidly, but bam5/1 was completely inactive in less than 5 min, whereas bam5/3 activity was less sensitive. A similar experiment using purified recombinant BAM1 and BAM3 held at 35°C, 45°C, and 55°C and then assayed at 22°C revealed a similar pattern, where BAM3 lost activity rapidly when held at 45°C, whereas BAM1 did not (Fig. 6, C and D). These observed differences in the thermostability of BAM1 and BAM3 are consistent with their reported roles during day and night, respectively, given the typical daily range of temperatures to which plants are exposed. We then wondered if there might be any structural evidence for this difference in thermostability.
Figure 6.
Residual BAM activity measured at 22°C after holding leaf extracts at 30°C, 40°C, or 50°C or purified recombinant proteins at 35°C, 45°C, or 55°C for up to 20 min. A, Extract from leaves of bam5/3. B, Extract from leaves of bam5/1. C, Purified recombinant BAM1. D, Purified recombinant BAM3. All assays were conducted at pH 6 with soluble starch as the substrate and are expressed as a percentage of the untreated activity.
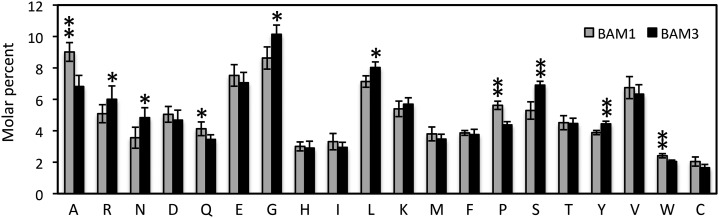
There is a well-known stability-flexibility tradeoff in homologous proteins adapted to function at different temperatures, in which enhanced thermostability conferred by restricted motion is often reflected in differences in amino acid composition (Zhou et al., 2008). To determine if there were any significant differences between the amino acid compositions of BAM1 and BAM3, we identified pairs of BAM1 and BAM3 orthologs from nine sequenced eudicot genomes, including Arabidopsis, Populus trichocarpa, Vitis vinifera, Ricinus communis, Fragaria vesca, Citrus sinensis, Cucumis sativus, soybean (Glycine max), and Solanum lycopersicum (Supplemental Fig. S1). These species were chosen from different taxonomic families to reduce sequence similarities caused by more recent speciation events. Chloroplast transit peptides were predicted using Target-P (Emanuelsson et al., 2007) and removed before calculating their amino acid composition. The mole percentage of each amino acid was averaged for each set of orthologs, and the orthologous sets were compared using a Student’s t test. Of 20 amino acids, 10 were significantly different between the two sets of orthologs (P < 1 × 10−2), and of those, five were highly significantly different (P < 1 × 10−4). BAM1 orthologs contained more Ala, Gln, Pro, and Trp, and BAM3 orthologs contained more Arg, Asn, Gly, Leu, Ser, and Tyr (Fig. 7). Some of these differences have been associated with thermostability or increased flexibility, respectively, in other organisms (Hochachka and Somero, 2002; Siddiqui and Cavicchioli, 2006). In addition, the BAM1 orthologs contained a small but significantly higher proportion of nonpolar residues (56.4% ± 1.1% versus 54.2% ± 0.9% mol) compared with the BAM3 orthologs (P < 1 × 10−3), which is also as expected for a thermostable protein.
Figure 7.
Amino acid composition of nine pairs of BAM1 and BAM3 orthologs from eudicots, including Arabidopsis (NP_189034.1 and NP_567523.1), C. sinensis (XP_006493994.1 and XP_006477060.1), C. sativus (XP_004134029.1 and XP_004147264.1), F. vesca (XP_004296549.1 and XP_004300297.1), soybean (XP_003534086.1 and XP_003524296.1), P. trichocarpa (XP_002314522.2 and XP_006385389.1), R. communis (XP_002518196.1 and XP_002517513.1), S. lycopersicum (NP_001234556.1 and XP_004244551.1), and V. vinifera (XP_002285569.1 and XP_002282871.1). Predicted chloroplast transit peptides were removed before the analysis. Values are means ± sd (n = 9). *, Significant difference (P < 1 × 10−2); **, highly significant difference (P < 1 × 10−4).
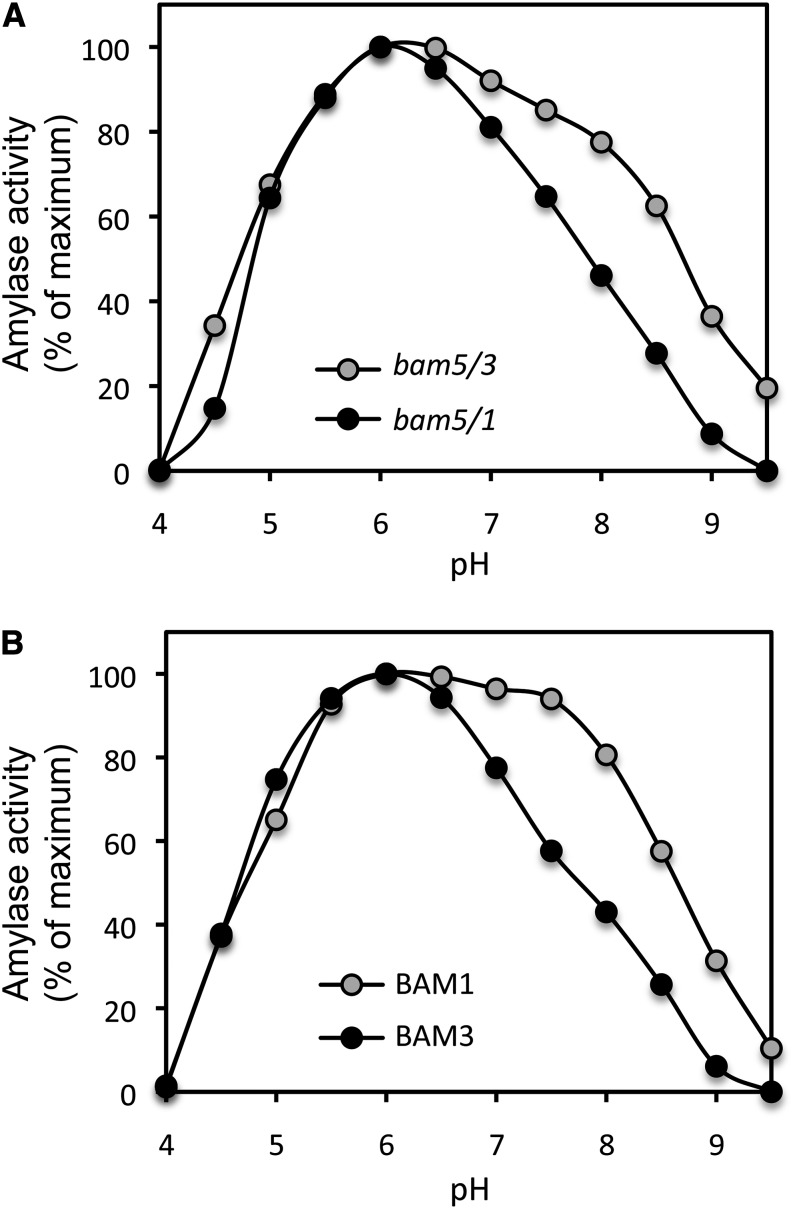
As plastid-localized enzymes, in addition to normally functioning in different thermal environments, BAM1 and BAM3 may also need to function at different pHs. During the night, when BAM3 should be active in mesophyll cells, the pH of the stroma is about 7, but during the day, when BAM1 should be active in guard cells, the pH of the stroma is about 8 (Werdan et al., 1975). Both Fulton et al. (2008) and Li et al. (2009) measured the activity of recombinant BAM1 and BAM3 at various pHs using the artificial substrate p-nitrophenyl maltopentaose (PNPG5). In each study, both enzymes were optimally active at pH 6, but the peaks were relatively narrow, and both enzymes had extremely low activity at pH 8. This raises the question of how BAM1 could be active in guard cell chloroplasts during the day, when it would normally be exposed to a high pH. We measured the effect of pH on extracts from bam5/3 and bam5/1 and recombinant BAM1 and BAM3 at 22°C using soluble starch as the substrate. Similar to the published pH curves, our results indicated that both BAM1 and BAM3 had maximal activity at pH 6, but our peaks were broader than those reported using PNPG5 as the substrate (Fig. 8). However, comparing the pH curves of BAM1 and BAM3, we also observed that, above pH 6.5, the activities of the recombinant BAM1 and the bam5/3 extract were much higher than those of BAM3 and the bam5/1 extract (Fig. 8). At pH 8, BAM1 still had over 80% of its maximum activity.
Figure 8.
Effect of pH on BAM1 and BAM3 activity. A, Activity in leaf extracts from two double mutants: bam5/3 (gray circles) and bam5/1 (black circles). B, Activity of purified recombinant BAM1 (gray circles) and BAM3 (black circles). Assays were conducted at 22°C with soluble starch as the substrate. Activity is expressed as a percentage of the maximum value.
The results described thus far establish that extracts from preflowering bam5/3 and bam5/1 plants probably contain primarily one of the plastid-localized BAM activities encoded by BAM1 or BAM3, respectively, and that these two enzymes have different thermal and pH profiles, consistent with BAM1 being active during the day in guard cells and BAM3 being active at night in mesophyll cells. We then examined the effects of abiotic stress on the expression of BAM1 and BAM3 in wild-type plants and the catalytic activity, sugar, and starch accumulation in bam5/3 and bam5/1 plants.
Activity of BAM1 and BAM3 in Abiotically Stressed Plants
Several different abiotic stresses, such as extreme temperatures, lack of water, and excess salt, cause changes in primary metabolism, leading to the accumulation of compatible solutes that promote survival (Chen and Murata, 2002). Stress-induced starch degradation provides carbon compounds for at least some of this solute synthesis (Lee et al., 2008), and maltose production from BAM activity is one of the first metabolites to appear after the onset of stress (Kaplan and Guy, 2004; Kempa et al., 2008).
Interestingly, several different BAM genes have been implicated in stress-induced starch degradation based on changes in mRNA levels. Osmotic and heat stress both lead to increases in BAM1 mRNA (Kaplan and Guy, 2004; Maruyama et al., 2009; Valerio et al., 2011), whereas cold stress leads to an increase in BAM3 mRNA (Kaplan and Guy, 2004, 2005; Maruyama et al., 2009; Sicher, 2011). Valerio et al. (2011) measured BAM activity after 4 h of exposure to 450 mm mannitol in wild-type Arabidopsis leaves and a mutant lacking BAM1 and observed an increase in total activity in wild-type leaves that was absent from the bam1 mutant. However, because osmotic stress leads to the accumulation of sugars, which in turn leads to elevated BAM5 expression (Mita et al., 1995), the osmotic stress-induced increase in BAM activity that Valerio et al. (2011) observed in wild-type plants may have been caused, in part, by increased BAM5 activity resulting from BAM1-induced starch degradation and not BAM1 activity directly.
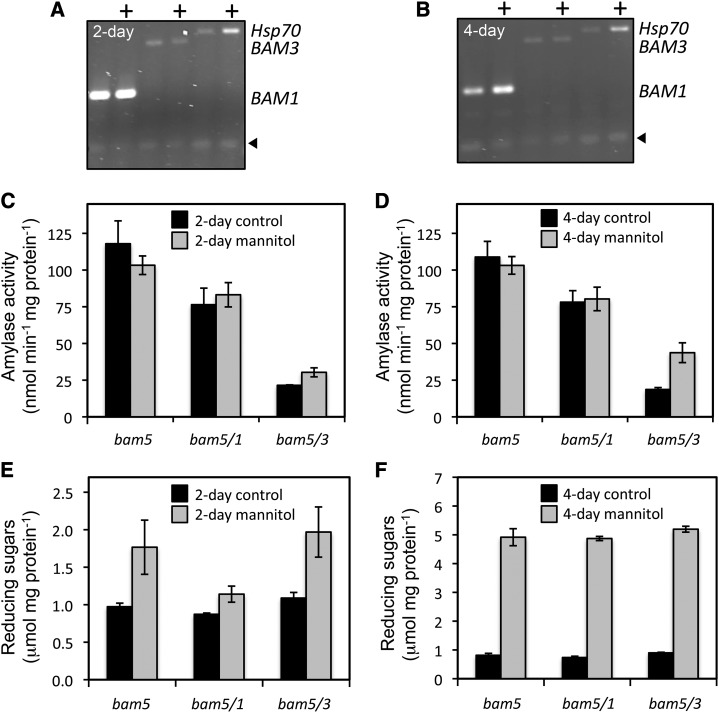
We conducted an experiment similar to one reported by Valerio et al. (2011), in which soil-grown plants were drenched with 450 mm mannitol in nutrient solution, but, after 4 h, there was only a slight increase in BAM activity in leaves of bam5/3 plants, suggesting that the activity of BAM1 was not significantly affected (data not shown). We suspected that this was because Valerio et al. (2011) used hydroponically grown plants that received a more immediate and even dose of the osmoticum. Our longer treatments with 450 mm mannitol caused wilting in all of the genotypes tested, and therefore, we then treated plants with a lower concentration of mannitol (200 mm) and measured mRNA and BAM activity after 2 and 4 d. After treatment for 2 d, growth was slowed in each genotype, but the leaves were not wilted (data not shown). RT-PCR data from wild-type plants showed that the BAM1 transcript was elevated but only at the 4-d time point compared with untreated plants, whereas the BAM3 transcript was unaffected (Fig. 9, A and B). BAM activity in leaf extracts after 2 and 4 d of osmotic stress was unchanged in bam5 and bam5/1 leaves, but in bam5/3 leaves, the activity increased by 40% after 2 d and over 2-fold after 4 d, suggesting that BAM1 activity was, indeed, elevated by this level of stress (Fig. 9, C and D). These results confirmed the finding by Valerio et al. (2011). It is possible that some of the elevated activity in bam5/3 could be encoded by BAM6; however, microarray data suggest that expression of BAM6 is not induced by osmotic stress (Winter et al., 2007).
Figure 9.
Effect of osmotic stress on BAM mRNA and activity and reducing sugars in crude leaf extracts from 5-week-old Arabidopsis plants. A and B, RT-PCR analysis of total RNA isolated from wild-type plants watered with 200 mL of nutrient solution alone or nutrient solution supplemented with 200 mm mannitol (+) and examined after 2 (A) or 4 (B) d. RNA was probed with primers specific for BAM1 or BAM3 mRNA (expected sizes for respective amplicon bands are indicated at the side of the gel). Hsp70 mRNA was also probed as a positive control for osmotic stress. Arrowheads indicate the internal control. C and D, Total BAM activity in bam5, bam5/1, and bam5/3 leaves from similarly treated plants. Assays were conducted at 22°C with soluble starch as the substrate. E and F, Total reducing sugars in the same leaf extracts used for the BAM assays. Black bars represent control plants, and gray bars represent treated plants. Each graph represents developmentally matched plants, but the 2- and 4-d experiments were conducted with different sets of plants. Each replicate included all of the leaves from three plants. Values are means ± sd (n = 3).
We also measured total reducing sugars in the mutants as a proxy for compatible solute accumulation and found that they increased by about 80% in bam5 and bam5/3 after 2 d of treatment with 200 mm mannitol, but in bam5/1, they increased by only 30%, suggesting that BAM1 and not BAM3 contributed to some of the sugar accumulation resulting from the osmotic stress treatment (Fig. 9E). However, after 4 d of osmotic stress, this difference disappeared, and each treated genotype contained 5- to 6-fold more reducing sugars than untreated plants (Fig. 9F). Clearly, factors other than BAM1 contribute to sugar accumulation under osmotic stress. The milder stress did not kill the plants as quickly as the 450 mm mannitol treatment, but we observed no consistent differences among the three genotypes in their sensitivity to the stress.
If BAM1 contributes to osmotic stress-induced sugar accumulation, then we reasoned that mutants lacking BAM1 may accumulate more starch under stress conditions. We used the iodine stain to examine leaves harvested at the start of the light period after 2 and 4 d of osmotic stress and could detect no differences among the genotypes at this level of sensitivity (Supplemental Fig. S2A). All of the genotypes tested had marginally more starch after 2 d of stress and considerably more starch after 4 d of stress.
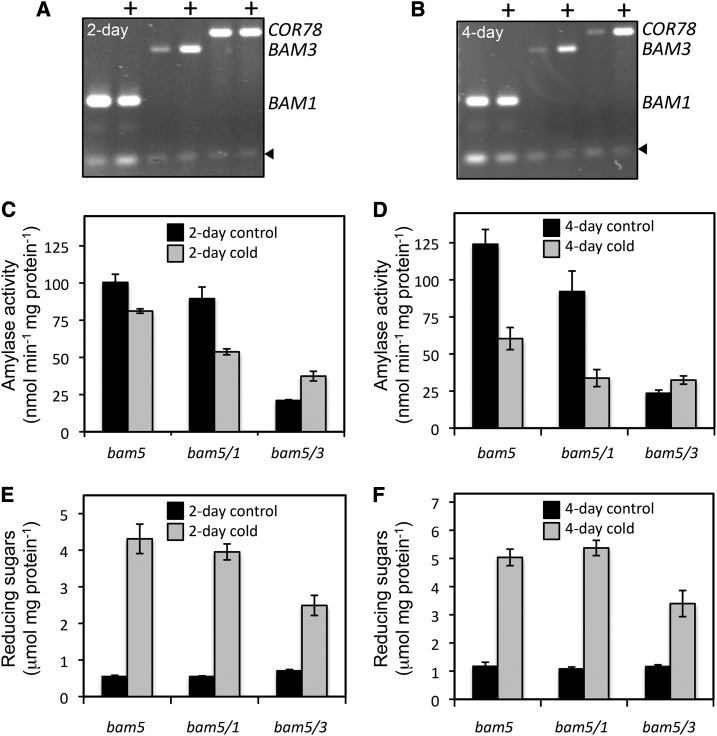
A similar set of experiments was conducted with the same mutants exposed for 2 and 4 d to 4°C under a 12-h-light/12-h-dark photoperiod. As observed by Kaplan and Guy (2004), we also found that the BAM3 transcript was elevated in cold-treated plants compared with untreated control leaves, whereas the level of BAM1 transcript was unchanged (Fig. 10, A and B). However, when we measured BAM activity in cold-stressed plants, there was a decline in activity in bam5 and bam5/1 leaves and an increase in activity in bam5/3 leaves. Activity in extracts from bam5/1, which contained mostly BAM3, was about 40% and 60% lower than that of control plants after 2 and 4 d of cold treatment, respectively, although activity in bam5/3 was about 80% and 60% higher than that of the controls after 2 and 4 d, respectively (Fig. 10, B and C). These results indicate that BAM3 activity declined and that BAM1 activity increased in the cold-treated plants. However, reducing sugars were elevated to the same extent in bam5 and bam5/1 leaves at both time points, whereas in bam5/3 leaves, reducing sugars increased by only about one-half as much (Fig. 10, E and F), suggesting that the accumulation of sugars during cold stress was probably facilitated by BAM3 and not by BAM1. As in the osmotic stress experiment, cold treatment caused starch levels to increase, but there were no obvious differences between the genotypes (Supplemental Fig. S2B).
Figure 10.
Effect of cold stress on BAM mRNA and activity and reducing sugars in crude leaf extracts from 5-week-old Arabidopsis plants. A and B, RT-PCR analysis of total RNA isolated from wild-type plants incubated at normal 22°C or stressed 4°C (+) under a 12-h-light/12-h-dark photoperiod for 2 (A) or 4 (B) d. RNA was probed with primers specific for BAM1 or BAM3 mRNA (expected sizes for respective amplicon bands are indicated at the side of the gel). Cor78 mRNA was also probed as a positive control for cold stress. Arrowheads indicate the internal control. C and D, Total BAM activity in bam5, bam5/1, and bam5/3 leaves from similarly treated plants. Assays were conducted at 22°C with soluble starch as the substrate. E and F, Total reducing sugars in the same leaf extracts used for the BAM assays. Black bars represent control plants maintained at 22°C, and gray bars represent plants treated at 4°C. Each graph represents developmentally matched plants, but the 2- and 4-d experiments were conducted with different sets of plants. Each replicate included all of the leaves from three plants. Values are means ± sd (n = 3).
DISCUSSION
BAMs are known to play an important role in chloroplast starch degradation, but little is known about the properties of these enzymes, in part because there are multiple isozymes but more importantly, because of the masking of their activity by a potentially abundant extrachloroplastic BAM (Caspar et al., 1989; Ghiena et al., 1993). Genetic evidence was used to show that BAM3 is the primary enzyme acting on starch at night, because mutants lacking or with diminished BAM3 have a starch-excess phenotype (Kaplan and Guy, 2005; Fulton et al., 2008). Mutants lacking BAM1 accumulate starch in guard cells, suggesting that it normally functions during the day, when guard cell starch degradation supports malate and Suc accumulation (Valerio et al., 2011). Another role for BAM1 in mesophyll cells was shown using mutants lacking both BAM1 and BAM3 (Fulton et al., 2008), which accumulate more starch than the mutant lacking only BAM3. Valerio et al. (2011) also showed that the BAM1 promoter was active in mesophyll cells but only after the onset of flowering. Our genetic evidence using single and double mutants showed that, in addition to BAM3 and BAM1, BAM2 and BAM6 may also play a role in leaf starch degradation after plants begin flowering (Fig. 2). This conclusion is supported by a microarray study showing that the expression of BAM2 and BAM6 increased in older leaves (Breeze et al., 2011).
To assess the relative contribution of four active or putatively active plastidic BAMs to plastidic activity, we generated double mutants lacking each active plastidic BAM (BAM1, BAM2, BAM3, and BAM6) and the cytosolic BAM5. Despite the potential of BAM5 to contribute most of the leaf activity under some conditions, such as high light (Caspar et al., 1989; Fig. 3), mutants lacking BAM5 have no obvious phenotype (Laby et al., 2001). Several lines of evidence indicate that, before flowering, most of the plastidic BAM activity is encoded by only BAM1 and BAM3. First, BAM4, BAM7, BAM8, and BAM9 are known to be catalytically inactive or nearly so (Fulton et al., 2008, Li et al., 2009; Reinhold et al., 2011; K. Fedkenheuer and J. Monroe, unpublished data). Second, compared with the BAM activity in bam5 leaves, the activity in bam5/2 and bam5/6 leaves was similar, suggesting that BAM2 and BAM6 do not contribute significantly to leaf BAM activity before flowering (Fig. 4). We also found that the BAM activities in bam5/3 and bam5/1 differed in terms of the effect of pH and temperature on their activity in a nearly identical manner to purified BAM1 and BAM3 expressed in E. coli (Figs. 5, 6, and 8). Together, these results indicate that BAM activities in bam5/3 and bam5/1 are encoded primarily by BAM1 and BAM3, respectively, and that these enzymes comprise most of the extractable plastidic activity in Arabidopsis leaves at this developmental stage.
Interestingly, we found that BAM1 and BAM3 activities were present in leaves harvested during the day and the night, but activities were slightly higher in leaves harvested at night (Fig. 4). Pongratz and Beck (1978) also observed more amylase activity at night than during the day in isolated spinach (Spinacia oleracea) chloroplasts. Diurnal microarray studies indicate that, other than BAM9, which has a large peak of expression at the night-to-day transition, none of the other BAM genes showed significant diurnal fluctuation in mRNA levels (Smith et al., 2004; Mockler et al., 2007); however, fluctuations in mRNA levels can be poor predictors of enzyme levels (Gibon et al., 2004).
The presence of BAM1 and BAM3 activities in leaf extracts harvested during the day and the night raises questions about how their activities might be controlled when starch degradation is not occurring: in guard cells at night (BAM1) or in mesophyll during the day (BAM3). At night, oxidizing conditions in the chloroplast may lead to the formation of the disulfide bond known to inhibit BAM1 activity (Sparla et al., 2006; Valerio et al., 2011). However, this does not explain how BAM1 can contribute to nighttime starch degradation in mesophyll cells that lack BAM3 or leaves of older plants. BAM1 can be reduced by thioredoxin f (Valerio et al., 2011), but this enzyme apparently does not function at night (Thormählen et al., 2013). An alternative pathway for BAM1 reduction in mesophyll cells at night might be from NADPH through NADP-thioredoxin reductase C, which is known to activate ADP Glc pyrophosphorylase in the dark and has the ability to reduce BAM1 in vitro (Michalska et al., 2009; Valerio et al., 2011). The mechanism by which BAM3 activity is limited in the light is not known; however, BAMs have little activity against starch granules (Lizotte et al., 1990) before the phosphorylation of the outer glucan chains by glucan/water dikinase and phosphoglucan/water dikinase (Edner et al., 2007), and therefore, regulation of the kinases may affect the daytime function of BAM3.
pH and Temperature Have Different Effects on BAM1 and BAM3 Activities
We observed clear differences in the responses of BAM1 and BAM3 to pH and temperature that are consistent with their putative functions. Regarding pH, BAM1 is more active than BAM3 at high pH (Fig. 7), consistent with its role during the day, when the pH of the guard cell stroma should be higher than it is at night. Pongratz and Beck (1978) characterized the diurnal amylase activity in spinach chloroplasts and discussed the role that diurnal changes in pH may play in starch degradation. Pongratz and Beck (1978) observed that, at pH 8, the amylase had only 20% of its maximum activity; however, it is not known which BAM was measured. The effects of pH on the activity of purified BAM1 and BAM3 from Arabidopsis were investigated previously, but in those experiments, PNPG5 was used as the substrate (Fulton et al., 2008; Li et al., 2009). No differences were observed between the enzymes, and both had low activity at pH 8. Unlike soluble starch, PNPG5 is a small molecule that would interact with fewer amino acids on an active BAM than starch, and because the binding pocket is rather deep (Mikami et al., 1994), it is conceivable that the p-nitrophenyl group of PNPG5 does not allow efficient binding at low or high pH. Indeed, Sparla et al. (2006) measured the effect of pH on the activity of recombinant BAM1 and found that the activity peak with soluble starch was much broader than one with PNPG5 as the substrate, which is in agreement with our findings.
Detailed structural investigations of the soybean ortholog of BAM5 revealed that it is a processive enzyme that remains bound to starch between successive rounds of catalysis (Mikami et al., 1994). Assuming that all active BAMs have this property, the differences between our results and those in Fulton et al., 2008 and Li et al., 2009 regarding the higher activity of BAM1 at high pH may be explained if the difference between BAM1 and BAM3 is caused by a pH sensitivity of starch binding and not catalysis. Because the activity of BAM1 with soluble starch at pH 8 is about 80% of the optimal activity (Fig. 8), its activity during the day should not be impaired by the high stromal pH.
The optimum temperature of activity for BAM1 was about 10°C higher than BAM3 (Fig. 5), which is consistent with the daily range of temperatures under which these enzymes would function. We are unaware of any reports of day- and night-active homologous enzymes from a single plant having different temperature optima. Importantly, our results indicate that the temperature commonly used to assay leaf BAM activity, namely 37°C, is too high to reliably report the activity of BAM3.
Involvement of BAM1 and BAM3 in Abiotic Stress-Induced Starch Degradation
Abiotic stress leads to changes in metabolism that result in the accumulation of metabolites, including sugars, that have a variety of beneficial effects (Levitt, 1980). For example, under cold stress, accumulated metabolites protect cells from subsequent freezing injury, whereas under heat or osmotic stress, accumulated metabolites act as osmolites, protecting cells from dehydration. In either case, starch degradation can be a source of sugars for metabolite synthesis (Kaplan and Guy, 2004; Kempa et al., 2008). At the level of transcription, BAM1 is induced by heat and osmotic stress, and BAM3 is induced by cold stress (Kaplan and Guy, 2005; Kaplan et al., 2006; Kempa et al., 2008; Maruyama et al., 2009; Sicher, 2011). Kaplan and Guy (2005) showed that cold stress-induced maltose accumulation and freezing tolerance in Arabidopsis was impaired in plants expressing a BAM3 RNA interference construct. Valerio et al. (2011) showed that strong osmotic stress leads to lower levels of starch in wild-type plants over a period of 4 to 8 h but not in plants lacking BAM1. In contrast, we observed starch accumulation in osmotically stressed plants, including those lacking BAM1, when leaves were sampled at the start of the photoperiod (Supplemental Fig. S2A). After a period of several days, minor differences in starch degradation caused by the lack of a single enzyme may be overwhelmed by large changes in overall metabolism.
We tested the hypothesis that elevated transcription of BAM1 or BAM3 under abiotic stress leads to higher levels of BAM1 or BAM3 activity, respectively, by comparing the activity measured at 22°C in leaves from bam5/3 and bam5/1 mutants with that from bam5. Treating plants with 200 mm mannitol led to increased transcription of BAM1 but only after 4 d of stress (Fig. 9, A and B). However, after just 2 d of stress, we observed elevated BAM1 activity in bam5/3 and impaired reducing sugar accumulation in bam5/1 (Fig. 9, C and E). After 4 d, the effect on activity in bam5/3 was increased, but the effect on reducing sugars was lost (Fig. 9, D and F). Several possible explanations for the apparent discrepancy between the levels of mRNA and enzyme activity at day 2 exist. The elevated BAM1 activity could arise from increased translation of preexisting BAM1 mRNA or a change in mRNA that was undetectable. It is also possible that a portion of the oxidized pool of BAM1 was activated by reduction caused by the stress treatment. Regardless of the mechanism, our observations are consistent with BAM1 playing a role in osmotic stress-induced starch degradation, at least transiently. Valerio et al. (2011) showed that the BAM1 promoter could drive GUS expression in mesophyll cells but only later in development after plants had begun flowering. However, osmotic stress induced the expression of BAM1 in mesophyll cells of young leaves, and therefore, we suspect that the increased BAM1 activity in our osmotically stressed plants was because of the BAM1 expression in mesophyll cells, adding to the existing activity in guard cells.
The effects of cold stress on BAM1 and BAM3 mRNAs and enzyme activity levels were different from the effects of osmotic stress, but they also differed from the expected results. Despite a clear increase in BAM3 mRNA after 2 and 4 d of cold stress (Fig. 10, A and B), we observed a surprising decline in BAM3 activity in bam5/1 by 40% and 60% after 2 and 4 d, respectively, in the stressed plants (Fig. 10, C and D). BAM1 activity appeared to be slightly elevated in the cold-stressed plants. The response of Arabidopsis leaf BAMs to cold stress is in stark contrast to the rapid induction of potato tuber BAM activity by cold stress that leads to cold sweetening in storage (Nielsen et al., 1997). Despite the lower total BAM3 activity present in the leaf extracts, the double mutant lacking BAM3 accumulated fewer reducing sugars than the other plants, and therefore, it is likely that BAM3 was responsible for some of the sugar accumulation caused by cold stress (Fig. 10).
Chiba et al. (2013) measured the effect of 4°C stress for 24 h on the rates of mRNA synthesis and degradation in Arabidopsis and found that mRNA degradation was much lower in cold-stressed plants, leading to 13-fold longer mRNA half-lives. Interestingly, Chiba et al. (2013) observed that the effects of cold stress on the relative stabilities of BAM3 and BAM1 mRNAs were the opposite of each other and correlated with the effect of cold stress on mRNA levels observed in our experiments. Cold stress had a stabilizing effect on BAM3 mRNA and a destabilizing effect on BAM1 mRNA, but the effects of cold stress on the relative rates of BAM3 and BAM1 transcription were comparatively small (Chiba et al., 2013). Given the increase in the amount of BAM3 mRNA in cold-stressed plants, it was unexpected that the level of BAM3 activity, as measured in the bam5/1 double mutant, was lower in cold-stressed plants (Fig. 10, C and D). Measurements of the rates of protein turnover or other means of posttranslational regulation would be useful to help explain this result. These results underscore the observation by Gibon et al. (2004) that there is often little correlation between the levels of gene transcripts and the proteins that they encode.
Thermal Adaptation of BAM1 and BAM3
Despite the lack of an increase in BAM3 activity after cold stress, based on the effects of eliminating BAM1 or BAM3 on the accumulation of reducing sugars after osmotic or cold stress, respectively, it is apparent that BAM1 and BAM3 are involved in stress-induced starch degradation to some extent. What appears to be gene redundancy may actually be explained by the different properties of these two enzymes. We observed that BAM1 and BAM3 differ in their thermal properties and that this difference is consistent with BAM1 functioning during the day and under heat stress and BAM3 functioning at night and under cold stress. Fulton et al. (2008) generated a phylogenetic tree showing that, of nine BAM genes in Arabidopsis, BAM1 and BAM3 are in the same clade and separate from all of the other BAMs. The fact that they are only 50% identical at the amino acid level and that all sequenced flowering plant genomes contain at least one of each of these two BAMs indicates that they arose before the evolution of flowering plants. We wondered if there might be discernable structural differences between BAM1 and BAM3 that could be the basis for their divergent thermal properties. BAM1 is known to contain a regulatory disulfide bond that inactivates the enzyme when oxidized (Sparla et al., 2006). If there is a pool of oxidized BAM1 in leaves, it is likely to be more thermostable than the reduced form because of the well-known role of disulfide bonds in stabilizing protein structure. However, because we only monitored the active enzymes, our results suggest that the reduced form of BAM1 is more thermostable than BAM3 (Figs. 5 and 6).
It is well known that proteins from organisms adapted to widely different temperatures differ in their thermal properties, with heat-adapted proteins having optimal activity at a higher temperature than cold-adapted proteins (Somero, 1995; Siddiqui and Cavicchioli, 2006). There is a flexibility-stability tradeoff limiting the range of temperatures at which a protein can work efficiently. At cold temperatures, heat-adapted proteins are overly rigid and have low activity, whereas at high temperatures, cold-adapted proteins lose activity as they become denatured.
There has been much interest in understanding the structural basis for heat- and cold-adapted enzymes, in part for the purpose of engineering thermally stable proteins (Kumar et al., 2000). Systematic comparisons of orthologous proteins from thermophilic, mesophilic, and psychrophilic prokaryotes have been conducted to identify residues that affect their thermal properties (Siddiqui and Cavicchioli, 2006). A number of consistent differences has been observed, but in general, no hard and fast rules have emerged; many different types of changes can affect the thermostability of proteins. For example, compared with cold-adapted proteins, thermophilic proteins tend to replace some uncharged polar amino acids, such as Ser, Thr, Asn, and Gln, with either charged amino acids or nonpolar amino acids (Hochachka and Somero, 2002). These changes can confer greater thermostability, because ionic bonds are the strongest of the noncovalent bonds and, at high temperatures, hydrogen bonds become weaker, and hydrophobic interactions become stronger. The proportions of two of these four amino acids, Ser and Asn, were significantly lower in the thermophilic BAM1, although the proportions of the other two amino acids were not different between the two BAMs (Fig. 7). Also, BAM1 contains significantly more nonpolar residues than BAM3, which is expected if BAM1 is more heat-adapted compared with BAM3.
Heat-adapted proteins also tend to be enriched in amino acids having larger side chains and thus, tend to have more Ala and less Gly among others. BAM1 contains significantly fewer Gly and significantly more Ala residues than BAM3 (Fig. 7). These changes lead to a tighter packing of side chains within the interior of the protein, resulting in a larger number of van der Waals interactions. If the Gly to Ala substitution occurs on the surface of a protein, the effect may be on the entropy of folding, because the methyl group of Ala hinders some conformations. Another amino acid that sometimes differs between cold- and heat-adapted proteins is Pro; because it reduces backbone flexibility, it tends to be less frequent in cold-adapted proteins (Siddiqui and Cavicchioli, 2006). We found that BAM3 contains significantly fewer Pro residues than BAM1 (Fig. 7). Although any of these differences between BAM1 and BAM3 might be the result of different selective pressures, the overall trend is consistent with thermal adaptation as a contributing factor.
The effects of heat and cold stress on plant proteins have been well studied, but the focus has been on those protein families that are dramatically influenced by changing temperature and specialized to protect cells from damage, including the heat shock protein (HSP) and cold-regulated (COR) proteins induced by heat and cold, respectively. Studies on the thermal properties of individual protein families have been focused on comparisons between different species that are adapted to cold and hot climates, such as Rubisco activase (Salvucci and Crafts-Brandner, 2004), or the thermal properties of enzymes from a species that grows across a wide temperature range, such as malate dehydrogenase in Typha latifolia (McNaughton, 1974). What has received less attention thus far is the idea that all plants, regardless of habitat, experience daily fluctuations of temperature (coolest at the end of the night and warmest in the late afternoon) that might have led to pairs of proteins, like BAM1 and BAM3, that are subtly adapted to work best at slightly different temperatures.
CONCLUSION
Arabidopsis contains nine BAM genes, but only four encode active plastidic enzymes. Two of these enzymes, BAM1 and BAM3, dominate the activity in 5-week-old leaves, and both enzymes are active in extracts prepared from day- and night-harvested leaves. The other two, BAM2 and BAM6, may have more function in leaves after plants begin flowering. Both BAM1 and BAM3 have optimum activity at pH 6, but BAM1 is more active than BAM3 at high pH, consistent with its role in guard cells during the day. BAM3 has a temperature optimum that is about 10°C lower than BAM1, consistent with BAM3 being active in mesophyll cells at night. Osmotic stress leads to elevated BAM1 mRNA and BAM1 activity, and leaves lacking BAM1 accumulate fewer reducing sugars, at least transiently, indicating that it has a secondary function in mesophyll cells of osmotically stressed plants. Cold stress leads to elevated BAM3 mRNA, and plants lacking BAM3 accumulate fewer reducing sugars. Surprisingly, BAM3 activity is much lower after 2 and 4 d of cold stress, perhaps because of posttranslational modification. Comparing BAM1 and BAM3 orthologs across the eudicots, BAM1 contains significantly more Ala and Pro residues and significantly fewer Asn, Ser, and Gly residues, suggesting that these two BAM proteins have undergone thermal adaptation for their day and night roles, respectively.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 plants were grown in 5-inch pots (five plants per pot) at 22°C with a 12-h-light/12-h-dark photoperiod and 130 μmol m−2 s−1 of illumination on a growth cart (Grower’s Supply Co.) or in a growth chamber (AR-22L; Percival). The growth medium was Sunshine Mix#3 (Sun Gro Horticulture) supplemented with macronutrients and micronutrients as described by LEHLE SEEDS. For one experiment, plants were grown in a greenhouse during the summer. T-DNA lines were obtained from the Arabidopsis Biological Resource Center and included bam1 (Salk_039895), bam2 (Salk_086084), bam3 (Salk 041214), bam5 (Salk_004259), and bam6 (Salk_023637). For osmotic stress experiments, 200 mL of the same nutrient solution with or without 200 mm mannitol was applied to each pot. For cold stress experiments, plants were transferred to a walk-in 4°C chamber with lighting as described above.
T-DNA Mutant Analysis
T-DNA lines from the Salk Institute were verified by PCR using the primers listed in Supplemental Table S1. Double mutants were generated by crossing homozygous single mutants and allowing self-pollination of confirmed double heterozygotes.
RNA Extraction and RT-PCR
Leaf blades from 5- to 6-week-old plants were ground under liquid nitrogen using a mortar and pestle, and total RNA was extracted using the GeneJET Plant RNA Purification Kit (Thermo Scientific). For cold-stressed and osmotically stressed plants and their respective controls, all leaves from three separate plants were combined. Total RNA was isolated from plants for at least two separate treatments. RNA was quantified using a UV spectrophotometer and DNase treated using the TURBO DNA-Free Kit (Ambion).
Illustra Ready-to-Go RT-PCR beads (GE Healthcare) were used for RT-PCR to confirm knockout of transcripts in T-DNA mutants and detect changes in transcription in cold-stressed and osmotically stressed plants. RT was accomplished by using 50 ng µL−1 pd(N)6 deoxynucleotide primer and 0.8 ng µL−1 total RNA for first strand synthesis. The reaction was incubated at 42°C for 30 min and then 95°C to inactivate the RT. Forward and reverse gene-specific primers and primers for protein phosphatase 2A (PP2A; At1g13320; internal control) were added at 0.44 µm each (Supplemental Table S1) along with 2% (v/v) dimethyl sulfoxide. PCR amplification was carried out with an initial incubation at 95°C for 5 min followed by a cycle of 95°C for 5 min, 50°C for 1 min, and 72°C for 1 min and a final incubation at 72°C for 5 min. For RT-PCR screening of T-DNA mutants, primers were designed to straddle the T-DNA insertion site and at least one intron, with the exception of BAM3, and the PCR cycle was repeated 30 times. RT-PCR for measuring transcription levels in stressed plants used primers that included at least one intron, and the PCR cycle was repeated 27 times for BAM3, Cor78, and Hsp70 and 30 times for BAM1. The number of PCR cycles was adjusted for each gene as previously described (King, 2010) to ensure that DNA amplification was within the linear phase at termination. PCR products were run on a 1.5% (w/v) agarose gel and stained with ethidium bromide.
Recombinant Protein Expression and Purification
Plasmids for expressing BAM1 and BAM3 in Escherichia coli were a gift from Heike Reinhold. Briefly, coding sequences lacking the predicted plastid transit peptide coding regions (BAM1; 41 amino acids and BAM3, 55 amino acids as described by Fulton et al. [2008]) were amplified from complementary DNAs using the primers BAM1, 5′-CCGATTCGCAATGAATCGAAACTACAAGG-3′ and 5′-TGCGGCCGCGTGAGTGAGAGCCACTGCAG-3′; and BAM3, 5′-TCGGATCCGAGAAGACCTTCACGCCAGA-3′ and 5′-TGCGGCCGCCACTAAAGCAGCCTCCTCCAC-3′. PCR products were cloned into pJET1.2 (Fermentas) and then transferred into pET29a using EcoRI and NotI (BAM1) or BamH1 and NotI (BAM3) so that the expressed proteins contained C-terminal His tags. Plasmids were transformed into BL21+ E. coli cells for protein expression. Cells were grown to an optical density of 0.6 in Luria-Bertani media supplemented with 50 μg mL−1 kanamycin at 37°C. Isopropylthio-β-galactoside was added to a final concentration of 1 mm, and flasks were shaken at 20°C overnight. Cells were lysed by sonication, and then, nickel-nitrilotriacetic acid agarose His-Bind Resin (QIAGEN) was used to purify each protein using the manufacturer’s recommendations. Eluted proteins were dialyzed against 20 mm MOPS, pH 7.2, 100 mm NaCl, and 2 mm DTT, concentrated using an Amicon Ultra-4 10K filter, and stored at −80°C with 25% (v/v) glycerol. Coomassie Blue-stained SDS-PAGE and His-tag western blots showed that the proteins were nearly pure (data not shown).
BAM Activity and Starch Content Assays
Leaves from three 5- to 6-week-old plants per replicate extract were ground in 3 volumes of extraction buffer (50 mm MOPS, pH 7.0, 5 mm EDTA, and 2 mm DTT) with sand. After centrifugation, amylase assays were conducted in 0.5 mL containing 50 mm MES (pH 6.0) and 5 mg of Lintner soluble starch (Pfansteihl Laboratories). For assays using purified proteins, 1 mg mL−1 bovine serum albumin was included in the reaction. Reactions were stopped after no more than 2 h by immersion in a boiling water bath for 3 min; then, reducing sugars were measured by the Somogyi-Nelson assay (Nelson, 1944) with maltose as the standard. Rates of activity of recombinant BAM1 and BAM3 and extracts from bam5/1 and bam5/3 were linear for 2 h when measured at 22°C. Protein was determined using the Bio-Rad Protein Assay Kit with bovine serum albumin as the standard. For starch content analysis, leaves were harvested at the end of the night period, decolorized with hot 80% (v/v) ethanol, and stained with iodine-potassium iodide solution as described by Caspar et al. (1991).
Sequence Composition Analysis
To compare the amino acid composition of BAM1 and BAM3 orthologs, all of the BAM reference sequences from nine eudicot genomes were identified using BLAST and aligned using Clustal Omega (Sievers et al., 2011). Truncated sequences or those deemed to be misannotated based on missing segments in otherwise identical sequences were removed, and the remaining sequences listed in Supplemental Table S2 were realigned and visualized using BoxShade v3.2. The variable N-terminal regions containing DNA-binding domains or targeting sequences N-terminal to the first highly conserved residue, Val-13 (numbering from AtBAM5; NP_567460.1), were removed, and the remaining sequences were realigned. A neighbor-joining phylogenetic tree was then generated and visualized using FigTree v1.4.0 (Supplemental Fig. S1). Sequences within subfamily II were identified as falling within either the BAM1 or the BAM3 clade. One member from each of the BAM1 and BAM3 clades in each genome was then selected randomly so as to eliminate redundancy arising from recent whole-genome duplications. The full-length sequences from these 18 proteins were then analyzed using TargetP (Emanuelsson et al., 2007), plastid transit peptide cleavage sites were predicted or approximated, and transit peptides were removed before calculating the amino acid composition of each protein. Statistical significance between each set was then determined using a two-tailed Student’s t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Radial phylogenetic tree based on an alignment of the BAM catalytic domains from all of the complete BAM sequences in nine eudicot genomes.
Supplemental Figure S2. Effect of 2 and 4 d of osmotic and cold stress on starch levels in Arabidopsis leaves detected using iodine stain.
Supplemental Table S1. List of primers used in PCR and RT-PCR reactions.
Supplemental Table S2. List of reference sequence numbers for BAMs used for generation of the phylogenetic tree shown in Supplemental Figure S1.
Supplementary Material
Acknowledgments
We thank Heike Reinhold for the BAM1 and BAM3 plasmids.
Glossary
- DTT
dithiothreitol
- PNPG5
p-nitrophenyl maltopentaose
- RT
reverse transcription
Footnotes
This work was supported by the National Science Foundation (Research at Undergraduate Institutions Grant to J.D.M.) and the James Madison University Program of Grants for Faculty Assistance.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Monroe J, Bernhard W, Spilatro S, Preiss J, Somerville C (1989) Altered regulation of β-amylase activity in mutants of Arabidopsis with lesions in starch metabolism. Proc Natl Acad Sci USA 86: 5830–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen THH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5: 250–257 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Mineta K, Hirai MY, Suzuki Y, Kanaya S, Takahashi H, Onouchi H, Yamaguchi J, Naito S (2013) Changes in mRNA stability associated with cold stress in Arabidopsis cells. Plant Cell Physiol 54: 180–194 [DOI] [PubMed] [Google Scholar]
- Cho MH, Lim H, Shin DH, Jeon JS, Bhoo SH, Park YI, Hahn TR (2011) Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol 190: 101–112 [DOI] [PubMed] [Google Scholar]
- Comparot-Moss S, Kötting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, et al. (2010) A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol 152: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EA, Lane AM, Sides JM, Mudgett MB, Monroe JD (2007) An α-amylase (At4g25000) in Arabidopsis leaves is secreted and induced by biotic and abiotic stress. Plant Cell Environ 30: 388–398 [DOI] [PubMed] [Google Scholar]
- Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, et al. (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial β-amylases. Plant Physiol 145: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiena C, Schulz M, Schnabl H (1993) Starch degradation and distribution of the starch-degrading enzymes in Vicia faba leaves:diurnal oscillation of amylolytic activity and starch content in chloroplasts. Plant Physiol 101: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G (2008) Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J 55: 323–334 [DOI] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN (2002) Biochemical Adaptation: Mechanism and Process in Physiological Evolution. Oxford University Press, New York [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2005) RNA interference of Arabidopsis β-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J 44: 730–743 [DOI] [PubMed] [Google Scholar]
- Kaplan K, Sung DY, Guy CL (2006) Roles of β−amylase and starch breakdown during temperatures stress. Physiol Plant 126: 120–128 [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C (2008) A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE 3: e3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. (2010) The use of comparative quantitative RT-PCR to investigate the effect of cysteine incubation on GPx1 expression in freshly isolated cardiomyocytes. Methods Mol Biol 630: 215–232 [DOI] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. (2009) STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, Nussinov R (2000) Factors enhancing protein thermostability. Protein Eng 13: 179–191 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kim D, Gibson SI (2001) The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mould RM, Hibberd JM, Gray JC, Kavanagh TA (1999) An Arabidopsis gene encoding a chloroplast-targeted β-amylase. Plant J 20: 519–527 [DOI] [PubMed] [Google Scholar]
- Lee BR, Jin YL, Jung WJ, Avice JC, Morvan-Bertrand A, Ourry A, Park CW, Kim TH (2008) Water-deficit accumulates sugars by starch degradation—not by de novo synthesis—in white clover leaves (Trifolium repens). Physiol Plant 134: 403–411 [DOI] [PubMed] [Google Scholar]
- Levitt J. (1980) Responses of Plants to Environmental Stress. Vol. I. Chilling, Freezing and High Temperature Stresses. Academic Press, New York [Google Scholar]
- Li J, Francisco P, Zhou W, Edner C, Steup M, Ritte G, Bond CS, Smith SM (2009) Catalytically-inactive β-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein. Arch Biochem Biophys 489: 92–98 [DOI] [PubMed] [Google Scholar]
- Lin TP, Spilatro SR, Preiss J (1988) Subcellular localization and characterization of amylases in Arabidopsis leaf. Plant Physiol 86: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizotte PA, Henson CA, Duke SH (1990) Purification and characterization of pea epicotyl β-amylase. Plant Physiol 92: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton SJ. (1974) Enzymic thermal adaptations. The evolution of homeostasis in plants. Am Nat 106: 165–172 [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami B, Degano M, Hehre EJ, Sacchettini JC (1994) Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis. Biochemistry 33: 7779–7787 [PubMed] [Google Scholar]
- Mita S, Suzuki-Fujii K, Nakamura K (1995) Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiol 107: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Monroe JD, Preiss J (1990) Purification of a β-amylase that accumulates in Arabidopsis thaliana mutants defective in starch metabolism. Plant Physiol 94: 1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Salminen MD, Preiss J (1991) Nucleotide sequence of a cDNA clone encoding a β-amylase from Arabidopsis thaliana. Plant Physiol 97: 1599–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem 153: 375–380 [Google Scholar]
- Nielsen TH, Deiting U, Stitt M (1997) A β-amylase in potato tubers is induced by storage at low temperature. Plant Physiol 113: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Outlaw WH Jr, Manchester J (1979) Guard cell starch concentration quantitatively related to stomatal aperture. Plant Physiol 64: 79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongratz P, Beck E (1978) Diurnal oscillation of amylolytic activity in spinach chloroplasts. Plant Physiol 62: 687–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold H, Soyk S, Simková K, Hostettler C, Marafino J, Mainiero S, Vaughan CK, Monroe JD, Zeeman SC (2011) β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 23: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related R1 protein is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004) Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol 134: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J (2002) Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J 30: 581–591 [DOI] [PubMed] [Google Scholar]
- Sicher R. (2011) Carbon partitioning and the impact of starch deficiency on the initial response of Arabidopsis to chilling temperatures. Plant Sci 181: 167–176 [DOI] [PubMed] [Google Scholar]
- Siddiqui KS, Cavicchioli R (2006) Cold-adapted enzymes. Annu Rev Biochem 75: 403–433 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Somero GN. (1995) Proteins and temperature. Annu Rev Physiol 57: 43–68 [DOI] [PubMed] [Google Scholar]
- Soyk S, Simková K, Zürcher E, Luginbühl L, Brand LH, Vaughan CK, Wanke D, Zeeman SC (2014) The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 26: 1746–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol 141: 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Eicke S, Zeeman SC (2012) The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J Biol Chem 287: 41745–41756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain RR, Dekker EE (1966) Seed germination studies. I. Purification and properties of an α-amylase from the cotyledons of germinating peas. Biochim Biophys Acta 122: 75–86 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P (2013) Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29 [DOI] [PubMed] [Google Scholar]
- Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, Sparla F (2011) Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J Exp Bot 62: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjölund RD (1995) Identification and characterization of a phloem-specific β-amylase. Plant Physiol 109: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Weber AP, Sharkey TD (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482 [DOI] [PubMed] [Google Scholar]
- Werdan K, Heldt HW, Milovancev M (1975) The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark. Biochim Biophys Acta 396: 276–292 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Thorneycroft D, Schupp N, Chapple A, Weck M, Dunstan H, Haldimann P, Bechtold N, Smith AM, Smith SM (2004) Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol 135: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler P. (1988) Partial purification and characterization of the major endoamylase of mature pea leaves. Plant Physiol 86: 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XX, Wang YB, Pan YJ, Li WF (2008) Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids 34: 25–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.