An l-cysteine desulfhydrase is a unique component of ABA signaling in guard cells, mediating H2S production and acting upstream of nitric oxide to induce stomatal closure.
Abstract
Abscisic acid (ABA) is a well-studied regulator of stomatal movement. Hydrogen sulfide (H2S), a small signaling gas molecule involved in key physiological processes in mammals, has been recently reported as a new component of the ABA signaling network in stomatal guard cells. In Arabidopsis (Arabidopsis thaliana), H2S is enzymatically produced in the cytosol through the activity of l-cysteine desulfhydrase (DES1). In this work, we used DES1 knockout Arabidopsis mutant plants (des1) to study the participation of DES1 in the cross talk between H2S and nitric oxide (NO) in the ABA-dependent signaling network in guard cells. The results show that ABA did not close the stomata in isolated epidermal strips of des1 mutants, an effect that was restored by the application of exogenous H2S. Quantitative reverse transcription polymerase chain reaction analysis demonstrated that ABA induces DES1 expression in guard cell-enriched RNA extracts from wild-type Arabidopsis plants. Furthermore, stomata from isolated epidermal strips of Arabidopsis ABA receptor mutant pyrabactin-resistant1 (pyr1)/pyrabactin-like1 (pyl1)/pyl2/pyl4 close in response to exogenous H2S, suggesting that this gasotransmitter is acting downstream, although acting independently of the ABA receptor cannot be ruled out with this data. However, the Arabidopsis clade-A PROTEIN PHOSPHATASE2C mutant abscisic acid-insensitive1 (abi1-1) does not close the stomata when epidermal strips were treated with H2S, suggesting that H2S required a functional ABI1. Further studies to unravel the cross talk between H2S and NO indicate that (1) H2S promotes NO production, (2) DES1 is required for ABA-dependent NO production, and (3) NO is downstream of H2S in ABA-induced stomatal closure. Altogether, data indicate that DES1 is a unique component of ABA signaling in guard cells.
Abscisic acid (ABA) regulates diverse physiological and developmental processes in plants, among which seed dormancy and stomatal movement are the most studied. Stomata are pores bordered by pairs of specialized cells named guard cells located in the epidermis of the aerial part of most land plants. Due to the waxy cuticle of plants, stomatal pores regulate approximately 90% of all the gas exchange (i.e. the uptake of CO2 required for photosynthesis and the loss of water vapor during transpiration) between the plant and the environment (Hetherington and Woodward, 2003). Thus, stomatal movement is a key process for the regulation of plant water status and biomass production. In guard cells, ABA induces an increase of intracellular calcium concentrations, which, in turn, induces an efflux of anions that causes membrane depolarization. In this voltage milieu, ion uptake is blocked through the inactivation of inward-rectifying potassium channels, and ion efflux is induced through activation of outward-rectifying potassium channels. This solute relocation drives water out of the guard cells and closes the stomatal pore as a result of a reduction in guard cell turgor (Blatt, 2000; Kim et al., 2010). The ABA-dependent signaling network in guard cells involves numerous second messengers, including calcium, K+, protein phosphatases (mainly PP2C), guanylate cyclase/cyclic ADP ribose, hydrogen peroxide, and nitric oxide (NO) among others. The complexity of the interactions resembles that of a scale-free network (Hetherington and Woodward, 2003). Numerous loci have been identified to be regulated by ABA in guard cells either upstream or downstream of the receptor complex (Leonhardt et al., 2004; Cutler et al., 2010); however, unique components are still emerging.
Hydrogen sulfide (H2S) is a small and reactive water-soluble gas. In aqueous solutions (pH 7.4), the ratio between the hydrosulfide anion (HS–) and H2S is 3 to 1 (Kabil and Banerjee, 2010). The active form of H2S in biological systems has not been specified yet; therefore, H2S usually stands for H2S/HS–. H2S can freely permeate lipid membranes, as its solubility is 5 times greater in lipophilic solvents than in water (Wang, 2002).
H2S has been proposed as the third gasotransmitter in animals cells, after NO and carbon monoxide, due to its high level of permeability through biological membranes, its effect at low concentrations, and toxicity at high doses (Mancardi et al., 2009). H2S has been implicated in different physiological processes such as blood vessel relaxation, neurotransmission (Li et al., 2006), insulin signaling (Yang et al., 2005), angiogenesis (Coletta et al., 2012), and inflammation (Szabó, 2007), among others. In mammals, most of the endogenous H2S is produced through the activity of two pyridoxal-5′-P-dependent enzymes cystathionine β-synthase (EC 4.2.1.22) and cystathionine γ-lyase (EC 4.4.1.1). It has long been known that plants have l-Cys desulfhydrase (DES) activity (Harrington and Smith, 1980; Papenbrock et al., 2007). However, no bona fide DES gene was reported until 2010, when the Cys synthase-like gene (At5G28030), a member of the O-acetyl-Ser(thiol)lyase family was identified as a true DES and named DES1 (Alvarez et al., 2010). DES1 releases H2S, pyruvate, and ammonia during l-Cys degradation (Alvarez et al., 2010). Early studies concerning H2S emission in plants were associated with the plant response to pathogens as part of a so-called Sulfur Induced Resistance (Bloem et al., 2004). Using a H2S-releasing compound, H2S was later reported to confer a protective effect against oxidative (Zhang et al., 2009b, 2010b) and cadmium (Sun et al., 2013) stresses, alleviate aluminum toxicity (Zhang et al., 2010c), increase antioxidant activity, and participate in root organogenesis (Zhang et al., 2009a). Several independent groups have recently reported the participation of H2S in ABA- and ethylene-dependent stomatal closure induction (García-Mata and Lamattina, 2010; Liu et al., 2011, 2012; Jin et al., 2013). In this study, we used the characterized DES1 knockout Arabidopsis (Arabidopsis thaliana) mutant plants des1-1 and des1-2 to obtain new insights on the cross talk between H2S and NO and further evidence supporting the involvement and requirements of H2S in ABA-induced signaling cascade leading to stomatal closure.
RESULTS
DES1 Is Required for ABA-Dependent Stomatal Closure
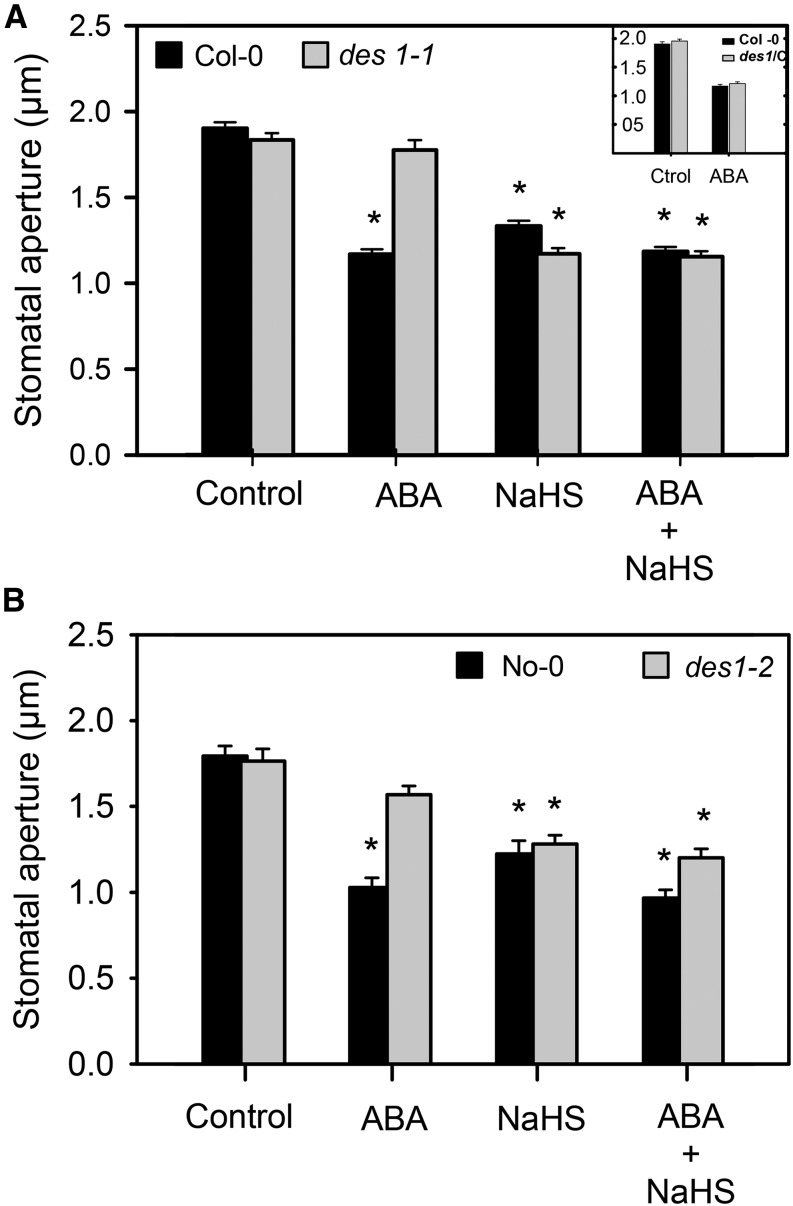
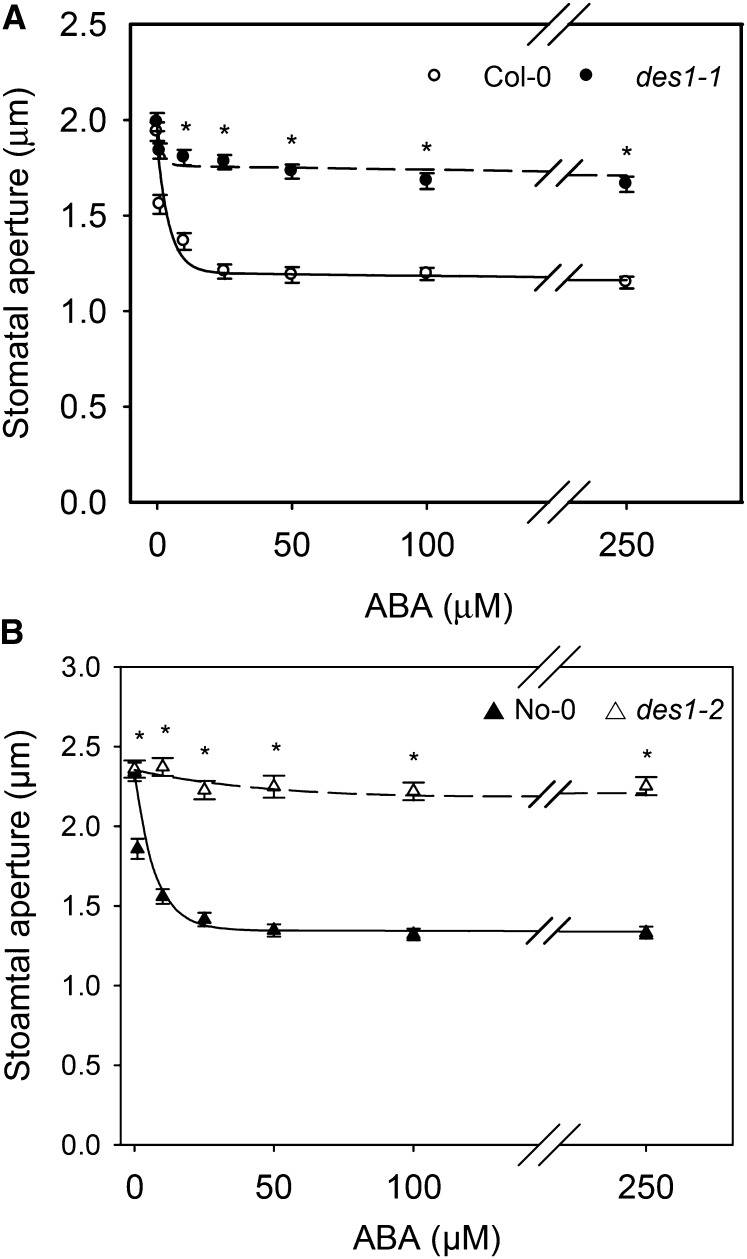
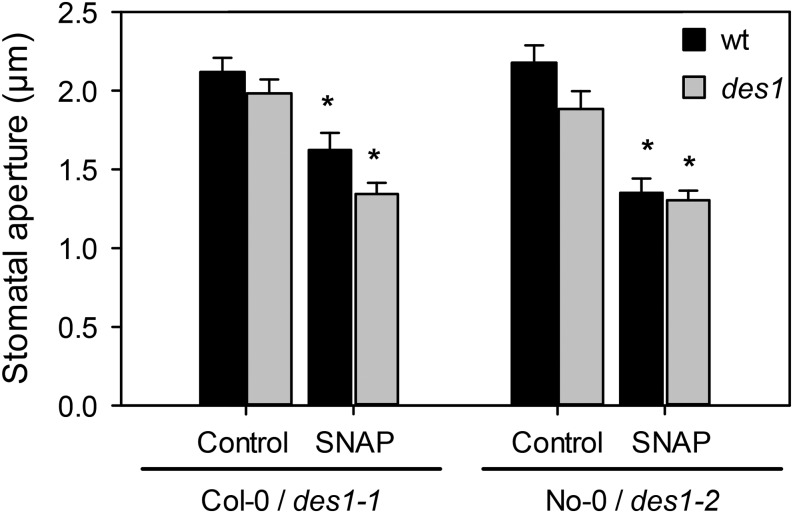
In a previous work, we presented pharmacological evidence showing that H2S might be part of the signaling network leading to ABA-dependent stomatal closure in different plant species (García-Mata and Lamattina, 2010). This was recently confirmed for Arabidopsis by Jin et al. (2013) using a transfer DNA insertion mutant of the gene AT3G62130 that codes for an l-Cys desulfhydrase. In this work, by contrast, we used two null mutants deficient in the DES1 protein to demonstrate the participation of DES1 in ABA signaling in stomata. Previously, the recombinant DES1 protein was expressed in bacteria and was enzymatically characterized as l-Cys desulfhydrase (Alvarez et al., 2010). With that aim, epidermal strips from Arabidopsis DES1 null mutant plants (des1-1 and des1-2) and their genetic backgrounds (ecotypes Columbia [Col-0] and Nössen [No-0], respectively) were prepared and treated with or without 50 µm ABA for 90 min. As expected, epidermal strips from the wild-type plants closed the stomata in response to exogenous ABA application (Fig. 1); however, ABA-dependent stomatal closure was strongly inhibited in both des1-1 (Fig. 1A) and des1-2 (Fig. 1B) epidermal strips, indicating that DES1 is required for ABA-dependent stomatal closure. The lack of response of des1-1 to ABA was restored in epidermal strips of des1-1 knockout mutant complemented with the full-length DES1 complementary DNA (cDNA; Fig. 1, inset). Moreover, the addition of exogenous H2S as 100 µm of the H2S donor sodium hydrosulfide (NaHS) together with ABA treatment also restored the stomatal response to ABA in both des1-1 and des1-2 mutants, suggesting that the lack of response was due to reduced levels of endogenous H2S (Fig. 1, A and B). Interestingly, an ABA dose-response experiment showed that des1 mutant plants remain insensitive, even when the epidermal strips were treated with 250 µm ABA, suggesting that this effect was not dependent on ABA concentration (Fig. 2). Consistent with our previous report (García-Mata and Lamattina, 2010), epidermal strips from both genetic backgrounds No-0 and Col-0 responded to the H2S donor in a dose-dependent manner, showing maximal stomatal closure induction at 100 µm of the donor NaHS (Supplemental Fig. S1A). Interestingly, high doses of the H2S donor (500 μm) did not induce stomatal closure, probably due to rather toxic effects (Supplemental Fig. S1A). The fact that morpholin-4-ium 4 methoxyphenyl(morpholino) phosphinodithioate (GYY 4137, another H2S donor) induced stomatal closure in both wild-type plants and des1 mutants (Supplemental Fig. S1B) and that the H2S scavenger hypotaurine (HT) blocked the effect of the donor (Supplemental Fig. S1C) confirms that the response was due to the released H2S and not by any by-product of the donor molecule.
Figure 1.
DES1 is involved in ABA-dependent stomatal closure. Epidermal strips, peeled from Arabidopsis mutants des1-1 (A), des1-2 (B), des1-1 complemented with DES1 full cDNA (des1/C, inset), and their genetic backgrounds, were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light. Strips were subsequently treated for 90 min under light with opening buffer (Control), 100 µm of the H2S donor NaHS, 50 µm ABA, or 100 µm NaHS plus 50 µm ABA. The values are expressed as the means ± se and represent the mean of 20 to 40 stomata per experiment from at least three independent experiments (n = 60–160). The asterisks denote significant differences with respect to the control treatments (Student’s t test, P < 0.05). Ctrol, Control (inset).
Figure 2.
Stomata from Arabidopsis des1 mutants show reduced sensitivity to ABA. Epidermal strips peeled from des1-1 (A) or des1-2 (B) plants and their genetic backgrounds were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light and subsequently treated with increasing concentrations of ABA (0, 1, 10, 25, 50, 100, and 250 µm) for 90 min under light. The curves were fitted to an exponential linear combination function. The values are expressed as the means ± se and represent the mean of 20 to 40 stomata per experiment from at least three independent experiments (n = 60–160). Asterisks denote significant differences with respect to the wild-type plants (Dunn’s post hoc test, P < 0.05).
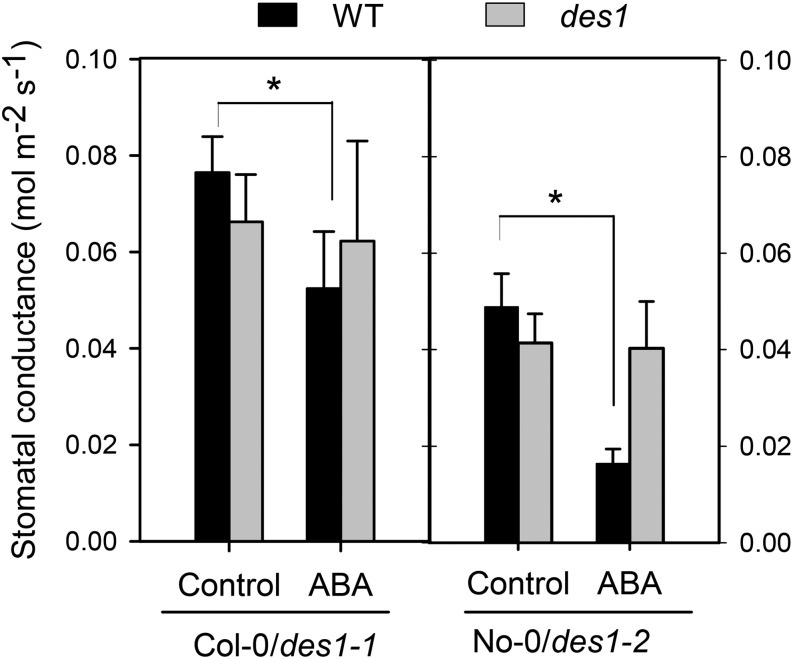
To determine if the des1 mutants also show reduced sensitivity to ABA at the whole-plant level, Arabidopsis des1 mutant plants and their respective genetic background were sprayed with water or 50 µm ABA for 3 h, and then stomatal conductance measurements were performed using an infrared gas analyzer (IRGA). Figure 3 shows that ABA treatment induced a significant reduction of stomatal conductance in wild-type plants, while des1 mutants were less sensitive to ABA treatment. This result indicates that the reduced response to ABA observed in stomata from des1 mutants correlates with the response at the whole-plant level.
Figure 3.
Stomatal conductance is less responsive to ABA in Arabidopsis des1 mutants. The effect of ABA on the stomatal conductance was measured in planta in leaves of both des1 mutant plants and in their respective genetic backgrounds. The plants were sprayed with water (Control) or 50 µm ABA, and the stomatal conductance was measured 3 h after treatment from a fully expanded leaf in a closed chamber under a constant CO2 concentration for 5 min using an IRGA. The values are expressed as the means ± sd of at least three independent experiments. The asterisks indicate significant differences with respect to each control (Student’s t test, P < 0.05). WT, Wild type.
ABA Increases DES1 Expression in Arabidopsis
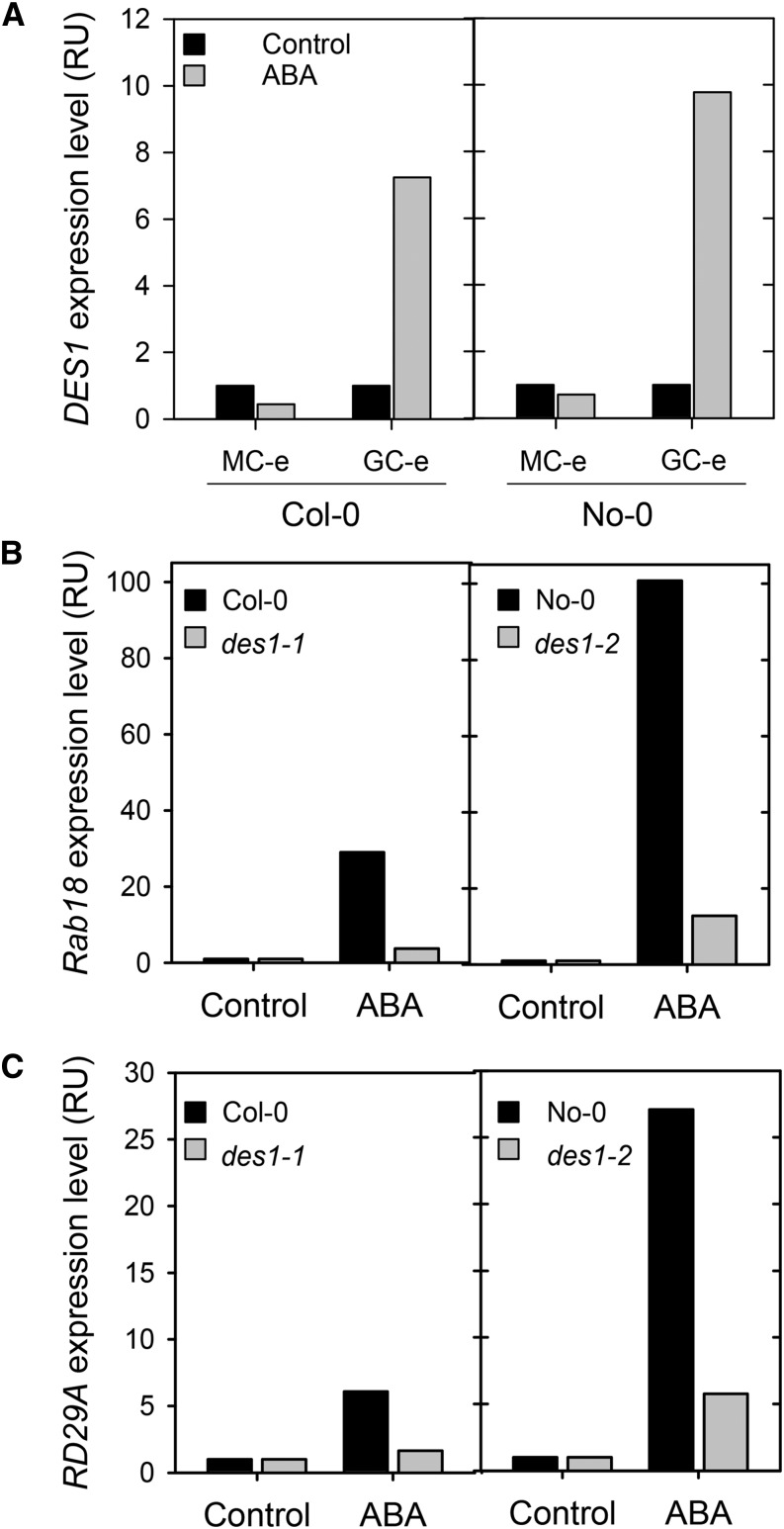
Both pharmacological and genetic data indicate that DES1 is involved in ABA-induced stomatal closure. To obtain further evidence for ABA-DES1 cross talk, we analyzed the expression levels of the DES1 gene in epidermal strips from Arabidopsis wild-type plants. The RNA extracted from the epidermal strips was considered as guard cell-enriched (GC-e) RNA because 90% to 95% of the living cells in epidermal strips are guard cells. We also extracted RNA from the rest of the leaf (without the abaxial epidermis layer), and we considered this extract as mesophyll cell-enriched (MC-e) RNA. Marker genes were used in reverse transcription PCR (RT-PCR) assays to confirm that we have both GC-e and MC-e extracts. ECERIFERUM2 (CER2; Leonhardt et al., 1999; Wang et al., 2011) was used as a marker gene for guard cells, and β-Carbonic anhidrase1 (Canh1; Pandey et al., 2002) as a marker gene for the mesophyll cells (Supplemental Fig. S2). To assess if DES1 is regulated by ABA at the transcriptional level, we treated the epidermal strips and mesophyll tissue from wild-type plants with stomatal opening buffer or 50 µm ABA for 90 min. Subsequently, GC-e and MC-e RNA were extracted from the samples, and DES1 gene expression was assessed using quantitative reverse transcription (qRT)-PCR analysis. The results shown in Figure 4A reveal only marginal variations in DES1 expression levels in MC-e; however, GC-e showed a dramatic increase of DES1 transcript levels upon ABA treatment in both ecotypes (7- and 9-fold as compared with the control treatment for Col-0 and No-0, respectively), indicating that DES1 was significantly regulated by ABA in the guard cells. Furthermore, we performed a qRT-PCR analysis of the expression levels of the ABA-responsive genes RESPONSIVE TO DEHYDRATION A (RD29A) and RESPONSIVE TO ABA18 (RAB18) in GC-e RNA extracts from wild-type and des1 plants. The results shown in Figure 4, B and C, reveal that both des1 mutant lines have a significant reduction of the expression of both RD29A and RAB18 genes in response to ABA treatments with respect to the wild types. Taken together, these results suggest that DES1 is regulated by ABA at the transcriptional level in Arabidopsis guard cells and that an active DES1 is required to attain a full expression of ABA-responsive genes.
Figure 4.
Analysis of the expression levels of DES1 and ABA-responsive genes in GC-e and MC-e RNA extracts of wild-type and des1 mutant plants. Epidermal strips and mesophyll tissue prepared from des1 plants and their respective genetic background were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) and then treated with or without 50 µm ABA under light. After 90 min of treatment, qRT-PCR analysis of DES1 gene expression was performed in GC-e RNA extracted from Col-0 and No-0 (A). qRT-PCR analysis of ABA-responsive genes RD29A (B) and RAB18 (C) was performed for GC-e RNA extracted from the wild type and des1 mutants. The values are expressed as percentages with respect to the control treatments. RU, Relative unit.
H2S Participates in ABA-Dependent NO Production during Stomatal Closure Induction
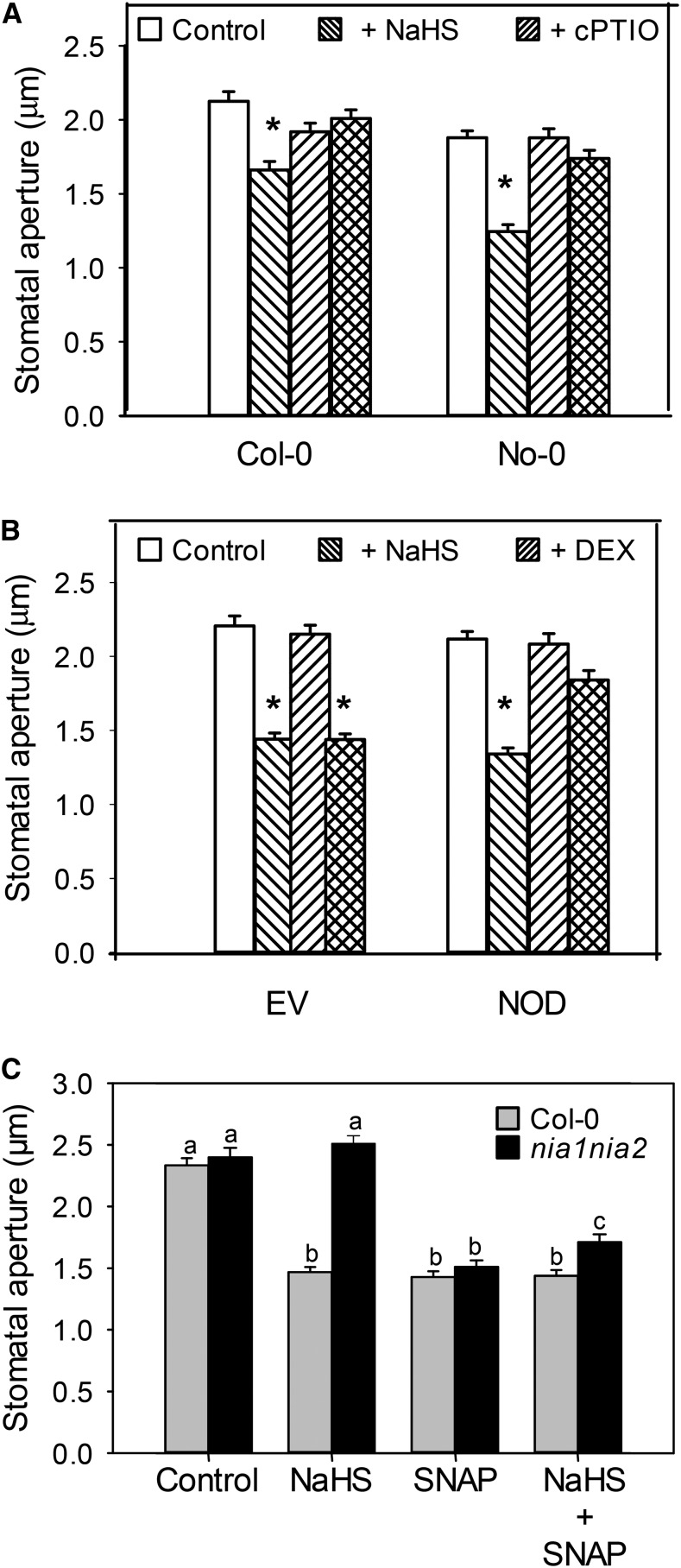
It has been demonstrated that, in animal systems, H2S and NO might interact either in an agonistic or antagonistic way, depending on the biological system and physiological process (Li et al., 2009; Kajimura et al., 2010; Yong et al., 2010). In plants, it has been recently reported that H2S acts downstream of NO in ethylene-induced stomatal closure (Liu et al., 2011, 2012). It was also reported that H2S reduces ABA-dependent NO production in Arabidopsis and Capsicum annuum guard cells (Lisjak et al., 2010, 2011). To add some knowledge to the NO-H2S cross talk in Arabidopsis guard cells, we first performed a pharmacological assay to assess whether NO is involved in H2S-dependent stomatal closure. Epidermal strips from Arabidopsis wild-type leaves were treated with or without the NO-specific scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide (cPTIO). In Figure 5A, it is shown that cPTIO impaired H2S-dependent induction of stomatal closure in both wild-type ecotypes. This result was supported by genetic assays using epidermal strips from Col-0 plants expressing the NO dioxygenase (NOD; Zeier et al., 2004), which were proven to have reduced levels of NO after inducing NOD expression with dexamethasone (DEX; Zeier et al., 2004; Tossi et al., 2011). Exogenous application of H2S induced stomatal closure in water-sprayed NOD plants, but no stomatal closure was observed in DEX-sprayed NOD plants, confirming the NO requirement for H2S-dependent stomatal closure (Fig. 5B). To further confirm the interaction of H2S and NO, we used NITRATE REDUCTASE1 / NITRATE REDUCTASE2 double mutant (nia1/nia2) plants, which produced very low levels of NO (Desikan et al., 2002; Lozano-Juste and León, 2010). As shown in Figure 5C, NaHS was not able to induce stomatal closure in the nia1/nia2 mutant, an effect that was restored by exogenous addition of the NO-specific donor S-nitroso-N-acetylpenicillamine (SNAP). All together, the pharmacological and genetic evidence indicate that the depletion of endogenous NO blocks H2S-mediated induction of stomatal closure, demonstrating the interaction of these two gasotransmitters in stomatal closure processes.
Figure 5.
NO is required for the H2S-induced stomatal closure. A, Epidermal strips peeled from Arabidopsis leaves were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light. Subsequently, strips were treated for 90 min under light with 100 µm NaHS in the absence or presence (+cPTIO) of 200 µm of the NO scavenger cPTIO. B, Arabidopsis Col-0 plants transformed with empty vector (EV) or NOD were sprayed with water (mock) or 0.01% (v/v) DEX. After 48 of the DEX induction, epidermal strips were peeled, preincubated for 3 h in opening buffer under light, and then treated for 90 min with opening buffer (Control) or 100 µm NaHS. C, Epidermal strips peeled from Arabidopsis double mutant nia1/nia2 leaves were preincubated for 3 h in opening buffer under light and then treated for 90 min with 100 µm NaHS, 100 µm of NO donor SNAP, or 100 µm NaHS plus 100 µm SNAP. The values are expressed as the means ± se and represent the mean of 20 to 40 stomata per experiments from at least three independent experiments (n = 60–160). The asterisks indicate significant differences with respect to each control, and different letters indicate significant differences between treatments (Dunn’s post hoc test, P < 0.05).
The lack of response to H2S in NO-depleted epidermal strips prompted us to investigate whether NO was able to induce stomatal closure in the des1 mutants. With that aim, stomatal aperture experiments were performed in epidermal strips from the wild type and both des1 mutants in presence or absence of 100 µm of the NO donor SNAP. Exogenous addition of NO induced stomatal closure to the same extent in epidermal strips from both wild-type and des1 mutant plants (Fig. 6), confirming again that NO is downstream of DES1 in the ABA signaling pathway, leading to stomatal closure.
Figure 6.
NO induces stomatal closure in Arabidopsis des1 mutants. Epidermal strips peeled from Arabidopsis mutants des1 and their genetic backgrounds were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light and subsequently treated for 90 min under light with 100 µm SNAP. The values of stomatal aperture are expressed as the means ± se and represent the mean of 20 to 40 stomata per experiments from at least three independent experiments (n = 60–160). Asterisks denote statistical differences with respect to the control treatment of each genetic background (Dunn’s post hoc test, P < 0.05). wt, Wild type.
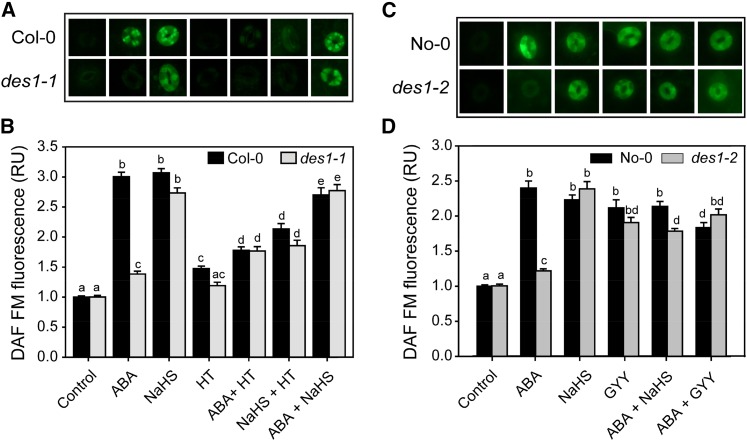
Several reports have shown that ABA increases endogenous NO production in guard cells (García-Mata and Lamattina, 2002; Neill et al., 2002). The results presented in this work indicate that H2S acts upstream of NO in ABA-dependent stomatal closure; therefore, we used the fluorescent dye 4,5-diaminoflorescein diacetate (DAF-FM-DA) to test whether H2S has any effect on endogenous NO production. Figure 7 shows that guard cells from wild-type epidermal strips loaded with DAF-FM-DA display significant increases in endogenous NO levels when treated with H2S donors NaHS or GYY (Fig. 7, A and C). However, this increase was not evident either in des1 mutants or in Col-0 plants treated with ABA together with the H2S scavenger HT (Fig. 7, A and B), indicating that DES1 is required for ABA-induced NO formation. Moreover, the impairment of des1-1 and des1-2 mutants to produce NO in response to ABA was rescued by the addition of exogenous H2S using either NaHS or GYY, supporting that DES1 acts upstream of NO in ABA-dependent stomatal closure (Fig. 7).
Figure 7.
H2S is required for the ABA-induced NO production in Arabidopsis guard cells. Epidermal peels from Arabidopsis des1 mutant and their genetic backgrounds were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light and subsequently loaded with 10 µm of the NO-specific fluorescent dye DAF-FM-DA. After washing, the strips were treated for 15 min under light with opening buffer (Control) or with the following treatments: 50 µm ABA, 100 µm NaHS, 200 µm of the H2S scavenger HT, ABA plus HT, NaHS plus HT, or ABA plus NaHS for Col-0 and des1-1 plants (A and B) and 50 µm ABA, 100 µm NaHS, 100 µm of the H2S donor GYY 4137, ABA plus NaHS, or ABA plus GYY for No-0 and des1-2 plants (C and D). Images depict one representative picture of stomata from epidermal peels corresponding to three independent experiments. The green fluorescence pixel intensity is expressed as relative unit (RU) with respect to the control treatment. The values are expressed as means ± se and represent the mean of 40 to 50 stomata per experiment from three independent experiments (n = 120–150). Different letters denote significant differences (Dunn’s post hoc test, P < 0.05). [See online article for color version of this figure.]
H2S Acts Upstream of ABI1 in ABA-Dependent Stomatal Closure
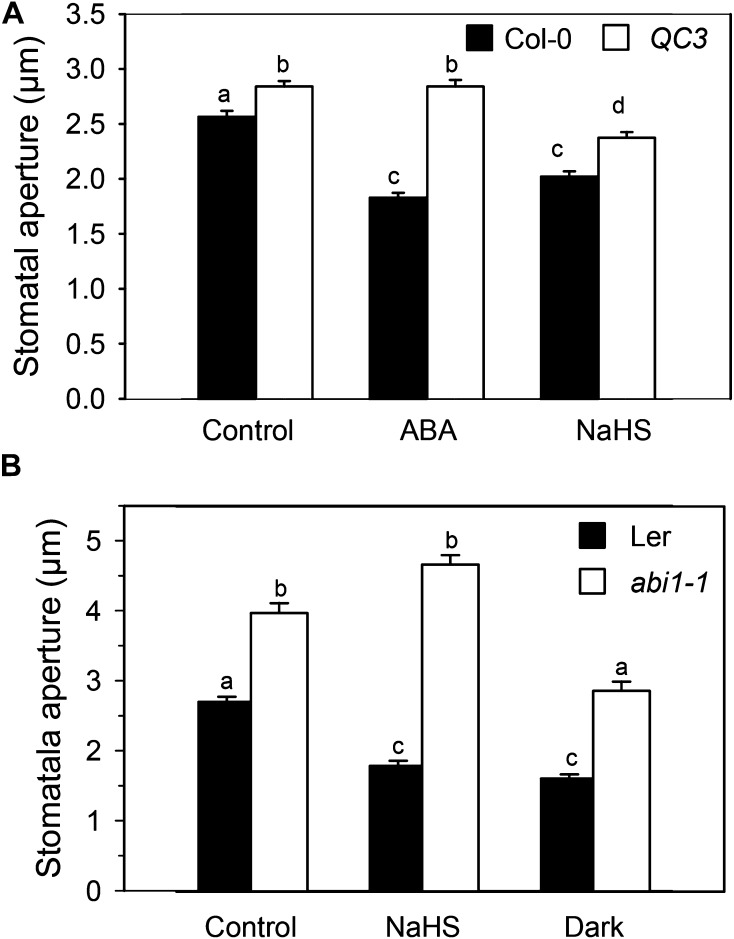
The mutants of the recently identified ABA receptor PYRABACTIN RESISTANT/PYRABACTIN-LIKE/REGULATORY COMPONENTS OF ABA RECEPTOR (PYR/PYL/RCAR) show a strong insensibility to ABA and failure to close stomata (Nishimura et al., 2010) but do not show insensitivity to downstream signaling components such as extracellular Ca2+ (Nishimura et al., 2010; Wang et al., 2013) or hydrogen peroxide (Wang et al., 2013). Therefore, we assayed H2S-dependent stomatal closure in epidermal strips of the quadruple mutant of the ABA receptor pyr1/pyl1/pyl2/pyl4 (hereafter, called QC3). Figure 8A shows that H2S was able to close the stomata both in the wild type and, to a lesser extent, in the QC3 quadruple mutant epidermal strips. These data suggest that H2S is acting downstream or independently of the ABA receptor. It is noteworthy that exogenous application of the H2S donor NaHS did not induce stomatal closure in epidermal strips of the ABA-insensitive abi1-1 mutant (Fig. 8B), but it does induce stomatal closure in its genetic background Landsberg erecta, indicating that a functional ABI1 is required for DES1/H2S action in ABA-dependent guard cell signaling.
Figure 8.
Position of H2S in a PYR/PYR/RCAR cascade. Epidermal strips, peeled from the Arabidopsis mutants in ABA receptor pyr1/pyl1/pyl2/pyl4 (QC3; A), the PP2C mutant abi1-1 (B), and their genetic backgrounds, were preincubated for 3 h in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) under light. Strips were subsequently treated for 90 min under light with opening buffer (Control), 100 µm of the H2S donor NaHS (A and B), or 50 µm ABA (A) or shifted to the dark for 90 min (B). The values are expressed as the means ± se and represent the mean of 20 to 40 stomata per experiment from at least three independent experiments (n = 60–160). Different letters indicate significant differences among treatments (Dunn’s post hoc test, P < 0.05). Ler, Landsberg erecta.
DISCUSSION
The gasotransmitter H2S has rapidly emerged as a hot topic in animal physiology, where it is known to have an active role in cardioprotection, neurotransmission, and O2 sensing, among others (Kabil and Banerjee, 2010; Peng et al., 2010). In plants, although originally associated with plant-pathogen interaction, it is now known to have an active role in diverse physiological processes such as oxidative stress, germination, and heat tolerance (Zhang et al., 2009a, 2009b, 2010a, 2010b, 2010d; Li et al., 2013a, 2013b). Recently, H2S has been proven to participate in ABA- or ethylene-induced stomatal closure in different plant species (García-Mata and Lamattina, 2010; Liu et al., 2011, 2012; Jin et al., 2013). Moreover, it has been reported also that H2S induces stomatal opening (Lisjak et al., 2010, 2011).
In this work, we present new insights into the regulation of the H2S-generating enzyme DES1 and DES1 gene expression by ABA, providing evidence supporting the participation of DES1 in ABA-induced stomatal closure. It is known that DES1 plays a role in leaf senescence and modulates the progression of autophagy (Alvarez et al., 2010; Álvarez et al., 2012); however, no data are available concerning the role of hormones in the regulation of DES1. In a previous report, it was shown that the pharmacological inhibition of DES results in a partial blockage of ABA-dependent stomatal closure (García-Mata and Lamattina, 2010). In this work, using a genetic approach, we show that two independent knockout mutants of the Arabidopsis DES1, des1-1 and des1-2, failed to close the stomata in response to increasing concentrations of ABA. The lack in the sensitivity to ABA of des1 mutants was restored through either the exogenous addition of H2S donors or by the complementation of the des1-1 mutant with the full-length DES1 cDNA, indicating that DES1 participates in ABA-dependent stomatal closure.
We demonstrate that, at the time scale used for stomatal aperture experiments (90 min of treatment), DES1 is up-regulated by ABA in guard cells but not in mesophyll cells, supporting the participation of DES1 in the signaling events leading to stomatal closure. Guard cell-specific gene expression has been already reported by microarray analysis using guard cell protoplasts (GCPs) versus mesophyll cell protoplasts (MCPs; Leonhardt et al., 2004) and epidermal strips versus leaf tissue (Wang et al., 2011). In both works, the authors have identified three groups of genes: (1) those equally regulated in GCP and MCP, (2) those with altered expression in MCPs and not in GCPs, and (3) those with modified expression in GCPs and not in MCPs. We have found that DES1 expression is selectively activated in guard cells by ABA. Transcriptional cell specificity regulation in response to ABA may reflect a differential distribution of ABA in the different cell types or to a differential utilization of ABA-specific promoters or transcription factors in each cell type (Leonhardt et al., 2004). Further measurements of enzyme activity in guard cells will be needed to unequivocally correlate the up-regulation of DES1 in GC-e extracts with an increase in DES-dependent H2S production in guard cells; however, we could not obtain enough mass of protein from the isolated epidermal strips to assay the DES1 activity. Nevertheless, we did analyze the effect of ABA treatments on DES activity levels in whole-leaf extracts from wild-type Arabidopsis plants, and we observed a positive correlation between DES1 expression and DES activity but at over longer periods of treatments (Supplemental Fig. S3, A and B). Although further work is needed to unveil the mechanism by which ABA activates DES1 transcription, the presence of a Dc3 Promoter-Binding Factor3 (DPBF3) and DPBF2 and a Dehydration-Responsive Element-Like in the promoter region of DES1 let us speculate on the possibility that ABA is directly regulating the expression of the gene (Supplemental Fig. S4).
Lately, the interest on the biology of gasotransmitters has been oriented toward the interactions between these three gases on the different biological systems (García-Mata and Lamattina, 2013; Kolluru et al., 2013). In animal systems, there are many reports on the interaction of H2S and NO, although the nature of this interaction is still poorly understood. Some reports have shown a positive interaction between H2S and NO, where H2S increases the rates of NO release from small nitrosothiols such as S-nitrosoglutathione (Ondrias et al., 2008) or even forms new nitrosothiol species with NO that might act as biological regulators of the availability of both gasotransmitters (Whiteman et al., 2006; Filipovic et al., 2013). Contrasting evidence, however, has come from observations that: (1) in vitro, H2S inhibits all the three isoforms of mammalian NO synthases (Kubo et al., 2007) and (2) H2S reacts with NO to form a new compound that exhibits contrasting effects in the heart (Yong et al., 2010). In addition, it was recently reported that both gasotransmitters are mutually required for regulation of vascular function (Coletta et al., 2012).
The data presented here indicate that there is a cross talk between H2S and NO during ABA-dependent stomatal closure. The depletion of NO, by chemical or genetic means, blocked the H2S-dependent stomatal closure, suggesting that NO is acting downstream of H2S in this particular physiological response. Accordingly, none of the des1 mutants produce NO in response to ABA treatment, although they do close the stomata in response to exogenous application of NO donors. This result is in agreement with a recently annotated microarray data where both NIA1 and NIA2 isoforms of the nitrate reductase are up-regulated upon H2S treatment (data accessible at National Center for Biotechnology Information Gene Expression Omnibus database, accession no. GSE32566), which supports the lack of response of the nia1/nia2 double mutant to the exogenous addition of H2S observed in this study. As for animal systems, in plants, there is controversial evidence about H2S-NO interaction. On one hand, it was shown that H2S acts upstream of NO in root organogenesis and heavy metal toxicity (Zhang et al., 2009a; Li et al., 2012). On the other hand, H2S was reported to act downstream of NO in the ethylene-induced stomatal closure and in the resistance to heat stress (Liu et al., 2012; Li et al., 2013b). Interestingly, Jin et al. (2011) have recently reported that, in Arabidopsis whole-plant extracts, the production rate of H2S increased upon drought stress treatment.
Lisjak et al. (2010, 2011) have proposed a negative interaction between H2S and NO in ABA-induced stomatal closure in Arabidopsis and C. annuum. Based in pharmacological evidence, the authors speculate that the NO produced by ABA is removed by H2S, and thus, the depletion of endogenous NO increases stomatal aperture in the dark (Lisjak et al., 2010). Differences in methodological procedures concerning the isolation of epidermal strips and the timing of the treatments may result in the differential response of the guard cells to the H2S donors between Lisjak et al. (2010) and this study.
Liu et al. (2011) showed that stomatal closure but not NO synthesis is impaired in an Arabidopsis mutant of the l-Cys desulfhydrase AtL-CDES gene. Therefore, they suggest that NO is acting upstream of H2S in ethylene-induced stomatal closure. However, some considerations must be taken into account. First, the locus mutated in Atl-cdes codes for a mitochondrial/chloroplastic Cys desulfurase (At5g65720), which is a NITROGEN FIXATION S (NIFS)-LIKE1 protein involved in the biosynthesis of different cofactors such as the iron-sulfur clusters (Van Hoewyk et al., 2008). This NifS-like protein catalyzes the conversion of Cys to Ala and elemental sulfur instead of the conversion to pyruvate, ammonium, and sulfide, as has been unequivocally established for DES1 (At5g28030; Alvarez et al., 2010). Second, despite it having been reported that a convergence exists between ABA-dependent and ethylene-dependent signaling pathways (Ribeiro et al., 2009), the interaction of these two players is still poorly understood. Therefore, the data presented by Liu et al (2011) is not enough to rule out an ABA-induced, H2S-independent NO production.
The characterization of the central ABA signaling pathway in stomata, which involves the participation of the PYR/PYL/RCAR receptors, Clade I PP2C (including ABI1, ABI2, and HOMOLOGYTOABI1), and ABA-activated SNF1-RELATED KINASE2 (SnRK2) (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Umezawa et al., 2009), has become a milestone in the study of ABA signaling. A challenge for identifying the new components that are emerging in guard cell ABA signaling, such as DES1, is to understand how these new pathways are associated with the central ABA signaling pathway. The quadruple mutant of the ABA receptor QC3 generated by Nishimura et al. (2010) was showed to be highly insensitive to ABA; however, it still will close the stomata in response to downstream elements (Nishimura et al., 2010; Wang et al., 2013). Accordingly, guard cells from QC3 plants do not produce endogenous reactive oxygen species or NO in response to ABA (Yin et al., 2013). Likewise, exogenous H2S is able to induce stomatal closure in the QC3 mutant, suggesting that it acts downstream of the ABA receptor, although a PYL/PYR/RCAR-independent pathway cannot be ruled out with this data. Our data show that exogenous H2S does not induce stomatal closure in the gain-of-function mutants of ABI1 that is a negative regulator of ABA-induced stomatal closure (Merlot et al., 2001; Saez et al., 2004; Kuhn et al., 2006; Rubio et al., 2009), suggesting that H2S is acting upstream of this ABA signaling element. Interestingly, previous reports show that (1) NO is produced in guard cells from Arabidopsis wild-type plants in response to ABA (García-Mata and Lamattina, 2002; Lozano-Juste and León, 2010) but not in the QC3 quadruple mutant (Yin et al., 2013), and (2) exogenous addition of NO induces stomatal closure in wild-type Arabidopsis plants but not in abi1-1 and abi1-2 (Desikan et al., 2002; Dubovskaya et al., 2011), suggesting that NO is acting downstream of the PYR/PYL/RCAR receptor and upstream of ABI1. Considering the evidence presented here indicating that DES1/H2S is acting upstream of NO, a simplified model is shown in Figure 9 to propose the position of DES1/H2S in the ABA signaling network. The nature of the interaction between NO and ABI1 is yet to be clarified. It can be speculated that NO might be regulating the binding of ABI1 to PYR/PYL/RCAR through the modification of either ABI1 protein or the ABI1 binding domain at the receptor. Another possibility is the existence of a signaling element positioned between NO and ABI1. A candidate for this signaling component might be the phospholipid signal phosphatidic acid, whose production is increased by NO in guard cells (Distéfano et al., 2008) and who was reported to regulate ABI1/PP2C activity preventing ABI1 translocation from the cytosol to the nucleus (Zhang et al., 2004).
Figure 9.
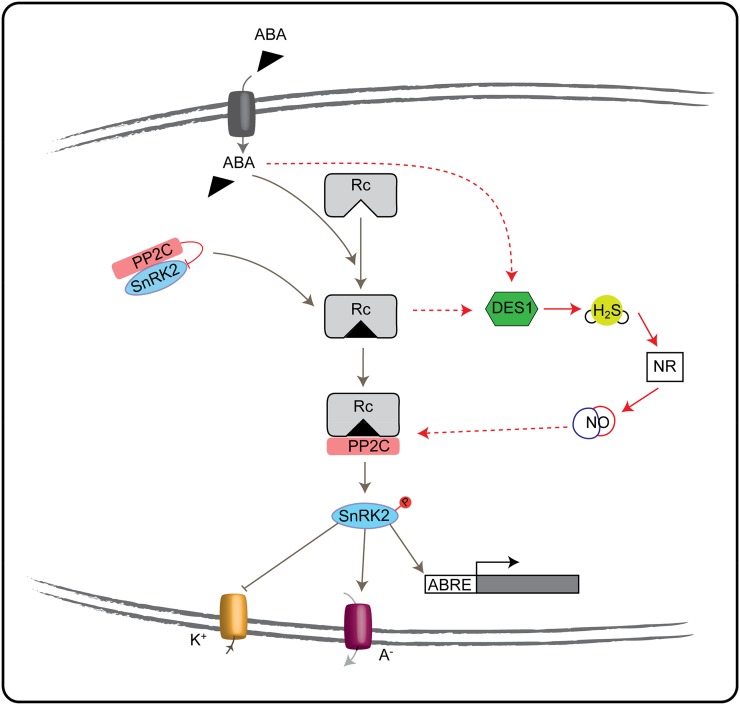
Simplified model showing the interaction of DES1/H2S with the core ABA signaling pathway in guard cells. ABA enters in the guard cells through ATP-binding cassette transporters. Once in the guard cells, it binds to the PYR/PYL/RCAR receptor (Rc). The ABA PYL/PYR/RCAR receptor binds the clade A PP2C inhibiting its phosphatase activity and, through it, releases the SnRKs protein kinases and the signaling elements downward. DES1 is up-regulated by ABA, releasing H2S, which increases endogenous NO via nitrate reductase (NR) activity. NO might regulate the binding of ABI1 to the PYR/PYL/RCAR receptor and through the downstream response. Arrow lines indicate activation, blunt lines indicate inhibition, dashed red lines indicate cross talk proposed in this work (hypothetical), gray lines indicate already published. K+, Potassium uptake; A–, anion efflux; ABRE, ABA-responsive element transcription factors.
Recent evidence demonstrates the H2S can regulate protein activity through the direct interaction of H2S with the thiol group of the target protein yielding a hydropersulfide moiety (Mustafa et al., 2009). Further studies will be needed to see if this is the mechanism by which H2S regulates ABA-signaling components. Overall, this study presents compelling evidence supporting DES1 as a unique component of ABA signaling in guard cells, mediating H2S production and acting upstream of NO to induce stomatal closure.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) wild-type Col-0 and No-0, des1-1 (SALK_103855) and des1-2 (RIKEN RATM13-27151_G; Alvarez et al., 2010; Álvarez et al., 2012; Supplemental Fig. S5), abi1-1 (Arabidopsis Biological Resource Center stock CS22), nia1nia2 (Arabidopsis Biological Resource Center stock CS6512), and pyr1/pyl1/pyl2/pyl4 (QC3, kindly provided by Sean Cutler, University of California, Riverside) mutants were used in this work. NOD and EV Arabidopsis seeds were kindly provided by Jurgen Zeier (University of Fribourg). NOD Arabidopsis plants expressing the bacterial flavohemoglobin (Col-0 background) were generated by Agrobacterium tumefaciens-mediated plant transformation using the DEX-inducible expression vector pTA7001 as described in Zeier et al. (2004). Plants transformed with empty expression vector pTA7001 EV were used as controls (EV). Plants were grown in soil:perlite:vermiculite: (1:1:1, v/v/v) at 25°C under a 16-h-light/8-h-dark photoperiod and watered with Arabidopsis salt (Wilson et al., 1990) nutritive medium.
To generate the des1-1 complementation line, a 972-bp cDNA fragment containing the full-length coding sequence of DES1 was obtained by RT-PCR amplification using the proofreading Platinum Pfx DNA polymerase (Invitrogen) and the primers DES1F and DES1R (Supplemental Table S1). The fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) and transferred into the pMDC32 vector (Curtis and Grossniklaus, 2003) using the Gateway system (Invitrogen) according to the manufacturer’s instructions. The final construct was generated by transformation into A. tumefaciens and then introduced into des1-1 null plants using the floral dip method (Clough and Bent, 1998). Four transgenic lines were generated, showing complementation, and the line used in this work is a homozygous T4.
Chemicals and Treatments
NaHS, HT, and ABA were purchased from Sigma, (p-methoxyphenyl)morpholino-phosphinodithioic acid (GYY 4137) was purchased from Cayman Chemicals, cPTIO and SNAP were purchased from Molecular Probes, and 3-amino-4-(N-methylamino)-2′, 7′-difluorofluorescein diacetate (DAF-FM-DA) was purchased from Calbiochem. The stomatal aperture treatments were performed on epidermal strips excised from the abaxial side of fully expanded Arabidopsis leaves. Immediately after stripping, the epidermal peels were floated in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) for 3 h. The strips were subsequently maintained in the same opening buffer and exposed to different treatments. After 90 min, stomata were digitized using a Nikon DS-Fi 1 camera coupled to a Nikon Eclipse Ti microscope. The stomatal aperture width was measured using ImageJ analysis software (National Institutes of Health).
Stomatal Conductance Measurements
Arabidopsis wild-type Col-0 and No-0 and des1 mutant plants were sprayed with water (control) or 50 μm ABA. After 4 h of treatment, the leaf gas exchange was measured in planta using a S151 IRGA (Qubit Systems), and the leaf temperature was measured using a S171 Leaf Chamber Thermistor (Qubit Systems) according to the manufacturer’s instructions.
RNA Isolation, RT Reaction, and PCR and Real-Time PCR Analysis
Whole Leaf
Three-week-old wild-type Col-0 and No-0 plants were floated for 1, 2, 5, 7, 9, and 24 h in water as controls or treated with different ABA concentrations. Total RNA was extracted from the leaves using an RNeasy Plant Mini Kit (Qiagen). The RNA was reverse transcribed using an oligo(dT) primer and the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the recommended protocols.
GC-e
Arabidopsis abaxial epidermal peels (GC-e) were floated in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) for 3 h and then treated in the same buffer with or without 50 μm ABA for 90 min. Samples of the epidermal peels were loaded with the viability fluorescent probe fluorescein diacetate. None of the analyzed samples showed stained mesophyll cells; however, epidermis showed roughly 10% of pavement cells that survived. Therefore, we state that we have a GC-e extraction. To assess if the epidermal cell contamination or any unnoticed mesophyll cell contamination has any effect on the expression levels of the selected transcripts, we collected leaf cuts after the extraction of the abaxial epidermal layer ( MC-e), we floated them in opening buffer (10 mm K-MES, pH 6.1, and 10 mm KCl) for 3 h, and then we subjected them to the same treatments assayed for GC-e. Total RNA was extracted using Trizol reagent (Invitrogen). Subsequently, 2 μg of total RNA was used for the RT reaction and 1 μg of total RNA for qRT-PCR using an oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Promega).
RT-PCR Reactions
An aliquot of the cDNA was amplified in subsequent PCR reactions using the following primers: ActinF/ActinR for the constitutive Actin (At4g05320) gene, CER2F/CER2R for the guard cell marker CER2 (At4G24510) gene, and Canh1F/Canh1R for the mesophyll cell marker Canh1 (At3g01500) gene (Supplemental Table S1). For the amplification of CER2 and Canh1, the following amplification conditions were used: a denaturation cycle of 5 min at 94°C; 32 amplification cycles of 30 s at 94°C, 50 s at 60°C, and 1 min 30 s at 72°C; and an extension cycle of 5 min at 72°C. For the Actin gene, 24 amplification cycles were performed.
Quantitative Real-Time RT-PCR to Analyze DES1 Gene Expression
First-strand cDNA was synthesized as described above. The specific primers for this gene and the constitutive POLYUBIQUITIN10 (UBQ10) used as a control were designed using the Vector NTI Advance 10 software (Invitrogen; Supplemental Table S1). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad), and the signals were detected on an iCYCLER (Bio-Rad) according to the manufacturer’s instructions. The cycling profile consisted of 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Subsequent to the PCR cycling, a melt curve from 60°C to 90°C was performed. The expression levels of DES1 were normalized to that of the constitutive UBQ10 gene by subtracting the cycle threshold value of UBQ10 from the cycle threshold value of the gene (ΔCT). The gene expression level was calculated as 2–ΔCT (López-Martín et al., 2008). The results shown are given as means ± sd of at least three independent RNA samples. For RD29A and Rab18 gene analysis, the specific primers for this gene and the constitutive ACTIN2 (ACT2) used as a control were designed using the Vector NTI Advance 10 software (Invitrogen; Supplemental Table S1). The Fast Universal SYBR Green Master mix from Roche was employed, using a Step-One Real-Time PCR machine from Applied Biosystems. The standard amplification program was used. The expression levels of the gene of interest were normalized to that of the constitutive ACT2 gene as described above.
DES Activity
Plant leaf material was ground in 20 mm Tris-HCl (pH 8) using a mortar and pestle with liquid nitrogen. After centrifugation at 15,000g for 15 min at 4°C, the resulting supernatant was used as plant-soluble extract for measuring DES activity. The total amount of protein in the extracts was determined by using the Bradford method. The DES activity was measured by the release of sulfide from l-Cys as described previously (Alvarez et al., 2010). The assay contained 1 mm dithiothreitol, 1 mm l-Cys, 100 mm Tris-HCl, pH 8.0, and enzyme extract in a total volume of 1 mL. The reaction was initiated upon the addition of l-Cys. After incubation for 15 min at 37°C, the reaction was terminated upon the addition of 100 μL of 30 mm FeCl3 dissolved in 1.2 n HCl and 100 μL of 20 mm N,N-dimethyl-p-phenylenediamine dihydrochloride dissolved in 7.2 n HCl. The formation of methylene blue was determined at 670 nm, and the enzyme activity was calculated using the extinction coefficient of 15 × 106 cm2 mol–1.
Fluorescence Microscopy
NO was visualized using the specific NO dye DAF-FM-DA. Arabidopsis epidermal strips preincubated in opening buffer for 3 h in the dark were loaded with 10 µm DAF-FM-DA for 30 min. The strips were washed three times with fresh opening buffer and exposed to different treatments for 15 min. Fluorescent images were obtained using a Nikon DS-Fi 1 digital camera coupled to a Nikon Eclipse Ti epifluorescence microscope (excitation, 495 nm; emission, 515–555 nm). The green fluorescence was quantified as the pixel intensity of a fixed area for all guard cells using ImageJ analysis software (National Institutes of Health). The fluorescence values are presented as relative units with respect to the control treatments and are expressed as the means ± se. Fifteen to 30 guard cells were observed per experiment in each treatment for at least four independent replicates.
Statistical Analysis
Data analyses were performed using Sigmaplot for Windows (Systat Software). The statistically significant differences were analyzed using one-way ANOVA, or the Student’s t test, as indicated in the figure legends.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At5G28030.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. H2S induces stomatal closure in a dose dependent manner in Arabidopsis.
Supplemental Figure S2. Gene expression analysis of Arabidopsis guard cell- vs. mesophyll cell-enriched extracts.
Supplemental Figure S3. ABA regulates DES1 transcript levels and DES enzyme activity in whole leaf.
Supplemental Figure S4. Promoter region of DES1 contains ABA-related motifs.
Supplemental Figure S5. Structure of the DES1 gene in the insertion mutants.
Supplemental Table S1. Sequences of oligonucleotides used in this work.
Supplementary Material
Glossary
- ABA
abscisic acid
- H2S
hydrogen sulfide
- NO
nitric oxide
- qRT
quantitative reverse transcription
- Col-0
ecotype Columbia
- No-0
ecotype Nössen
- cDNA
complementary DNA
- HT
hypotaurine
- IRGA
infrared gas analyzer
- GC-e
guard cell-enriched
- MC-e
mesophyll cell-enriched
- RT
reverse transcription
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide
- NOD
nitric oxide dioxygenase
- DEX
Dexamethasone
- SNAP
S-nitroso-N-acetylpenicillamine
- DAF-FM-DA
4,5-diaminoflorescein diacetate
- GCP
guard cell protoplast
- MCP
mesophyll cell protoplast
- EV
empty vector
- NaHS
sodium hydrosulfide
Footnotes
This work was supported by grants from the Universidad Nacional de Mar del Plata, Consejo Nacional de Investigaciones Científicas y Técnicas, and Agencia Nacional de Promoción Científica y Tecnológica and by the European Regional Development Fund through the Ministerio de Economia y Competitividad (grant no. BIO2013–44648), the Junta de Andalucía (grant no. CVI–7190), and fellowship support through the program Junta para la Ampliación de Estudios (to C.Á.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alvarez C, Calo L, Romero LC, García I, Gotor C (2010) An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol 152: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Moreno I, Pérez-Pérez ME, Crespo JL, Romero LC, Gotor C (2012) Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24: 4621–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt MR. (2000) Cellular signaling and volume control in stomatal movements in plants. Annu Rev Cell Dev Biol 16: 221–241 [DOI] [PubMed] [Google Scholar]
- Bloem E, Riemenschneider A, Volker J, Papenbrock J, Schmidt A, Salac I, Haneklaus S, Schnug E (2004) Sulphur supply and infection with Pyrenopeziza brassicae influence l-cysteine desulphydrase activity in Brassica napus L. J Exp Bot 55: 2305–2312 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coletta C, Papapetropoulos A, Erdelyi K, Olah G, Módis K, Panopoulos P, Asimakopoulou A, Gerö D, Sharina I, Martin E, et al. (2012) Hydrogen sulfide and nitric oxide are mutually dependent in the regulation of angiogenesis and endothelium-dependent vasorelaxation. Proc Natl Acad Sci USA 109: 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distéfano AM, García-Mata C, Lamattina L, Laxalt AM (2008) Nitric oxide-induced phosphatidic acid accumulation: a role for phospholipases C and D in stomatal closure. Plant Cell Environ 31: 187–194 [DOI] [PubMed] [Google Scholar]
- Dubovskaya LV, Bakakina YS, Kolesneva EV, Sodel DL, McAinsh MR, Hetherington AM, Volotovski ID (2011) cGMP-dependent ABA-induced stomatal closure in the ABA-insensitive Arabidopsis mutant abi1-1. New Phytol 191: 57–69 [DOI] [PubMed] [Google Scholar]
- Filipovic MR, Eberhardt M, Prokopovic V, Mijuskovic A, Orescanin-Dusic Z, Reeh P, Ivanovic-Burmazovic I (2013) Beyond H2S and NO interplay: hydrogen sulfide and nitroprusside react directly to give nitroxyl (HNO). A new pharmacological source of HNO. J Med Chem 56: 1499–1508 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2002) Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol 128: 790–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2010) Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New Phytol 188: 977–984 [DOI] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2013) Gasotransmitters are emerging as new guard cell signaling molecules and regulators of leaf gas exchange. Plant Sci 201-202: 66–73 [DOI] [PubMed] [Google Scholar]
- Harrington HM, Smith IK (1980) Cysteine metabolism in cultured tobacco cells. Plant Physiol 65: 151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Jin Z, Shen J, Qiao Z, Yang G, Wang R, Pei Y (2011) Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem Biophys Res Commun 414: 481–486 [DOI] [PubMed] [Google Scholar]
- Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y (2013) Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol Biochem 62: 41–46 [DOI] [PubMed] [Google Scholar]
- Kabil O, Banerjee R (2010) Redox biochemistry of hydrogen sulfide. J Biol Chem 285: 21903–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura M, Fukuda R, Bateman RM, Yamamoto T, Suematsu M (2010) Interactions of multiple gas-transducing systems: hallmarks and uncertainties of CO, NO, and H2S gas biology. Antioxid Redox Signal 13: 157–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluru GK, Shen X, Kevil CG (2013) A tale of two gases: NO and H2S, foes or friends for life? Redox Biol 1: 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo S, Kurokawa Y, Doe I, Masuko T, Sekiguchi F, Kawabata A (2007) Hydrogen sulfide inhibits activity of three isoforms of recombinant nitric oxide synthase. Toxicology 241: 92–97 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Vavasseur A, Forestier C (1999) ATP binding cassette modulators control abscisic acid-regulated slow anion channels in guard cells. Plant Cell 11: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bhatia M, Moore PK (2006) Hydrogen sulphide: a novel mediator of inflammation? Curr Opin Pharmacol 6: 125–129 [DOI] [PubMed] [Google Scholar]
- Li L, Hsu A, Moore PK (2009) Actions and interactions of nitric oxide, carbon monoxide and hydrogen sulphide in the cardiovascular system and in inflammation: a tale of three gases! Pharmacol Ther 123: 386–400 [DOI] [PubMed] [Google Scholar]
- Li L, Wang Y, Shen W (2012) Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots. Biometals 25: 617–631 [DOI] [PubMed] [Google Scholar]
- Li ZG, Ding XJ, Du PF (2013a) Hydrogen sulfide donor sodium hydrosulfide-improved heat tolerance in maize and involvement of proline. J Plant Physiol 170: 741–747 [DOI] [PubMed] [Google Scholar]
- Li ZG, Yang SZ, Long WB, Yang GX, Shen ZZ (2013b) Hydrogen sulphide may be a novel downstream signal molecule in nitric oxide-induced heat tolerance of maize (Zea mays L.) seedlings. Plant Cell Environ 36: 1564–1572 [DOI] [PubMed] [Google Scholar]
- Lisjak M, Srivastava N, Teklic T, Civale L, Lewandowski K, Wilson I, Wood ME, Whiteman M, Hancock JT (2010) A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol Biochem 48: 931–935 [DOI] [PubMed] [Google Scholar]
- Lisjak M, Teklić T, Wilson ID, Wood M, Whiteman M, Hancock JT (2011) Hydrogen sulfide effects on stomatal apertures. Plant Signal Behav 6: 1444–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hou L, Liu G, Liu X, Wang X (2011) Hydrogen sulfide induced by nitric oxide mediates ethylene-induced stomatal closure of Arabidopsis thaliana. Chin Sci Bull 56: 3547–3553 [Google Scholar]
- Liu J, Hou Z, Liu G, Hou L, Liu X (2012) Hydrogen sulfide may function downstream of nitric oxide in ethylene-induced stomatal closure in Vicia faba L. J Integr Agric 11: 1644–1653 [Google Scholar]
- López-Martín MC, Becana M, Romero LC, Gotor C (2008) Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol 147: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J (2010) Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol 152: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mancardi D, Penna C, Merlino A, Del Soldato P, Wink DA, Pagliaro P (2009) Physiological and pharmacological features of the novel gasotransmitter: hydrogen sulfide. Biochim Biophys Acta 1787: 864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH (2009) H2S signals through protein S-sulfhydration. Sci Signal 2: ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrias K, Stasko A, Cacanyiova S, Sulova Z, Krizanova O, Kristek F, Malekova L, Knezl V, Breier A (2008) H2S and HS– donor NaHS releases nitric oxide from nitrosothiols, metal nitrosyl complex, brain homogenate and murine L1210 leukaemia cells. Pflugers Arch 457: 271–279 [DOI] [PubMed] [Google Scholar]
- Pandey S, Wang XQ, Coursol SA, Assmann SM (2002) Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol 153: 517–526 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Riemenschneider A, Kamp A, Schulz-Vogt HN, Schmidt A (2007) Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants: from the field to the test tube and back. Plant Biol (Stuttg) 9: 582–588 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YJ, Nanduri J, Raghuraman G, Souvannakitti D, Gadalla MM, Kumar GK, Snyder SH, Prabhakar NR (2010) H2S mediates O2 sensing in the carotid body. Proc Natl Acad Sci USA 107: 10719–10724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro DM, Desikan R, Bright J, Confraria A, Harrison J, Hancock JT, Barros RS, Neill SJ, Wilson ID (2009) Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ 32: 46–57 [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Sun J, Wang R, Zhang X, Yu Y, Zhao R, Li Z, Chen S (2013) Hydrogen sulfide alleviates cadmium toxicity through regulations of cadmium transport across the plasma and vacuolar membranes in Populus euphratica cells. Plant Physiol Biochem 65: 67–74 [DOI] [PubMed] [Google Scholar]
- Szabó C. (2007) Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov 6: 917–935 [DOI] [PubMed] [Google Scholar]
- Tossi V, Amenta M, Lamattina L, Cassia R (2011) Nitric oxide enhances plant ultraviolet-B protection up-regulating gene expression of the phenylpropanoid biosynthetic pathway. Plant Cell Environ 34: 909–921 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Pilon M, Pilon-Smits EAH (2008) The functions of NifS-like proteins in plant sulfur and selenium metabolism. Plant Sci 174: 117–123 [Google Scholar]
- Wang R. (2002) Two’s company, three’s a crowd: Can H2S be the third endogenous gaseous transmitter? FASEB J 16: 1792–1798 [DOI] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM (2011) Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen ZH, Zhang B, Hills A, Blatt MR (2013) PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl– channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol 163: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman M, Li L, Kostetski I, Chu SH, Siau JL, Bhatia M, Moore PK (2006) Evidence for the formation of a novel nitrosothiol from the gaseous mediators nitric oxide and hydrogen sulphide. Biochem Biophys Res Commun 343: 303–310 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Yang W, Yang G, Jia X, Wu L, Wang R (2005) Activation of KATP channels by H2S in rat insulin-secreting cells and the underlying mechanisms. J Physiol 569: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Adachi Y, Ye W, Hayashi M, Nakamura Y, Kinoshita T, Mori IC, Murata Y (2013) Difference in abscisic acid perception mechanisms between closure induction and opening inhibition of stomata. Plant Physiol 163: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong QC, Hu LF, Wang S, Huang D, Bian JS (2010) Hydrogen sulfide interacts with nitric oxide in the heart: possible involvement of nitroxyl. Cardiovasc Res 88: 482–491 [DOI] [PubMed] [Google Scholar]
- Zeier J, Delledonne M, Mishina T, Severi E, Sonoda M, Lamb C (2004) Genetic elucidation of nitric oxide signaling in incompatible plant-pathogen interactions. Plant Physiol 136: 2875–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dou W, Jiang CX, Wei ZJ, Liu J, Jones RL (2010a) Hydrogen sulfide stimulates β-amylase activity during early stages of wheat grain germination. Plant Signal Behav 5: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jiao H, Jiang CX, Wang SH, Wei ZJ, Luo JP, Jones RL (2010b) Hydrogen sulfide protects soybean seedlings against drought-induced oxidative stress. Acta Physiol Plant 32: 849–857 [Google Scholar]
- Zhang H, Tan ZQ, Hu LY, Wang SH, Luo JP, Jones RL (2010c) Hydrogen sulfide alleviates aluminum toxicity in germinating wheat seedlings. J Integr Plant Biol 52: 556–567 [DOI] [PubMed] [Google Scholar]
- Zhang H, Tang J, Liu XP, Wang Y, Yu W, Peng WY, Fang F, Ma DF, Wei ZJ, Hu LY (2009a) Hydrogen sulfide promotes root organogenesis in Ipomoea batatas, Salix matsudana and Glycine max. J Integr Plant Biol 51: 1086–1094 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang MJ, Hu LY, Wang SH, Hu KD, Bao LJ, Luo JP (2010d) Hydrogen sulfide promotes wheat seed germination under osmotic stress. Russ J Plant Physiol 57: 532–539 [Google Scholar]
- Zhang H, Ye Y, Wang SH, Luo J, Tang J, Ma DF (2009b) Hydrogen sulfide counteracts chlorophyll loss in sweet potato seedling leaves and alleviates oxidative damage against osmotic stress. Plant Growth Regul 58: 243–250 [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase D α 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.