Natural variation provides clues to essential chloroplast gene functions and the significance of a duplicated nuclear gene that targets homomeric acetyl-CoA carboxylase to plastids.
Abstract
Mutations that eliminate chloroplast translation in Arabidopsis (Arabidopsis thaliana) result in embryo lethality. The stage of embryo arrest, however, can be influenced by genetic background. To identify genes responsible for improved growth in the absence of chloroplast translation, we examined seedling responses of different Arabidopsis accessions on spectinomycin, an inhibitor of chloroplast translation, and crossed the most tolerant accessions with embryo-defective mutants disrupted in chloroplast ribosomal proteins generated in a sensitive background. The results indicate that tolerance is mediated by ACC2, a duplicated nuclear gene that targets homomeric acetyl-coenzyme A carboxylase to plastids, where the multidomain protein can participate in fatty acid biosynthesis. In the presence of functional ACC2, tolerance is enhanced by a second locus that maps to chromosome 5 and heightened by additional genetic modifiers present in the most tolerant accessions. Notably, some of the most sensitive accessions contain nonsense mutations in ACC2, including the “Nossen” line used to generate several of the mutants studied here. Functional ACC2 protein is therefore not required for survival in natural environments, where heteromeric acetyl-coenzyme A carboxylase encoded in part by the chloroplast genome can function instead. This work highlights an interesting example of a tandem gene duplication in Arabidopsis, helps to explain the range of embryo phenotypes found in Arabidopsis mutants disrupted in essential chloroplast functions, addresses the nature of essential proteins encoded by the chloroplast genome, and underscores the value of using natural variation to study the relationship between chloroplast translation, plant metabolism, protein import, and plant development.
Embryo development in Arabidopsis (Arabidopsis thaliana) requires the coordinated expression of a large number of essential genes (Muralla et al., 2011). Recessive mutations that disrupt these nuclear genes result in an embryo-defective (emb) mutant phenotype (Meinke, 2013). Many EMB genes of Arabidopsis encode chloroplast-localized proteins involved in basic metabolism, protein import, and chloroplast gene expression (Hsu et al., 2010; Bryant et al., 2011; Savage et al., 2013). Functional plastids are therefore required for embryo development in Arabidopsis. Mutations that disrupt photosynthesis alone interfere with embryo and seedling pigmentation, not embryo development. Multiple examples of EMB genes that encode chloroplast-localized aminoacyl-tRNA synthetases, RNA-binding proteins, translation factors, and ribosomal proteins have been described in the literature (Berg et al., 2005; Bryant et al., 2011; Muralla et al., 2011; Romani et al., 2012; Tiller and Bock, 2014). Translation of some chloroplast-encoded mRNAs is therefore essential for seed development. This raises a basic question: which chloroplast genes are required? In this report, we used natural variation and genetic analysis to evaluate the model (Bryant et al., 2011) that a single chloroplast gene, acetyl-coenzyme A carboxylase D (accD), needed for the initial stages of fatty acid biosynthesis, underlies the requirement for chloroplast translation during heterotrophic growth and embryo development in Arabidopsis.
Targeted gene disruptions in tobacco (Nicotiana tabacum) have identified four chloroplast genes with essential functions that extend beyond photosynthesis: accD, caseinolytic protease P1 (clpP1), hypothetical chloroplast open reading frame1 (ycf1), and ycf2 (Drescher et al., 2000; Kuroda and Maliga, 2003; Kode et al., 2005). Comparative genomics have shown that all four genes are retained in the plastid genomes of most angiosperms, including chlorophyll-deficient, parasitic species (dePamphilis and Palmer, 1990; Funk et al., 2007; Jansen et al., 2007). Several examples of essential chloroplast genes that relocated to the nucleus have also been described (Magee et al., 2010; Rousseau-Gueutin et al., 2013). The absence of ycf1 and ycf2 in grasses (Jansen et al., 2007) and the replacement of accD with a nuclear gene that targets functional protein back to the chloroplast (Konishi and Sasaki, 1994; Chalupska et al., 2008) remain to be explained.
The accD gene in Arabidopsis (AtCg00500) encodes one subunit of the chloroplast-localized heteromeric acetyl-coenzyme A carboxylase (ACCase), an essential enzyme in fatty acid biosynthesis that converts acetyl-CoA to malonyl-CoA. Three other subunits are encoded by nuclear genes, one of which is also known to be required for embryo development (Li et al., 2011). Disruptions of three additional genes (At3g25860, At1g34430, and At2g30200) associated with the reactions that precede and follow the step catalyzed by heteromeric ACCase also result in embryo lethality (Lin et al., 2003; Bryant et al., 2011; Muralla et al., 2011). Embryo lethality is also encountered in auxotrophic mutants unable to produce biotin, an essential vitamin required for ACCase function (Schneider et al., 1989; Patton et al., 1998; Muralla et al., 2008). The conversion of acetyl-CoA to malonyl-CoA during fatty acid biosynthesis within the plastid is therefore required for embryo development in Arabidopsis.
In addition to the chloroplast-localized, heteromeric ACCase found in most angiosperms, there is also a cytosolic, homomeric ACCase involved in later stages of fatty acid biosynthesis. In both Arabidopsis and Brassica napus, the gene that encodes this homomeric enzyme is duplicated (Yanai et al., 1995; Schulte et al., 1997). One copy (ACC1; At1g36160) encodes an essential protein localized to the cytosol. Disruption of this gene in Arabidopsis (EMB22, GURKE, and PASTICCINO3 [PAS3]) results in an embryo-defective phenotype distinct from that seen following a loss of chloroplast translation (Meinke, 1985; Baud et al., 2004). Weak alleles exhibit cold sensitivity and glossy inflorescence stems resulting from changes in cuticular wax composition (Lü et al., 2011; Amid et al., 2012). The adjacent copy (ACC2; At1g36180) is expressed at low levels and is predicted to encode a chloroplast-localized protein (Yanai et al., 1995; Baud et al., 2003; Babiychuk et al., 2011). Knockouts of this gene exhibit no obvious phenotype under normal growth conditions (Babiychuk et al., 2011).
In Brassica spp., plants with albino leaves devoid of chloroplast ribosomes have been produced by germinating seeds on spectinomycin, an inhibitor of chloroplast translation, and then transplanting the young seedlings to basal medium (Zubko and Day, 1998). This experimental approach was initially described as a promising system for generating stable albinism without mutagenesis. However, different results were obtained with tobacco and Arabidopsis seedlings, which were much more sensitive to spectinomycin. In light of this reported variation in seedling responses to spectinomycin and the known duplication of ACC1 in the Brassicaceae, we decided to explore whether natural accessions of Arabidopsis differed in their ability to tolerate a loss of chloroplast translation and whether genetic analysis in Arabidopsis could uncover some of the genes involved. The results described here confirm the value of this approach, provide insights into the phenotypes of mutants defective in essential chloroplast functions, and help to explain the requirement of chloroplast translation for plant growth and development.
RESULTS
Arabidopsis Accessions Differ in Seedling Sensitivity to Spectinomycin
Several factors were considered before deciding which accessions to evaluate on spectinomycin: geographical location, genetic diversity (McKhann et al., 2004; Nordborg et al., 2005; Clark et al., 2007), inclusion among mutants defective in chloroplast translation (Bryant et al., 2011), availability of genomic sequence data (Weigel and Mott, 2009), and flowering time. Emphasis was placed on early-flowering accessions to facilitate genetic analysis. A list of accessions tested is presented in Supplemental Table S1. Spectinomycin was chosen to inhibit chloroplast translation based on past work (Zubko and Day, 1998; Dudas et al., 2012), known mode of action (Wirmer and Westhof, 2006), and limited impact on mitochondrial translation. The extent of seedling development was evaluated based on the size and number of leaves produced.
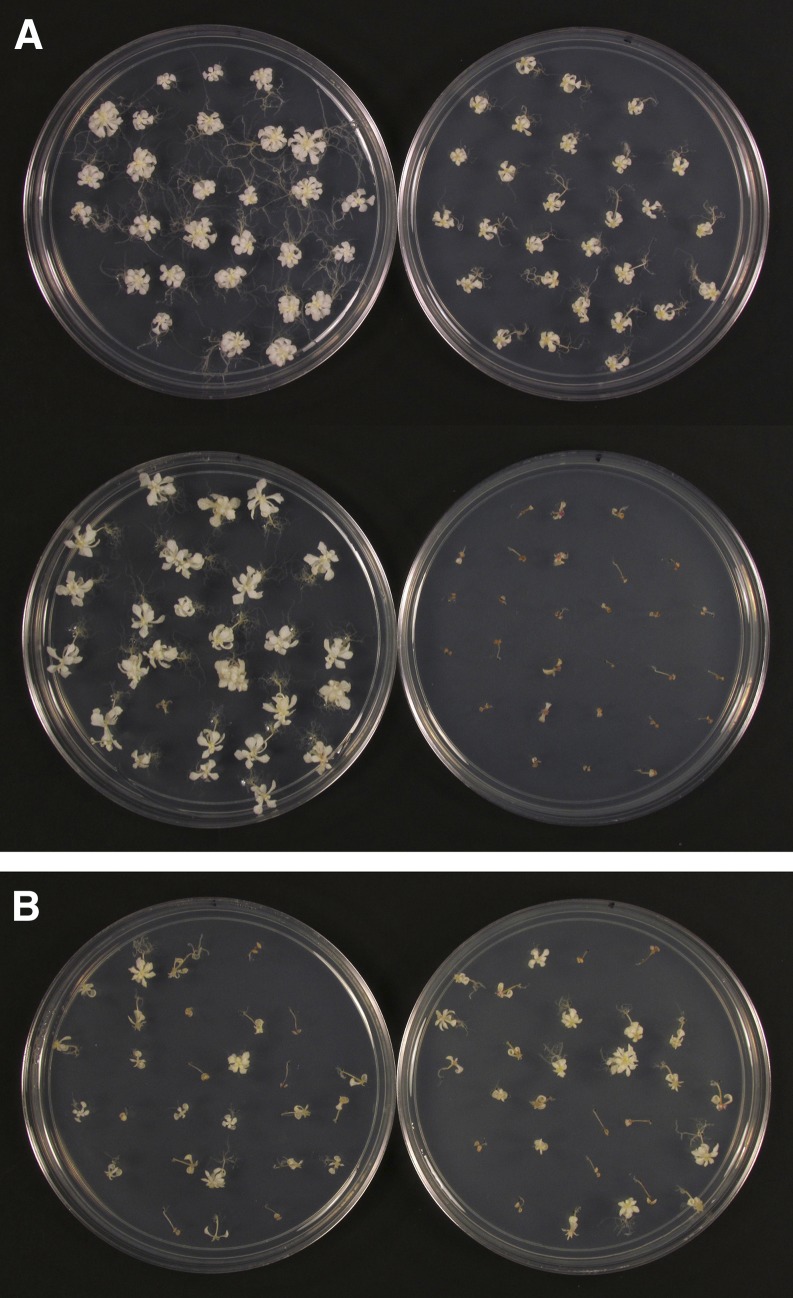
Seedling responses of 52 accessions after 5 weeks on medium containing spectinomycin and Glc are presented in Supplemental Figure S1. Because only 20 seedlings were initially tested for each accession, minor differences in growth responses are not significant. We focused instead on striking differences observed between the most and least tolerant accessions identified. Three tolerant accessions (Jl-3, Bensheim-1 [Be-1], and Tsu-0), three sensitive accessions (Oystese-0 [Oy-0], Nie1-2, and “Nossen”), and one intermediate accession (Columbia) were chosen for thorough analysis, including crosses with embryo-defective mutants disrupted in chloroplast translation. Seedlings from tolerant accessions developed albino rosettes with multiple pairs of leaves after 5 weeks on spectinomycin. By contrast, seedlings from sensitive accessions developed at most rudimentary leaf initials along with expanded cotyledons. Examples of seedling phenotypes are shown in Figure 1. The consistency of growth responses is evident in Figure 2A. Spectinomycin tolerance did not correlate with seedling growth rates on basal medium or with overall plant vigor in soil. Greening was restricted to seedlings with poor root contact with the growth medium. To rule out the possibility that accession-specific differences were unique to spectinomycin, we evaluated the effect of another inhibitor of chloroplast translation with a different mode of action (lincomycin) on tolerant and sensitive accessions. Similar results were found with four accessions examined in detail (Supplemental Fig. S2). Spontaneous resistance, reduced uptake, increased detoxification, and enhanced compartmentalization of antibiotics are unlikely explanations for these differences in seedling responses because, as noted later, the most tolerant accessions also supported the most advanced growth of mutant embryos defective in chloroplast translation, where antibiotics were not involved. Based on these results, we conclude that natural accessions of Arabidopsis differ in their ability to tolerate a loss of chloroplast translation.
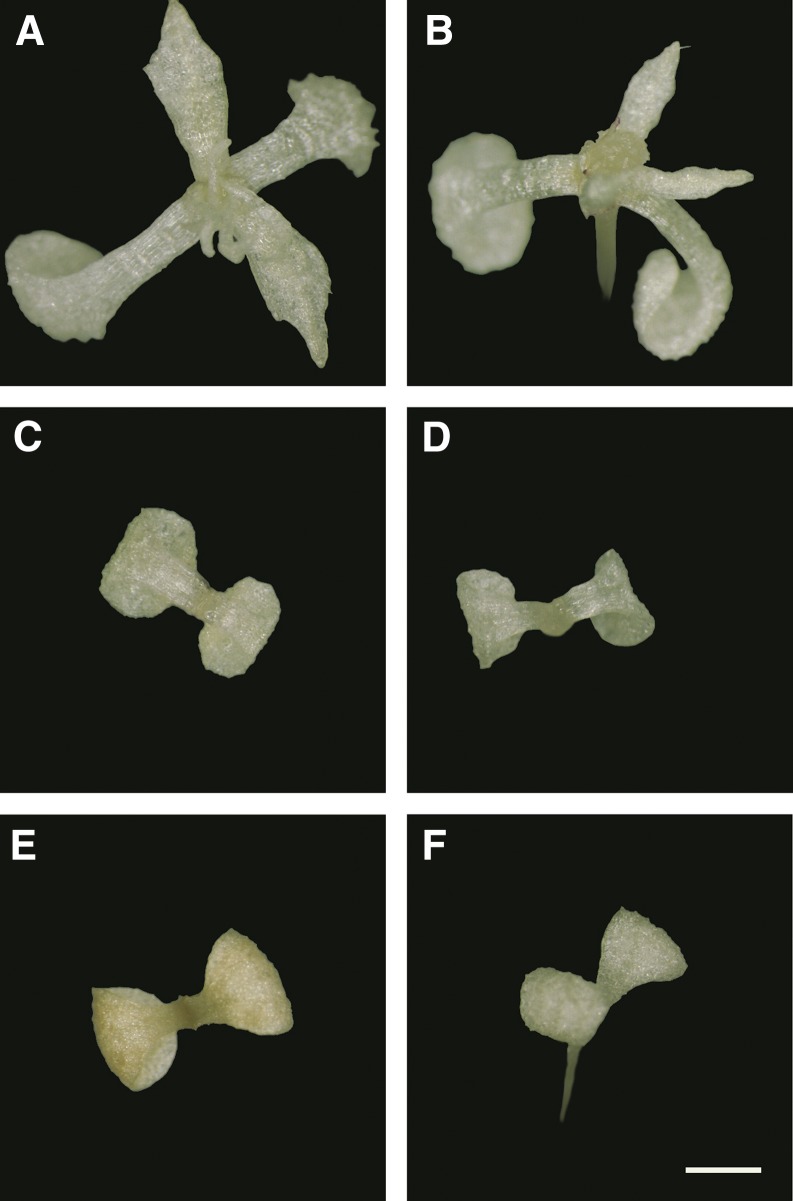
Figure 1.
Differential seedling responses of Arabidopsis accessions on spectinomycin. A and B, Tolerant accessions (Jl-3 and Be-1). C and D, Intermediate accession (Columbia). E and F, Sensitive accessions (“Nossen” and Oy-0). Bar = 1 mm.
Figure 2.
Consistent seedling responses of parental accessions on spectinomycin compared with segregating phenotypes in the F2 generation. A, Parental accessions (clockwise from lower left): tolerant, Jl-3, Be-1, and Tsu-0; and sensitive, “Nossen”. B, Segregating F2 seedlings from a Tsu-0 cross with “Nossen.” Plate diameter = 9 cm.
Crossing Tolerant and Sensitive Accessions Results in Phenotypic Segregation in the F2 Generation
To explore the genetic basis of differences in spectinomycin tolerance, wild-type plants from three tolerant accessions (Jl-3, Be-1, and Tsu-0) were crossed with a sensitive accession (“Nossen”) derived from segregating populations of RIKEN insertion mutants (Bryant et al., 2011). We designated this sensitive accession “Nossen” because it differs from the sequenced No-0 accession (Kuromori et al., 2004). In all crosses examined, sensitive and tolerant seedlings segregated in the F2 generation (Supplemental Table S2). Many F2 seedlings also exhibited intermediate phenotypes. We focused on the Tsu-0 accession for subsequent analyses because its F2 response on spectinomycin was most consistent with a single, semidominant locus conferring tolerance (Fig. 2B). However, F1 seedling phenotypes seemed to indicate that tolerance was determined by multiple genes instead. Overall, these studies revealed a genetic component to accession-specific differences in seedling growth in the absence of chloroplast translation. But the overlapping phenotypes observed suggested that identifying the genes involved would be difficult. We therefore pursued a second, complementary approach that involved crosses between tolerant accessions and embryo-defective mutants disrupted in chloroplast ribosomal proteins in a sensitive background. This approach led to the identification of distinct genetic loci that impacted the extent of embryo development in the absence of chloroplast translation and later to the demonstration that variation in these same genes contributed to the tolerance of seedlings from different accessions on spectinomycin.
Spectinomycin-Tolerant Accessions Partially Rescue Mutant Embryos Defective in Chloroplast Translation
Most studies on embryo-defective mutants of Arabidopsis have overlooked the impact of genetic background (accession) on mutant phenotype. We took the opposite approach here once we realized that embryo phenotypes of mutants defective in chloroplast translation are sensitive to genetic background. Information on 33 mutant alleles disrupted in chloroplast translation is presented in Supplemental Table S3. Embryos from 19 Salk and Syngenta mutants (Columbia accession) reach a late globular stage. Embryos from six RIKEN mutants (“Nossen” accession) arrest at a preglobular stage. Embryos from two other mutants (Landsberg erecta [Ler] accession) reach an intermediate (early globular) stage. Most importantly, after discounting four weak alleles and mutations in less essential genes, where screening for embryo rescue is ineffective because the original mutant allele supports continued growth, differences in the extent of embryo development in the absence of chloroplast translation correlate with differences in seedling growth on spectinomycin. In other words, the smallest mutant embryos are found in accessions that are most sensitive to spectinomycin. We therefore tested whether the tolerant Tsu-0 accession could rescue mutants in the sensitive “Nossen” accession. Two nuclear genes (EMB3126 and EMB3137) encoding chloroplast ribosomal proteins (L1 and S13) with mutant alleles in different genetic backgrounds (“Nossen”, Columbia, and Ler) and different embryo phenotypes were chosen for analysis (Table I). Work on the Ler mutant (emb3126-3) was later discontinued because of variable seed size in that accession.
Table I. Mutant alleles chosen for initial crosses with spectinomycin-tolerant accessions.
| Allele Symbol | Ribosomal Protein | Insertion Linea | Background Accession | Embryo Phenotype | Embryo Sizeb |
|---|---|---|---|---|---|
| μm | |||||
| emb3126-1 | L1 | RIKEN | “Nossen” | Preglobular | 25 |
| emb3126-3 | L1 | JICc | Ler | Small globular | 60 |
| emb3137-1 | S13 | RIKEN | “Nossen” | Preglobular | 25 |
| emb3137-2 | S13 | Salk | Columbia | Large globular | 90 |
Additional details are presented in Supplemental Table S3.
Values are rounded here to highlight major differences and to simplify comparisons with values reported elsewhere.
JIC, John Innes Centre.
To screen for dominant suppressors of early embryo arrest in RIKEN mutants (emb3126-1 and emb3137-1), we crossed Tsu-0 plants with emb/EMB heterozygotes. A dominant Tsu-0 suppressor unlinked to the EMB locus should enable 75% of the mutant seeds in siliques of selfed F1 heterozygotes to reach a later stage of development. In addition, three classes of F2 heterozygotes are expected in the next generation: those with a late seed phenotype, those with an early seed phenotype, and those with a mixture of both (Supplemental Fig. S3). Plants with more advanced embryos should be included among the late class if additional modifiers are present. Using this approach, we identified a single dominant suppressor in the Tsu-0 accession that significantly increases the size of mutant seeds and supports embryo development to a globular stage. As expected, 75% of the mutant seeds in F1 siliques reach a later (globular) stage of development (Table II, rows 1 and 2). Increased seed size is consistent with continued endosperm development. The remaining 25% of mutant seeds resemble those found in the parental RIKEN mutant. Three classes of F2 plants were found in the expected 1:2:1 ratio: TT plants with all rescued (globular and beyond) mutant seeds; SS plants with all parental (preglobular) mutant seeds; and ST plants with a 3:1 ratio of rescued to parental mutant seeds (Table III). Because emb3126-1 and emb3137-1 are both rescued to some extent by the dominant suppressor, the effect is not limited to a specific ribosomal protein. Partial rescue was also observed, although not examined in detail, when emb3126-1 and emb3137-1 were crossed with the tolerant Jl-3 and Be-1 accessions. The ability to rescue mutant seeds defective in chloroplast translation is therefore not unique to the Tsu-0 accession. As expected, rescue was less striking when emb3126-1 and emb3137-1 were crossed with two sensitive accessions (Oy-0 and Nie1-2); mutant seeds in F1 siliques failed to progress beyond an early globular stage and were reduced in size when compared with those from crosses involving the tolerant Tsu-0 accession (Table II, rows 3–6). Surprisingly, some F2 mutant seeds reached a much later (transition or cotyledon) stage of development when Tsu-0 was crossed with emb3126-1 but not with emb3137-1. These results are explained later, in the context of mapping a second locus known as the enhancer of the suppressor.
Table II. Partial embryo rescue in F1 siliques from crosses between natural accessions and embryo-defective mutants.
| Mutant Allelea | Wild-Type Accession | Siliques Screened | Seeds Screened | Percentage Mutant Seeds | Percentage Mutant Seeds Exhibiting Embryo Rescue | Phenotype of Rescued Embryob | Average Size of Rescued Embryo |
|---|---|---|---|---|---|---|---|
| μm | |||||||
| emb3126-1 | Tsu-0 | 40 | 1,842 | 24.1 | 71.4 | Most large globularc | 84 |
| emb3137-1 | Tsu-0 | 40 | 1,939 | 24.3 | 75.4 | Large globular | 78 |
| emb3126-1 | Oy-0 | 11 | 474 | 24.5 | 72.4 | Small globular | 55 |
| emb3137-1 | Oy-0 | 20 | 965 | 26.5 | 75.6 | Small globular | 55 |
| emb3126-1 | Nie1-2 | 11 | 550 | 24.2 | Not determinedd | Tiny globular | 49 |
| emb3137-1 | Nie1-2 | 10 | 491 | 27.1 | Not determinedd | Tiny globular | 50 |
Embryo arrest in parental (“Nossen”) lines occurs at the preglobular stage.
Embryo rescue was more pronounced in crosses with a spectinomycin-tolerant accession (Tsu-0) than in crosses with spectinomycin-sensitive accessions (Oy-0 and Nie1-2).
Some embryos reached a later stage.
Rescued mutant seeds did not differ sufficiently in size from parental mutant seeds.
Table III. Classes of F2 plants identified from Tsu-0 crosses with mutants in a sensitive (“Nossen”) background.
| Parental Mutant | F2 Class Symbol | Description of F2 Plant Phenotype | Total Plants Identified | Total Seeds Screened | Percentage Mutant Seeds | Percentage Embryo Visible |
|---|---|---|---|---|---|---|
| emb3126-1 | SS | No embryo rescue | 21 | 3,199 | 27.0 | 0.8 |
| ST | Partial rescue segregating | 49 | 6,103 | 26.3 | 76.7 | |
| TT | Partial rescue consistent | 31 | 7,862 | 25.3 | 99.4 | |
| Wild type | Wild-type plants | 45 | 1,549 | 0.2 | – | |
| emb3137-1 | SS | No embryo rescue | 9 | 1,491 | 24.5 | 0.5 |
| ST | Partial rescue segregating | 30 | 5,259 | 24.7 | 72.8 | |
| TT | Partial rescue consistent | 19 | 3,144 | 26.6 | 99.4 | |
| Wild type | Wild-type plants | 37 | 3,993 | 0.9 | – |
The Dominant Tsu-0 Suppressor of Embryo Arrest Maps to the ACC2 Region of Chromosome 1
Because a variety of changes in ACC2 itself might lead to the phenotypes observed, we pursued a candidate gene approach to determine whether ACC2 corresponds to the suppressor. Three classes of F2 plants from Tsu-0 × emb3126-1 and Tsu-0 × emb3137-1 populations were analyzed (Table III): SS plants with two sensitive (“Nossen”) alleles of the suppressor; TT plants with two tolerant (Tsu-0) alleles; and ST plants with one copy of each. PCR primers were designed to amplify DNA adjacent to ACC2 in one accession but not the other (Fig. 3). PCR genotyping of 80 F2 plants (49 TT, 29 SS, and 2 wild type) revealed perfect linkage between ACC2 and the suppressor. This corresponds to a genetic distance of less than 1 centimorgan (cM). To provide further evidence that ACC2 is the suppressor, we analyzed seedling growth responses on spectinomycin of two insertion mutants disrupted in ACC2 (Salk lines, Columbia accession). Both of these mutants exhibited hypersensitivity to spectinomycin. None of the wild-type accessions examined had a more severe phenotype. We then crossed acc2 knockout homozygotes, which exhibit no visible phenotype on soil, with a Salk mutant (emb3137-2) in the Columbia background, which exhibits embryo arrest at the late globular stage, to determine whether some mutant embryos in F1 siliques arrested at an earlier stage of development. As expected, 25% of the mutant seeds in these F1 siliques arrested at the preglobular stage instead of the globular stage (Supplemental Table S4). Collectively, these results support the conclusion that the Tsu-0 suppressor represents an allele of ACC2.
Figure 3.
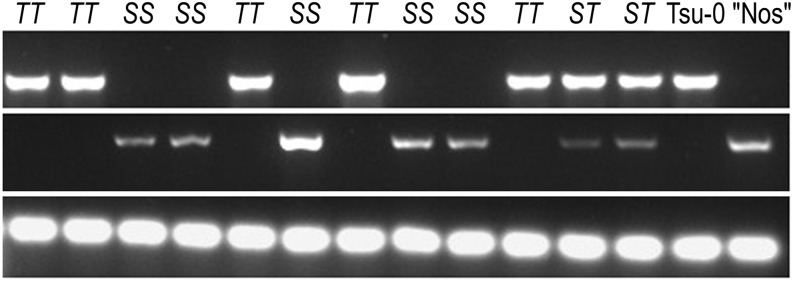
PCR genotyping of a segregating population of F2 plants from a Tsu-0 cross with emb3126-1. Primers were designed to amplify sequences adjacent to the ACC2 coding region in either Tsu-0 (top row) or “Nossen” (middle row). The bottom row shows the genomic region encoding 18S ribosomal RNA. SS, Plants homozygous for the “Nossen” allele of the suppressor; ST, heterozygotes; TT, plants homozygous for the Tsu-0 suppressor.
The Tsu-0 Suppressor Improves Seedling Growth in the Absence of Chloroplast Translation
To determine whether the suppressor impacts both embryo development in the absence of chloroplast translation and seedling response to spectinomycin, we PCR genotyped sensitive and tolerant F2 seedlings from Tsu-0 × “Nossen” populations using the ACC2-linked markers noted above. The results (Table IV) indicated that seedlings with a sensitive phenotype similar to the “Nossen” parent were typically homozygous for the “Nossen” allele of the suppressor and that both highly tolerant and somewhat tolerant seedlings were either heterozygous or homozygous Tsu-0 for the suppressor, consistent with a dominant pattern of inheritance. To determine whether the suppressor found in the Tsu-0 accession is also present in other tolerant accessions, we genotyped F2 seedlings on spectinomycin derived from a “Nossen” cross with the tolerant Be-1 accession (Table IV). The same dominant suppressor appears to mediate spectinomycin tolerance. We conclude that the suppressor likely functions across multiple accessions to improve growth in the absence of chloroplast translation.
Table IV. Suppressor genotypes of F2 seedlings from “Nossen” crosses with tolerant accessions.
| Tolerant Accession | F2 Seedling on Spectinomycin | ACC2 Genotypea | Seedlings | Seedlings Genotyped |
|---|---|---|---|---|
| % | ||||
| Tsu-0 | Tolerant | T, H | 100 | 9 |
| Intermediate | T, H | 100 | 13 | |
| Sensitive | N | 94b | 17 | |
| Be-1 | Tolerant | T, H | 100 | 7 |
| Intermediate | T, H | 95b | 19 | |
| Sensitive | N | 100 | 10 |
H, Heterozygous; N, homozygous “Nossen” allele; T, homozygous tolerant allele. bRare exceptions had marginal seedling phenotypes that were likely misclassified.
ACC2 Expression Levels in Seedlings Are Not Correlated with Spectinomycin Tolerance
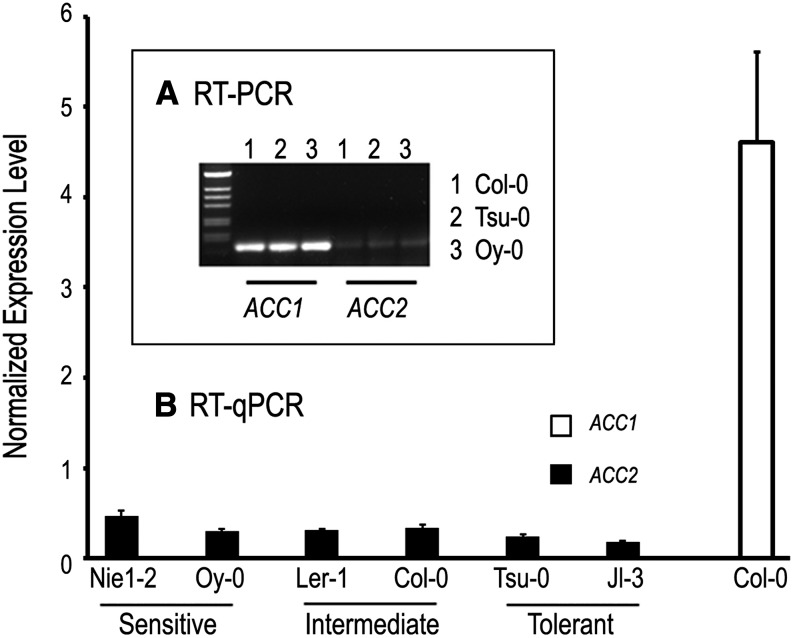
Arabidopsis microarray data indicate that ACC2 is expressed at low levels throughout growth and development. Thus, one mechanism for improving spectinomycin tolerance might be to increase the amount of ACC2 transcript produced. This model was not supported by repeated reverse transcription (RT)-PCR and RT-quantitative PCR (qPCR) experiments. ACC2 transcripts in wild-type seedlings were consistently less abundant than ACC1, and tolerant accessions did not produce more ACC2 transcript than sensitive or intermediate accessions (Fig. 4). This does not eliminate the possibility that some tolerant accessions overexpress ACC2 at the seedling stage. But increased ACC2 transcription does not underlie the suppressor effects analyzed here. We then searched for variations in ACC2 protein sequences that might explain the differences observed between tolerant and sensitive accessions and between late and early stages of embryo arrest in the absence of chloroplast translation. This ultimately led to the discovery that some of the most sensitive alleles of the suppressor found in natural accessions are associated with a complete loss of ACC2 protein function.
Figure 4.
Homomeric ACCase transcript levels in seedlings from Arabidopsis accessions that differ in spectinomycin tolerance. Note that ACC2 transcripts are less abundant than ACC1 and that their level does not increase with tolerance.
ACC2 Nonsense Mutations Are Present in Several Sensitive Accessions
Initially, we assumed that ACC2 was functional even in the most sensitive accessions and that the Tsu-0 suppressor allele of ACC2 improved its enzymatic activity, stability, or transport into chloroplasts. After sequencing the ACC2 genomic region from the sensitive “Nossen” accession used in crosses, we realized that this assumption was incorrect. The “Nossen” allele of ACC2 is interrupted by a nonsense mutation that removes two essential transcarboxylase domains located in the C-terminal half of the protein. Additional examples of ACC2 nonsense mutations (Supplemental Table S5) were then found among sequenced accessions at the Salk 1001 Genomes Web site (http://signal.salk.edu/atg1001). Two accessions with deletions or rearrangements that eliminate essential domains were also uncovered. Similar mutations are not found in the adjacent ACC1 locus, which is comparable in length but is essential for growth and development. Seedlings from accessions with major ACC2 disruptions are hypersensitive to spectinomycin (Supplemental Table S5), consistent with the knockout mutant phenotype. These results demonstrate that loss of ACC2 function is associated with heightened sensitivity to a loss of chloroplast translation in Arabidopsis and that functional ACC2 protein is not always required in natural environments, where heteromeric ACCase encoded in part by the chloroplast genome can function instead.
ACC2 Sequences Are More Variable Than ACC1 Sequences
Protein sequences from 855 accessions with sequenced genomes were then searched for evidence of reduced ACC2 function. First, we examined amino acid residues altered in lethal acc1 missense mutants from Arabidopsis and elsewhere. One accession with a substitution (E1689G) at the same location as the pas3-1 mutant was identified. However, seedling responses of this accession on spectinomycin were inconclusive. We then analyzed 416 amino acid residues (out of 2,355 total) perfectly conserved in a multikingdom alignment of 20 different ACC1/ACC2 sequences (Supplemental Fig. S4). We reasoned that substitutions at these locations were most likely to reduce protein function. More variation was found in ACC2 (8.2% of the conserved residues differed in at least one accession) than in ACC1 (2.2% differed; z test P < 0.001). Similar results were obtained when six members of the Brassicaceae with sequenced genomes were compared: 17 residues differed in ACC2 compared with just two for ACC1. When the frequencies of synonymous (Ks) versus nonsynonymous (Ka) substitutions were evaluated, the Ka/Ks ratios obtained (ACC1, 0.08; ACC2, 0.20) suggested a relaxation of purifying selection for ACC2. These analyses support the conclusion that ACC2 exhibits more variation in sequence and function than ACC1. We then examined the distribution of ACC1 and ACC2 in the Brassicaceae. Tandem gene duplications are present in the sequenced genomes of Arabidopsis, Arabidopsis lyrata, Capsella rubella, and Eutrema parvulum. Unlinked copies of ACC1 and ACC2 are found in Brassica rapa. A nonsense mutation is located in the third exon of ACC2 in Leavenworthia alabamica. No evidence of ACC2 was found in Aethionema arabicum or Boechera stricta. Overall, these results confirm that ACC2 structure, function, and retention differ widely throughout the Brassicaceae.
A Semidominant Enhancer of ACC2 Function Is Present in the Tsu-0 Accession
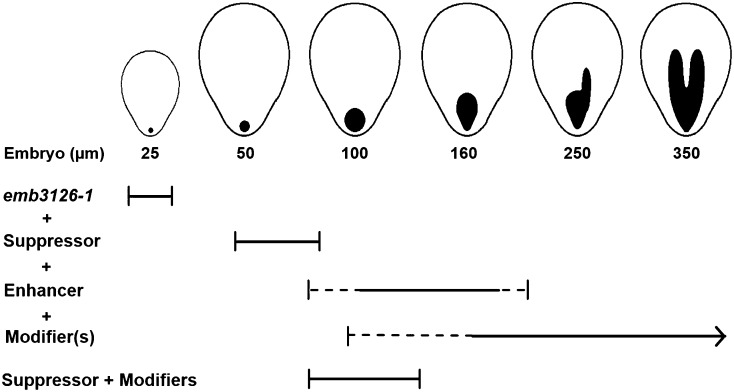
While screening F2 plants from crosses between Tsu-0 and emb3126-1 heterozygotes, we noticed that plants homozygous for the Tsu-0 suppressor differed in the extent of embryo rescue beyond the globular stage. One group (early TT) produced mutant embryos that consistently arrested at the globular stage and rarely exceeded 100 μm in diameter. A second group (late TT) produced mutant embryos that were typically 100 μm or larger and often progressed to a later (elongated or cotyledon) stage of development. A third group (intermediate TT) produced a mixture of both types of mutant embryos. Information on these phenotype classes is summarized in Table V. The number of progeny plants assigned to each class (26:46:26) is consistent with the 1:2:1 ratio expected for a second locus impacting the extent of embryo development. We named this locus the enhancer of the suppressor because its effect required the presence of the Tsu-0 suppressor and enhanced the ability of that suppressor to promote continued embryo development. Progeny testing of selected TT plants in the F3 generation confirmed that late plants were homozygous for the Tsu-0 allele of the enhancer, early plants were homozygous for the “Nossen” allele, and intermediate plants were heterozygous (Table VI). Furthermore, when F3 progeny of late TT plants were examined, it appeared that at least two additional modifiers increased the extent of embryo rescue. Once again, three phenotypic categories (late advanced, late moderate, and late reduced) were recognized (Supplemental Table S6). Plants in the first category produced mutant embryos that averaged 255 μm in length, a dramatic increase beyond the preglobular (25 μm) embryo observed in the parental mutant. Embryo rescue was even more extensive in F5 mutant seeds found in siliques of F4 plants derived from the most advanced F3 parental plants. All of these mutant embryos were elongated and more than 70% reached a cotyledon stage of development, frequently surpassing 300 μm in length. Cumulative effects of the Tsu-0 suppressor, enhancer, and modifier alleles on the development of emb3126-1 mutant embryos are presented in Figure 5. Examples of arrested embryo phenotypes are shown in Figure 6. Collectively, these results demonstrate that multiple genes from the spectinomycin-tolerant Tsu-0 accession impact the extent of embryo development in the absence of chloroplast translation.
Table V. Enhancer phenotype classes of TT plants from a Tsu-0 cross with emb3126-1.
| Plants Analyzeda |
Mutant Embryos Analyzed |
Embryo Lengths |
Embryo Phenotypes |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Enhancer Class | No. Screened | No. Measured | Average Length | Less Than 100 μm | Greater Than 200 μm | Globular | Triangular | Linear | Cotyledon |
| µm | % | ||||||||
| Late | 26 | 1,220 | 154 | 3.6 | 11.3 | 9.5 | 47.8 | 33.9 | 8.8 |
| Intermediate | 46 | 1,928 | 92 | 62.7 | 3.5 | 74.6 | 14.9 | 8.7 | 1.8 |
| Early | 26 | 965 | 66 | 94.8 | 0.1 | 99.4 | 0.5 | 0.1 | 0.0 |
Limited to F2 plants and F3 plants derived from F2 plants assigned to the intermediate enhancer class.
Table VI. Progeny testing of TT plants from different enhancer classes from a Tsu-0 cross with emb3126-1.
| Parental F2 Plants Enhancer Class |
EMB Genotypes of F3 Plants |
Seeds in Siliques of F3 Heterozygotes |
Enhancer Classes of F3 Plants |
||||
|---|---|---|---|---|---|---|---|
| Heterozygote | Wild Type | Total Screened | Percentage Mutant | Late | Intermediate | Early | |
| Late | 24 | 11 | 6,359 | 26.2 | 24 | 0 | 0 |
| Intermediate | 19 | 13 | 2,716 | 24.7 | 6 | 8 | 5 |
| Early | 35 | 12 | 2,004 | 23.7 | 0 | 0 | 35 |
Figure 5.
Combined effects of the Tsu-0 suppressor, enhancer, and modifier on seed and embryo rescue in emb3126-1 (“Nossen” background). Ellipses represent mutant seeds, filled images depict mutant embryos, and bars define the stage of arrest.
Figure 6.
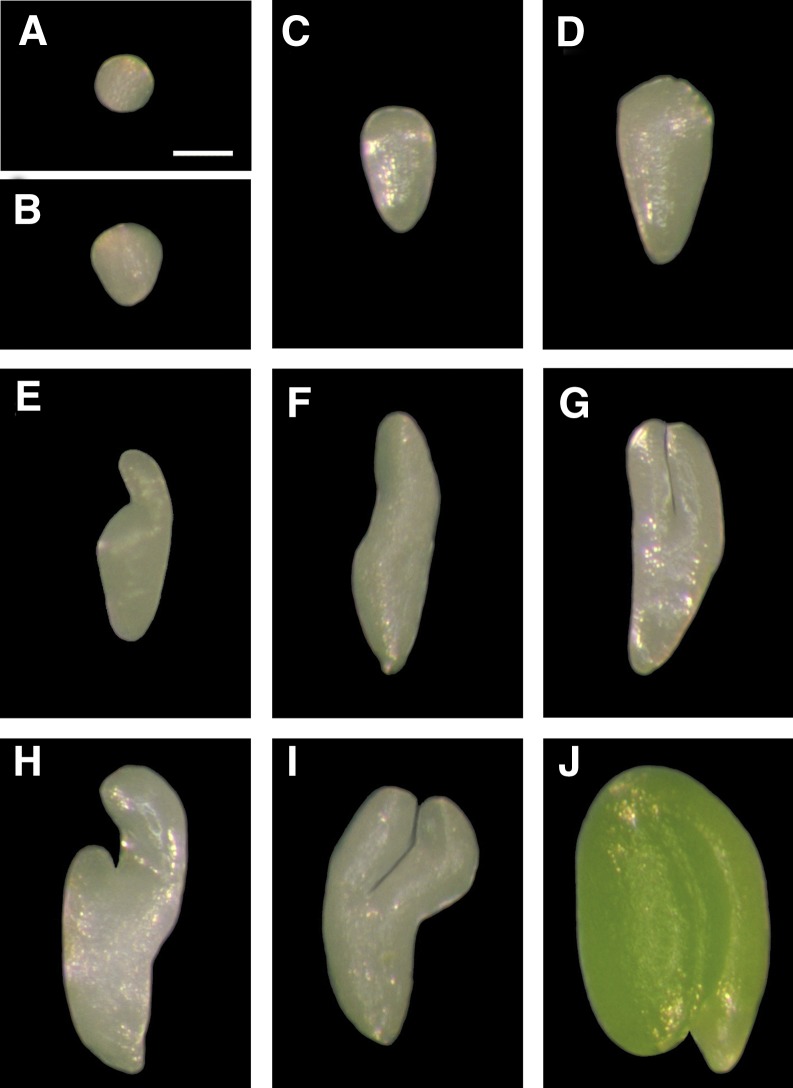
Examples of embryos in siliques of plants homozygous for the Tsu-0 suppressor. A, Late globular embryo. B, Triangular embryo. C and D, Elongated linear embryos with an expanded basal region and a small apical dome. E to I, Cotyledon stage embryos with one or two cotyledons. J, Sibling wild-type embryo. Bar = 100 μm.
The Tsu-0 Enhancer Maps Near the Top of Chromosome 5
One feature of Tsu-0 crosses with emb3137-1 was at first difficult to explain: F2 plants homozygous (TT) for the Tsu-0 allele of the enhancer lacked the late-embryo (elongated and cotyledon) phenotypes characteristic of emb3126-1 crosses (Supplemental Table S7). It seemed unlikely that this reflected differences in protein function because the Salk emb3137-2 allele had the same globular embryo phenotype found in other Columbia mutants defective in chloroplast translation. Instead, we thought the difference might result from linkage between the enhancer locus and EMB3137. In that case, most homozygous mutant embryos should also be homozygous for the “Nossen” allele of the enhancer. To explore this possibility, we assembled information on the map locations of known genes associated with chloroplast protein import, which represented logical candidates for an enhancer that improved the ability of ACC2 to compensate for a loss of heteromeric ACCase (Supplemental Fig. S5). Several candidate genes linked to EMB3137 were identified. When late TT and early TT plants from Tsu-0 crosses with emb3126-1 were genotyped (Supplemental Fig. S6) using PCR primers that revealed accession-specific DNA sequence polymorphisms, linkage between the enhancer and EMB3137 was confirmed. We then progeny tested plants with questionable phenotype assignments, adjusted their classifications as needed, and genotyped 110 plants of interest using PCR primers for EMB3137, OUTER ENVELOPE PROTEIN80 (OEP80), located 10 cM below EMB3137, and TRANSLOCON OUTER CHLOROPLAST34 (TOC34), located 10 cM above EMB3137. The results demonstrated that the enhancer is tightly linked to EMB3137, in a region devoid of genes that represent logical candidates based on known protein function.
To confirm that linkage between EMB3137 and the enhancer was responsible for the absence of late-embryo phenotypes in crosses between Tsu-0 and emb3137-1, we analyzed F2 seeds from crosses between Tsu-0 and two additional mutants, one disrupted in a closely linked gene (EMB3136) and the other (EMB1473) unlinked. Both genes encode chloroplast ribosomal proteins. As expected, late embryos were found with emb1473, which is unlinked to the enhancer, but not with emb3136, which is linked (Supplemental Tables S8 and S9).
PCR genotyping of sensitive and tolerant F2 seedlings from Tsu-0 × “Nossen” populations using linked markers revealed that the Tsu-0 enhancer is semidominant. The most tolerant seedlings were homozygous Tsu-0 for the enhancer, somewhat less tolerant seedlings were typically heterozygous, and the least tolerant (but not sensitive) seedlings tended to be homozygous “Nossen”. Sensitive seedlings, which are homozygous “Nossen” for the suppressor, had variable enhancer genotypes, as predicted. This pattern of inheritance was more difficult to document at the embryo stage because mutant embryos were not genotyped. When combined with corresponding data for ACC2, these results demonstrate that both the suppressor and enhancer function at two different stages of development (embryo and seedling) in the Tsu-0 accession, validating our decision to use seedling responses to spectinomycin as a rapid assay for differences between accessions that might impact seed development. Analysis of arrested embryos, however, was a more sensitive method for identifying the genes involved, as both genotype and phenotype information could be confirmed in subsequent generations.
Knockout Mutants of a Chloroplast Protein Import Gene Exhibit Hypersensitivity to Spectinomycin
To explore the role of chloroplast import proteins in modulating the ability of ACC2 to compensate for a loss of chloroplast translation, two knockout mutants disrupted in the TRANSLOCON INNER CHLOROPLAST20-IV (TIC20-IV) gene required for the import of housekeeping proteins through the inner chloroplast membrane were tested on spectinomycin. As shown in Figure 7, mutant seedlings were much more sensitive to spectinomycin than seedlings from the parental Columbia accession, resembling the response of acc2 knockouts. Because tic20-IV null mutants lack an obvious phenotype when grown on soil, the paralogous TIC20-1 gene, which targets photosynthetic proteins, must be less effective at importing ACC2 in the absence of chloroplast translation than other housekeeping proteins essential for growth and development in the presence of chloroplast translation. A different result was obtained when a toc34 knockout was tested on spectinomycin. Mutant seedlings were not more sensitive than the parental Columbia accession. In this case, the TOC33 paralog appears to compensate for the loss of TOC34 in both the presence and absence of chloroplast translation.
Figure 7.
Responses of selected mutant and wild-type seedlings on spectinomycin. A, Parental Columbia accession. B, toc34-1 (ppi3-2). C, tic20-IV-1 (SAIL_97_F10). D, tic20-IV-2 (Koncz 11324). E, acc2-1 (Salk_148966c). F, Most sensitive accession (Sav-0) identified in the initial screen. Bar = 1 mm.
DISCUSSION
Natural variation among wild-type accessions of Arabidopsis has been widely used to address fundamental questions in plant biology, from physiology and development to molecular ecology and plant-pathogen interactions (Alonso-Blanco et al., 2009; Weigel, 2012). We took a different approach here by analyzing natural variation in the developmental consequences of disrupting a basic cellular function: chloroplast translation. Our interest was driven by reports of differential responses of Arabidopsis, Brassica spp., and maize (Zea mays) to a loss of chloroplast translation (Zubko and Day, 1998; Asakura and Barkan, 2006) and by intriguing correlations noted between the extent of embryo development in Arabidopsis mutants defective in chloroplast translation (Bryant et al., 2011; Muralla et al., 2011) and the genetic background (accession) in which those mutations were generated. We demonstrate here that heterotrophic growth in the absence of chloroplast translation in Arabidopsis is mediated by ACC2, a duplicated nuclear gene that targets homomeric ACCase to plastids, and enhanced by genetic modifiers present in the most tolerant accessions. Further analysis of this experimental system could lead to a better understanding of essential chloroplast gene functions in flowering plants and to improved delivery of homomeric ACCase to plastids during seed development.
Comparative Genomics and the Loss of Essential Chloroplast Genes
Two approaches have been used to identify essential chloroplast genes in plants: targeted gene disruption in tobacco and comparison of sequenced chloroplast genomes, especially from parasitic, nonphotosynthetic plants. The first approach identified four candidate genes that seemed to explain the importance of chloroplast translation: accD (Kode et al., 2005); ycf1 (Drescher et al., 2000), which functions in protein import (Kikuchi et al., 2013); ycf2 (Drescher et al., 2000), whose function remains unknown; and clpP1 (Kuroda and Maliga, 2003), which encodes part of the ClpP protease that regulates protein degradation (Kim et al., 2013). All four genes are retained in the plastids of most flowering plants, consistent with their proposed essential functions. Nevertheless, repeated examples of selective gene loss or transfer to the nucleus have been documented in the literature.
In Trachelium caeruleum and Trifolium subterraneum, a truncated accD variant is found in the nucleus, where it likely targets functional protein back to the chloroplast (Haberle et al., 2008; Magee et al., 2010; Rousseau-Gueutin et al., 2013). In other species, accD appears to be nonfunctional or missing altogether (Chumley et al., 2006; Guisinger et al., 2011; Straub et al., 2011; Li et al., 2013). This implies that a redundant nuclear gene remains to be identified because fatty acid biosynthesis is essential. In the Poaceae, accD is replaced by a nuclear gene that encodes a chloroplast-localized homomeric ACCase (Goremykin et al., 2005; Jansen et al., 2007; Guisinger et al., 2010). Several common herbicides target this novel enzyme (Kaundun, 2014). The chloroplast ycf1 gene is also absent from the Poaceae and is missing or disrupted in selected members of other families (Chumley et al., 2006; Magee et al., 2010; Guisinger et al., 2011; Straub et al., 2011; Li et al., 2013). The ycf2 gene is lost in many of these lineages as well. Both of these genes are large, and when curated genomic sequences are available, related genes are not found in the nucleus. Examples of clpP1 loss or disruption have been noted in several angiosperm families (Haberle et al., 2008; Guisinger et al., 2011; Straub et al., 2011) and in Japanese cedar (Cryptomeria japonica), a gymnosperm (Hirao et al., 2008).
These examples seem to indicate that ycf1, ycf2, and clpP1 are dispensable. However, their retention in the reduced plastid genomes of parasitic plants (dePamphilis and Palmer, 1990; Funk et al., 2007; McNeal et al., 2007; Delannoy et al., 2011; Logacheva et al., 2011) argues that they are indeed essential, with functions not limited to photosynthesis. This appears to conflict with the model (Bryant et al., 2011) that accD alone is required for embryo development in Arabidopsis. How can ACC2 uptake by plastids defective in translation extend growth and development when Ycf1, Ycf2, and ClpP1 are not produced? One possibility is that these proteins are needed later in development than AccD. This might explain the failure of the Tsu-0 enhancer and modifier alleles described here to bring about complete embryo rescue. Alternatively, the consequences of clpP1 single gene disruption in transgenic tobacco, where other chloroplast proteins are still being made, might be more severe than when all chloroplast-encoded proteins are missing. Kikuchi et al. (2013) recently demonstrated that ycf1 is part of an essential 1-MD complex that facilitates chloroplast protein import. Understanding how mutant embryos defective in chloroplast translation continue development in the absence of ycf1 function, therefore, requires a consideration of how chloroplast proteins are imported from the cytosol.
Essential Components of Chloroplast Protein Import
Multiple components of the chloroplast protein import system have been identified in Arabidopsis (Jarvis, 2008; Kessler and Schnell, 2009; Shi and Theg, 2013): (1) cytosolic chaperones that participate in protein folding and targeting (Flores-Pérez and Jarvis, 2013); (2) Toc receptor GTPases in the outer chloroplast membrane that interact with these chaperones and the transit peptide of incoming proteins (Kubis et al., 2004; Inoue et al., 2010); (3) Toc75 outer membrane channel proteins (Baldwin et al., 2005); (4) inner membrane Tic20 channel proteins and associated Tic complex members (Hirabayashi et al., 2011; Kasmati et al., 2011; Kikuchi et al., 2013); (5) stromal chaperones, motor proteins, and heat-shock proteins such as Hsp93 (Kovacheva et al., 2007; Chu and Li, 2012), Hsp90C (Inoue et al., 2013), and chloroplast Hsc70 (Su and Li, 2010); and (6) a stromal processing peptidase (SPP) that cleaves the transit peptide (Trösch and Jarvis, 2011). Features of the N-terminal transit peptide that target proteins to plastids have also been analyzed (Chotewutmontri et al., 2012; Li and Teng, 2013).
Housekeeping and photosynthetic proteins are reportedly directed to different import complexes (Inoue et al., 2010; Hirabayashi et al., 2011). Complexes with Toc132/120, Toc34, and Tic20-IV are thought to associate with housekeeping proteins, whereas those with Toc159, Toc33, and Tic20-1 appear to import photosynthetic proteins. These differences are relevant here because disruption of complexes that import photosynthetic proteins should have less impact on embryo development in the absence of chloroplast translation than those involved with housekeeping proteins. Some overlap must still exist between these complexes, as both toc34 and tic20-IV insertion mutants produce viable plants in soil (Constan et al., 2004; Kasmati et al., 2011). The increased sensitivity of tic20-IV mutant seedlings to spectinomycin described here is consistent with a nonredundant, essential role for TIC20-IV in plastid import of ACC2 protein.
Many genes required for chloroplast protein import in Arabidopsis exhibit an embryo-defective mutant phenotype: TOC75-III (Baldwin et al., 2005), OEP80/EMB213 (Patel et al., 2008; Meinke et al., 2009), Hsp90C/EMB1956 (Tzafrir et al., 2004; Inoue et al., 2013), TIC100/EMB1211 (Tzafrir et al., 2004; Liang et al., 2010), TIC56 (Kikuchi et al., 2013), TIC32 (Hörmann et al., 2004), and SPP (Trösch and Jarvis, 2011). Several double knockouts of genes with overlapping functions also exhibit embryo or gametophyte lethality: toc159 toc132 (Kubis et al., 2004), toc33 toc34 (Constan et al., 2004), tic20-I tic20-IV (Hirabayashi et al., 2011), hsp93-III hsp93-V (Kovacheva et al., 2007), and cphsc70-1 cphsc70-2 (Su and Li, 2008). Based on the work presented here, the most severe embryo phenotypes should be found among knockouts of genes most essential for ACC2 import. The early embryo arrest observed in toc75-III single mutants (Baldwin et al., 2005) and in the double mutants toc33 toc34 (Constan et al., 2004), hsp93-V hsp93-III (Kovacheva et al., 2007), and possibly tic20-I tic20-IV (Hirabayashi et al., 2011) indicate that these proteins are essential for chloroplast protein import in general and ACC2 import in particular. The toc159 toc120 toc132 triple mutant would likely exhibit a similar phenotype if construction were not limited by lethality. The late phenotypes of tic100 and tic56 mutants defective in the 1-MD complex are consistent with a limited role for ycf1, a component of the same complex, in the uptake of housekeeping proteins late in development.
Potential Functions of the Tsu-0 Enhancer and Modifier Alleles
We propose two contrasting models for how the Tsu-0 enhancer improves growth in the absence of chloroplast translation. Both models are consistent with the requirement that a tolerant (functional) allele of the suppressor (ACC2) be present. According to the first model, the Tsu-0 enhancer facilitates the import of ACC2 protein into plastids. Candidate genes include some of the protein import components detailed above. Enhanced uptake of inactive ACC2 protein should have no effect, consistent with observations. One problem with this model is the absence of promising candidate genes in the enhancer map location. With the second model, the Tsu-0 enhancer increases the amount or stability of functional ACC2 protein made, for example by improving translational efficiency or decreasing protein degradation. Other modifiers in tolerant accessions could have similar, complementary functions. Another possibility for modifier action involves partial compensation for the absence of Ycf1, Ycf2, or ClpP1. This explanation is less compatible with enhancer function because it is more independent of suppressor activity. Two different types of gene disruption are consistent with our genetic analysis. In one case, the Tsu-0 enhancer acts as a gain-of-function mutant allele when combined with the “Nossen” allele. Alternatively, the Tsu-0 enhancer may be a loss-of-function mutant allele of a locus that exhibits haploinsufficiency. These alternative mechanisms predict different types of enhancer functions, with the Tsu-0 allele promoting ACC2 function in the first case, as detailed above, and the “Nossen” allele decreasing ACC2 function in the second. One practical application of identifying the enhancer and modifiers described here concerns improved targeting of overexpressed homomeric ACCase to plastids in transgenic plants. The modest increase in fatty acid biosynthesis observed with this approach in the past (Roesler et al., 1997; Klaus et al., 2004) may reflect inefficient stabilization or uptake of functional ACCase into plastids.
Chloroplast Translation and Embryo Development in Maize and Arabidopsis
The ability of some angiosperms to survive a localized loss of chloroplast translation was first revealed through studies with the maize iojap and barley (Hordeum vulgare) albostrians mutants deficient in chloroplast ribosomes (Walbot and Coe, 1979; Siemenroth et al., 1981). Subsequent work on chloroplast splicing mutants provided further evidence that grasses tolerate disruptions in chloroplast translation that cause embryo lethality in Arabidopsis (Asakura and Barkan, 2006). Extensive research on embryo-defective mutants of Arabidopsis was consistent with this conclusion (Bryant et al., 2011). At first, it appeared that differences in fatty acid biosynthesis alone could explain the phenotypes observed. However, at least five mutants with severe defects in maize embryogenesis are disrupted in chloroplast translation: lem1 (Ma and Dooner, 2004), prpl35-1 (Magnard et al., 2004), ppr8522 (Sosso et al., 2012), emb16/why1 (Zhang et al., 2013), and emb12 (Shen et al., 2013). Thus, a loss of chloroplast translation in maize is sometimes associated with embryo lethality. How can this observation be reconciled with the survival of maize iojap leaves devoid of chloroplast ribosomes? Interestingly, all of the embryo mutants noted above belong to the embryo-specific class in which development of the mutant endosperm tissue is normal. Embryo lethality is therefore not the result of a metabolic defect that limits growth. Furthermore, some of the phenotypes are sensitive to genetic background, with the same mutation resulting in embryo or seedling lethality, depending on the inbred line (Zhang et al., 2013).
This background effect in maize is different from the natural variation described here for Arabidopsis because it does not involve fatty acid biosynthesis. Instead, the model proposed for maize (Shen et al., 2013; Zhang et al., 2013) invokes the loss of a retrograde signal produced in functional plastids (Woodson et al., 2011; Terry and Smith, 2013) that activates the expression of nuclear genes, including some required for embryogenesis but not for endosperm or leaf development. The embryo-specific phenotype in maize may therefore reflect the loss of an essential chloroplast function that extends beyond photosynthesis. This raises the possibility that disruption of a retrograde signal during embryo development in Arabidopsis is also associated with a loss of chloroplast translation and that this defect contributes to the inability of the Tsu-0 enhancer and modifier alleles to bring about complete rescue of mutants defective in chloroplast translation.
Significance of the ACC1-ACC2 Tandem Gene Duplication in Arabidopsis
One question that remains to be addressed concerns the function of ACC2 in natural populations of Arabidopsis, where chloroplast translation is not blocked. Clearly, ACC2 function is not required in all environments or genetic backgrounds, because multiple accessions contain nonsense mutations or deletions that remove essential protein domains. Furthermore, most plant species lack ACC2 altogether. Whether ACC2 contributes to fatty acid biosynthesis in chloroplasts under selected conditions and what impact such contributions have on plant growth and development in natural environments remain to be evaluated. One possibility is that ACC2 might lack the feedback regulation system described for heteromeric ACCase in the Brassicaceae (Andre et al., 2012; Bates et al., 2014). In such a case, ACC2 might contribute to fatty acid biosynthesis when the heteromeric enzyme is down-regulated. Alternatively, ACC2 may contribute to one or more metabolic pathways in the cytosol, especially in genotypes or environments where such a function is advantageous and where plastid import of ACC2 protein is restricted.
A different pattern of genetic redundancy is encountered in mammals, where a duplicated ACCase (ACC2) performs an essential function in fatty acid oxidation in mitochondria (Abu-Elheiga et al., 2005). Yeast (Saccharomyces cerevisiae) also contains a duplicated ACCase that functions in mitochondria (Hoja et al., 2004). Notably, that duplication is not widespread among yeast genomes examined to date. Drosophila melanogaster and zebrafish also contain ACCase proteins with N-terminal extensions consistent with mitochondrial localization. Duplication and diversification of ACCase genes has therefore occurred multiple times during the evolution of eukaryotes. In Arabidopsis, many duplicated genes appear to lack a knockout phenotype. One common explanation is that appropriate growth conditions were not evaluated. The work presented here documents an intriguing and informative phenotype that appears only under specialized laboratory conditions. Why functional ACC2 protein has been retained in selected members of the Brassicaceae, and in some Arabidopsis accessions but not others, remains to be explained.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The embryo-defective mutants of Arabidopsis (Arabidopsis thaliana) used for crosses with natural accessions have been described elsewhere (Bryant et al., 2011; Muralla et al., 2011) and are included in the SeedGenes database of essential genes (www.seedgenes.org). Seeds for emb3126-1 (RATM-53-3245-1), emb3126-3 (GT-5-101962), emb3137-1 (RATM-15-0663-1), and emb3136 (RATM-51-2522-3) were obtained from Kazuo Shinozaki at the RIKEN Plant Science Center. Seeds for emb3137-2 (Salk-133412), acc2-1 (Salk-148966c), acc2-2 (Salk-110264), and wild-type accessions were obtained from the Arabidopsis Biological Resource Center. Stock numbers for each accession are listed in Supplemental Tables S1 and S5. Paul Jarvis provided seeds for tic20-IV-1 (SAIL_97_F10), tic20-IV-2 (Koncz 11324), and toc34 (ppi3-2; Salk-059206), all in the Columbia background. Seeds for the “Nossen” accession were obtained from wild-type plants segregating in populations of mutants (emb3126-1 and emb3137-1) grown in our laboratory. These plants were designated “Nossen” because they differ from the sequenced Nossen accession (Kuromori et al., 2004). Internal seed stocks were used for emb1473 (Syngenta 24154) in the Columbia background, and duplicates are available through the Arabidopsis Biological Resource Center. Mature seeds were germinated on agar plates (Meinke et al., 2009), and seedlings were transplanted to pots containing a mixture of soil, sand, and vermiculite. Plants were grown under fluorescent lights (16-h-light/8-h-dark cycle) in a growth room maintained at room temperature (23°C ± 1°C) and watered daily with a nutrient solution as described by Berg et al. (2005).
Seedling Responses on Spectinomycin
Antibiotics known to inhibit chloroplast translation were added to a basal germination medium containing Murashige and Skoog salts, 3% (w/v) Glc, and 0.8% (w/v) agar. Spectinomycin (50 mg L−1) was used for most experiments; lincomycin (200 mg L−1) was tested with several accessions to confirm the spectinomycin results. Seedling responses were characterized 5 weeks after plating. The following scale was used to classify F1 and F2 seedlings from crosses between accessions, with specified lengths corresponding to the maximal distance from leaf tip to leaf tip: A, cotyledons only; B, first pair of leaves only (less than 1.5 mm); C, multiple leaves (less than 1.5 mm); D, multiple leaves (1.5 to less than 2.5 mm); E, first pair of leaves only (more than 1.5 mm); F to J, multiple leaves: F, 2.5 to less than 4 mm; G, 4 to less than 6 mm; H, 6 to less than 9 mm; I, 9 to less than 12 mm; and J, 12 mm or more. For the initial screen of 52 accessions, a numerical score with fewer categories was used to facilitate the averaging of results: 1, A; 2, B; 3, C + D; 4, E; 5, F + G; and 6, H and above. Seedlings with evidence of greening due to limited root contact with the medium were excluded.
Crosses and Embryo Phenotyping
Crosses between wild-type accessions and plants heterozygous for an embryo-defective mutation were performed in both directions. When a heterozygous (emb/EMB) plant was the female parent, the harvested silique was expected to lack aborted seeds, unlike adjacent siliques produced from selfing. When the accession was the female parent, successful crosses were confirmed by segregation of mutant seeds in F1 plants. Crosses between two different accessions were confirmed by PCR genotyping. Methods used to phenotype mutant seeds and embryos are described at the tutorial section of the SeedGenes Web site. Heterozygous plants were identified by screening siliques for the presence of 25% aborted seeds. Seed and embryo measurements were performed with a stage micrometer using a dissecting microscope. The estimated accuracy of each measurement was 10 µm for embryos and deflated seeds and 20 µm for inflated seeds. Average lengths were calculated without sd; attention was focused instead on the frequency and distribution of size classes observed. Mutant embryos were removed from aborted seeds prior to desiccation using fine-tipped (Dumont no. 4) forceps. The smallest embryos measured were globular and 50 μm in diameter. Overall, embryos were less susceptible than seeds to a reduction in length resulting from seed desiccation.
Four categories of embryo phenotypes were recognized: globular (round), triangular (pointed basal region of the embryo visible), linear (further elongation of the basal region; sometimes accompanied by extension of the apical region without cotyledon formation), and cotyledon (presence of one or two cotyledons). Triangular embryos were typically more than 100 μm in diameter, and linear embryos were 150 μm or more in length. The extent of embryo development did not appear to be influenced by silique position along the stem. Most siliques analyzed contained aborted seeds that were slightly deflated and had begun to turn brown, which ensured that mutant embryos had reached a terminal stage of development. When siliques contained a mixture of aborted seeds at the globular and preglobular stages, the preglobular seeds were often collapsed and more highly desiccated than those with globular embryos. Even at maturity, globular embryos could often be detected as a small bump that distorted the shape of the seed.
PCR Genotyping of Plants
ACC2, TOC34, EMB3137, and OEP80 primers were designed by aligning sequences of relevant accessions from the 1001 Genomes Project (http://signal.salk.edu/atg1001). Additional sequencing and PCR were required for the “Nossen” accession examined here. A complete listing of all primers is presented in Supplemental Table S10. Primers were purchased from Integrated DNA Technologies. Accession-specific primers used for mapping the suppressor (ACC2) were designed to amplify a product from either the Tsu-0 or “Nossen” accession. Other primers used for mapping the enhancer locus were designed to amplify products from both accessions, followed by DNA sequencing of PCR products to distinguish between heterozygotes and homozygotes (Supplemental Fig. S5). Genomic DNA was isolated from seedlings and inflorescences using a modified cetyl trimethyl ammonium bromide protocol (Lukowitz et al., 2000). Salk insertion mutants (toc34-1, acc2-1, and acc2-2) were genotyped to confirm that the appropriate seed stock had been received. A standard transfer DNA left border primer (LB1a) was used in combination with gene-specific primers for TOC34 and ACC2. PCR was performed using the Qiagen PCR Master Mix and a Biometra Uno II thermocycler. PCR products were separated on 1% agarose gels containing GelRed Nucleic Acid Stain (Phenix). The AlphaImager HP system (Proteinsimple) was used to visualize the bands. For DNA sequencing, PCR products were purified with the Qiagen MinElute PCR Purification Kit and sequenced at the Oklahoma State University Recombinant DNA/Protein Resource Facility.
RT-qPCR and Subsequent Analysis
Total RNA was isolated from seedlings grown on a basal germination medium (described above) using an RNeasy plant mini kit (Qiagen) and reverse transcribed with a QuantiTect reverse transcription kit (Qiagen) that included elimination of genomic DNA for RT-qPCR. Reactions were performed using a SYBR Green PCR kit (Roche) with a Roche480 Light Cycler (Roche). PCR amplifications were completed with a melt curve analysis to confirm the specificity of amplification and the lack of primer dimers. PCR efficiencies were calculated based on serial dilution. The ACTIN2 and 18S ribosomal RNA genes were used as references. LightCycler 480 software version 1.5 was used for analysis. Three RNA samples, independently isolated from each accession, were used for RT-qPCR analysis. Reactions were performed four times for each sample. Primer sequences are shown in Supplemental Table S10.
Homomeric ACCase Sequence Analyses
To identify conserved amino acids important for ACCase function, 20 ACC1 and ACC2 protein sequences were analyzed from representative plant, animal, and fungal species: Arabidopsis (two), Brassica rapa (two), Medicago truncatula (one), wheat (Triticum aestivum; two), maize (Zea mays; two), Homo sapiens (two), Mus musculus (two), Danio rerio (two), Drosophila melanogaster (one), yeast (Saccharomyces cerevisiae; two), Schizosaccharomyces pombe (one), and Neurospora crassa (one). Details are presented in Supplemental Table S11. Sequences were aligned using ClustalW2 (Larkin et al., 2007). In several cases, small gaps were identified that spanned conserved residues. These were assumed to be annotation errors and were corrected based on genomic data. Homomeric ACCase sequences from 855 Arabidopsis accessions were obtained from the Salk Web site for the Arabidopsis 1001 Genomes project (http://signal.salk.edu/atg1001). For determination of Ka/Ks ratios, ACC1 and ACC2 coding sequences were obtained for six members of the Brassicaceae: Arabidopsis, Arabidopsis lyrata (Hu et al., 2011), B. rapa (Cheng et al., 2011), Capsella rubella (Slotte et al., 2013), Leavenworthia alabamica, and Sisymbrium irio, along with Theobroma cacao (Motamayor et al., 2013). Sequences were downloaded from the Phytozome (www.phytozome.net) and CoGe (www.genomevolution.org/CoGe/) Web sites (Lyons et al., 2008; Goodstein et al., 2012). Ka/Ks ratios were calculated using MEGA version 6 (Tamura et al., 2013).
Genomic DNA spanning the ACC2 locus from “Nossen” plants grown in our laboratory was sequenced at the Oklahoma State University Recombinant DNA/Protein Resource Facility. PCR primers were designed to avoid amplification of the adjacent ACC1 locus (Supplemental Table S10). Takara (Clontech) PrimeSTAR GXL DNA polymerase was used to amplify large fragments of ACC2. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced with primers within the ACC2 gene. A similar approach was used to confirm ACC2 nonsense mutations and deletions present in other accessions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Seedling responses of 52 Arabidopsis accessions on spectinomycin.
Supplemental Figure S2. Seedling responses of selected accessions on lincomycin.
Supplemental Figure S3. Expected F2 progeny from a selfed F1 plant heterozygous for an embryo-defective mutation and a second locus conferring sensitivity or tolerance to a loss of chloroplast translation.
Supplemental Figure S4. Conserved amino acid residues in eukaryotic ACCases and mutations analyzed in model genetic organisms.
Supplemental Figure S5. Chromosome locations of Arabidopsis genes encoding known components of the chloroplast protein import system and EMB genes encoding chloroplast ribosomal proteins.
Supplemental Figure S6. DNA sequence polymorphisms used to genotype plants for ACC2, TOC34, EMB3137, and OEP80 in Tsu-0 crosses with emb3126-1.
Supplemental Table S1. Natural accessions of Arabidopsis examined on spectinomycin.
Supplemental Table S2. Seedling responses on spectinomycin of parental accessions and F2 progeny from crosses between accessions.
Supplemental Table S3. Phenotypes and genetic backgrounds (accessions) of embryo-defective mutants of Arabidopsis disrupted in chloroplast translation.
Supplemental Table S4. Reduced embryo development in F1 siliques from acc2 (Columbia) crosses with emb3137-2 (Columbia).
Supplemental Table S5. ACC2 gene disruptions identified in natural accessions of Arabidopsis.
Supplemental Table S6. Modifier phenotype classes of late TT plants from a Tsu-0 cross with emb3126-1.
Supplemental Table S7. Differences in the extent of embryo rescue in TT plants from Tsu-0 crosses with emb3137-1 and emb3126-1.
Supplemental Table S8. Partial embryo rescue in F1 siliques from a Tsu-0 cross with emb1473 (Columbia)
Supplemental Table S9. Limited embryo rescue in F1 siliques from a Tsu-0 cross with emb3136 (“Nossen”).
Supplemental Table S10. PCR primer sequences used for RT-qPCR and plant genotyping.
Supplemental Table S11. Sequences used for ACC1/ACC2 alignments and determination of Ka/Ks ratios.
Supplementary Material
Acknowledgments
We thank Paul Jarvis (University of Oxford) for providing tic20-IV and toc34 seed stocks, Janette Steets (Oklahoma State University) and the Arabidopsis Biological Resource Center (Ohio State University) for information on Arabidopsis natural accessions, and Ian Small (University of Western Australia), Johnny Lloyd (Michigan State University), and Andrew Doust (Oklahoma State University) for helpful discussions.
Glossary
- ACCase
acetyl-coenzyme A carboxylase
- cM
centimorgan
- RT-qPCR
quantitative reverse transcription-PCR
- Ler
Landsberg erecta
Footnotes
This work was supported by the Developmental Systems Cluster, Biological Sciences Directorate, National Science Foundation.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abu-Elheiga L, Matzuk MM, Kordari P, Oh W, Shaikenov T, Gu Z, Wakil SJ (2005) Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proc Natl Acad Sci USA 102: 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amid A, Lytovchenko A, Fernie AR, Warren G, Thorlby GJ (2012) The sensitive to freezing3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies. J Exp Bot 63: 5289–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Haslam RP, Shanklin J (2012) Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc Natl Acad Sci USA 109: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P (2005) A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol 138: 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Johnson SR, Cao X, Li J, Nam JW, Jaworski JG, Ohlrogge JB, Browse J (2014) Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc Natl Acad Sci USA 111: 1204–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep 5: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P (2008) Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA 105: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Reddick LE, McWilliams DR, Campbell IM, Bruce BD (2012) Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell 24: 3040–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Li HM (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol 158: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK (2006) The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol 23: 2175–2190 [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Constan D, Patel R, Keegstra K, Jarvis P (2004) An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis CW, Palmer JD (1990) Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348: 337–339 [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Dudas B, Jenes B, Kiss GB, Maliga P (2012) Spectinomycin resistance mutations in the rrn16 gene are new plastid markers in Medicago sativa. Theor Appl Genet 125: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340 [DOI] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22: 1813–1822 [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK (2011) Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol 28: 583–600 [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66: 350–361 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kikuchi S, Oishi M, Nakai M (2011) In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant Cell Physiol 52: 469–478 [DOI] [PubMed] [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K (2008) Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol 8: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoja U, Marthol S, Hofmann J, Stegner S, Schulz R, Meier S, Greiner E, Schweizer E (2004) HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J Biol Chem 279: 21779–21786 [DOI] [PubMed] [Google Scholar]
- Hörmann F, Küchler M, Sveshnikov D, Oppermann U, Li Y, Soll J (2004) Tic32, an essential component in chloroplast biogenesis. J Biol Chem 279: 34756–34762 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Belmonte MF, Harada JJ, Inoue K (2010) Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr Genomics 11: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Li M, Schnell DJ (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc Natl Acad Sci USA 110: 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Rounds C, Schnell DJ (2010) The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P (2011) Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J 66: 877–889 [DOI] [PubMed] [Google Scholar]
- Kaundun SS. (2014) Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag Sci 70: 1405–1417 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell D (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kim J, Olinares PD, Oh SH, Ghisaura S, Poliakov A, Ponnala L, van Wijk KJ (2013) Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol 162: 157–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus D, Ohlrogge JB, Neuhaus HE, Dörmann P (2004) Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase. Planta 219: 389–396 [DOI] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y (1994) Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA 91: 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, et al. (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li HM, Teng YS (2013) Transit peptide design and plastid import regulation. Trends Plant Sci 18: 360–366 [DOI] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ (2011) Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol 155: 293–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang TC, Qiao Q, Ren Z, Zhao J, Yonezawa T, Hasegawa M, Crabbe MJC, Li J, Zhong Y (2013) Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 8: e58747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Lu X, Jiang L, Wang C, Fan Y, Zhang C (2010) EMB1211 is required for normal embryo development and influences chloroplast biogenesis in Arabidopsis. Physiol Plant 140: 380–394 [DOI] [PubMed] [Google Scholar]
- Lin M, Behal R, Oliver DJ (2003) Disruption of plE2, the gene for the E2 subunit of the plastid pyruvate dehydrogenase complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Mol Biol 52: 865–872 [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA (2011) Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol 3: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Zhao H, Parsons EP, Xu C, Kosma DK, Xu X, Chao D, Lohrey G, Bangarusamy DK, Wang G, et al. (2011) The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol 157: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Wang X, Bowers J, Paterson A, Lisch D, et al. (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol 148: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Dooner HK (2004) A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J 37: 92–103 [DOI] [PubMed] [Google Scholar]
- Magee AM, Aspinall S, Rice DW, Cusack BP, Sémon M, Perry AS, Stefanović S, Milbourne D, Barth S, Palmer JD, et al. (2010) Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res 20: 1700–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard JL, Heckel T, Massonneau A, Wisniewski JP, Cordelier S, Lassagne H, Perez P, Dumas C, Rogowsky PM (2004) Morphogenesis of maize embryos requires ZmPRPL35-1 encoding a plastid ribosomal protein. Plant Physiol 134: 649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann HI, Camilleri C, Bérard A, Bataillon T, David JL, Reboud X, Le Corre V, Caloustian C, Gut IG, Brunel D (2004) Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J 38: 193–202 [DOI] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW (2007) Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D. (2013) Large-scale mutant analysis of seed development in Arabidopsis. In PW Becraft, ed, Seed Genomics. Wiley-Blackwell, Ames, IA, pp 5–20 [Google Scholar]
- Meinke D, Sweeney C, Muralla R (2009) Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE 4: e7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. (1985) Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor Appl Genet 69: 543–552 [DOI] [PubMed] [Google Scholar]
- Motamayor JC, Mockaitis K, Schmutz J, Haiminen N, Livingstone D III, Cornejo O, Findley SD, Zheng P, Utro F, Royaert S, et al. (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol 14: r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Chen E, Sweeney C, Gray JA, Dickerman A, Nikolau BJ, Meinke D (2008) A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in Arabidopsis. Plant Physiol 146: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Hsu SC, Bédard J, Inoue K, Jarvis P (2008) The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol 148: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J (1997) Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol 113: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Huang X, Higginson E, Ayliffe M, Day A, Timmis JN (2013) Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol 161: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage LJ, Imre KM, Hall DA, Last RL (2013) Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS ONE 8: e73291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Dinkins R, Robinson K, Shellhammer J, Meinke DW (1989) An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev Biol 131: 161–167 [DOI] [PubMed] [Google Scholar]
- Schulte W, Töpfer R, Stracke R, Schell J, Martini N (1997) Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA 94: 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Li C, McCarty DR, Meeley R, Tan BC (2013) Embryo defective12 encodes the plastid initiation factor 3 and is essential for embryogenesis in maize. Plant J 74: 792–804 [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta 1833: 314–331 [DOI] [PubMed] [Google Scholar]
- Siemenroth A, Wollgiehn R, Neumann D, Börner T (1981) Synthesis of ribosomal RNA in ribosome-deficient plastids of the mutant “albostrians” of Hordeum vulgare L. Planta 153: 547–555 [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, et al. (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45: 831–835 [DOI] [PubMed] [Google Scholar]
- Sosso D, Canut M, Gendrot G, Dedieu A, Chambrier P, Barkan A, Consonni G, Rogowsky PM (2012) PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. J Exp Bot 63: 5843–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SCK, Fishbein M, Livshultz T, Foster Z, Parks M, Weitemier K, Cronn RC, Liston A (2011) Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R, Jarvis P (2011) The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS ONE 6: e23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V, Coe EH Jr (1979) Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc Natl Acad Sci USA 76: 2760–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Mott R (2009) The 1001 Genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirmer J, Westhof E (2006) Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol 415: 180–202 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Zhang YF, Hou MM, Tan BC (2013) The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize. PLoS ONE 8: e67369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Day A (1998) Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J 15: 265–271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.