Bioactive GAs can be synthesized in the cytosol and the nucleus.
Abstract
The maize (Zea mays) gibberellin (GA)-deficient mutant dwarf1 (d1) displays dwarfism and andromonoecy (i.e. forming anthers in the female flower). Previous characterization indicated that the d1 mutation blocked three steps in GA biosynthesis; however, the locus has not been isolated and characterized. Here, we report that D1 encodes a GA 3-oxidase catalyzing the final step of bioactive GA synthesis. Recombinant D1 is capable of converting GA20 to GA1, GA20 to GA3, GA5 to GA3, and GA9 to GA4 in vitro. These reactions are widely believed to take place in the cytosol. However, both in vivo GFP fusion analysis and western-blot analysis of organelle fractions using a D1-specific antibody revealed that the D1 protein is dual localized in the nucleus and cytosol. Furthermore, the upstream gibberellin 20-oxidase1 (ZmGA20ox1) protein was found dual localized in the nucleus and cytosol as well. These results indicate that bioactive GA can be synthesized in the cytosol and the nucleus, two compartments where GA receptor Gibberellin-insensitive dwarf protein1 exists. Furthermore, the D1 protein was found to be specifically expressed in the stamen primordia in the female floret, suggesting that the suppression of stamen development is mediated by locally synthesized GAs.
The plant hormone GAs are diterpenoid compounds found in plants, fungi, and bacteria. Among more than 130 species of GAs discovered in nature, only a few are bioactive; the rest are either biosynthetic intermediates or catabolites (Yamaguchi, 2008; Sponsel and Hedden, 2010). GA has multiple biological functions including promoting seed germination, stem elongation, flowering, pollen development, and fruit growth (Sponsel and Hedden, 2010). The stem elongation function of GA contributed to the Green Revolution in which mutations in GA signaling (Reduced height protein1) or biosynthesis (Gibberellin 20-oxidase) founded the semidwarf wheat (Triticum durum) and rice (Oryza sativa), respectively (Peng et al., 1999; Sasaki et al., 2002). In maize (Zea mays), GA also plays an important role in sex determination. The immature flowers of maize are bisexual in the ear and the tassel. The unisexual flowers are achieved by selective arrest and abortion of the stamen in the ear and the pistil in the tassel (Dellaporta and Calderon-Urrea, 1994). GA-deficient mutants (dwarf1 [d1], d3, d5, and anther ear1) and GA-insensitive mutants (d8 and d9) carry bisexual flowers in the ear although they maintain male flowers in the tassel, indicating that GA mediates suppression of stamen primordium development in the female flower.
GA biosynthesis can be divided into three phases, which are believed to occur in three separate compartments (Yamaguchi, 2008). The first phase is the cyclization of geranylgeranyl diphosphate in plastids (Supplemental Fig. S1). The common diterpenoid precursor geranylgeranyl diphosphate is converted to ent-kaurene through the functions of ent-copalyl diphosphate synthase and ent-kaurene synthase. These two enzymes are localized in plastids (Sun and Kamiya, 1994; Helliwell et al., 2001 ). The second phase is the formation of GA12 in the endoplasmic reticulum. ent-Kaurene is oxidized to form GA12 by cytochrome P450 monooxygenases, which is located on the plastid envelope and endoplasmic reticulum (Helliwell et al., 2001). The third phase is the formation of bioactive GAs. This involves the activities of two classes of 2-oxoglutarate-dependent dioxygenases: GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox). GA20ox catalyzes multiple oxygenation reactions at C-20 of GA12 to produce GA9, GA53 to GA20. GA9 is then converted to bioactive GA4, and GA20 to GA1 or GA3 by GA3ox (for review, see Yamaguchi, 2008). The third phase is widely believed to occur in the cytosol because GA3ox and GA20ox are soluble proteins and do not appear to contain any targeting sequences (Yamaguchi, 2008). However, direct experimental proof is lacking.
The GA signal transduction pathway involves a GA receptor (gibberellin-insensitive dwarf protein1 [GID1]) and several repressor proteins (DELLA). When the GA level is low, DELLA proteins accumulate and interact with specific transcription factors that function to promote GA-responsive gene expression (de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Hong et al., 2012). GA binding to GID1 facilitates the binding of GID1-GA with DELLA, triggering degradation of DELLA proteins by 26S proteasomes and release of transcription factors (Sun, 2011). DELLA proteins were localized in the nucleus (Peng et al., 1997; Silverstone et al., 1998; Ikeda et al., 2001). GID1 was found mostly in the nucleus, with substantial abundance in the cytosol (Ueguchi-Tanaka et al., 2005; Willige et al., 2007). Thus, this raises the question of how GA synthesis coordinates with the receptor in terms of compartmentation.
The d1 allele is a long-known GA-deficient mutant in maize (Phinney, 1956; Fujioka et al., 1988) and was studied biochemically and physiologically. The mutant was rescued by the application of GA1, but not by GA20 (Spray et al., 1996). It accumulated 10 times more GA20 and GA29 than the wild type (Fujioka et al., 1988). Metabolic analysis indicated that the mutation blocked three steps in GA biosynthesis (i.e. GA20 to GA5, GA5 to GA3, and GA20 to GA1; Spray et al., 1996). It was speculated that D1 either encodes a GA3ox, or a regulator required for GA3ox expression. However, the identity of the mutation in the d1 allele is not revealed.
In this study, we cloned the D1 gene and demonstrated that it encodes a GA3ox that catalyzes at least four reactions: GA20 to GA3, GA20 to GA1, GA5 to GA3, and GA9 to GA4. In contrast with the wide belief that bioactive GAs are synthesized in the cytosol, we prove that D1 as well as the upstream GA20ox proteins are dual localized to the nucleus and cytosol, providing strong evidence that GA can be synthesized in the cytosol and the nucleus where the GA receptor GID1 is localized. By using a highly specific D1 antibody, we revealed that specific GA production in the stamen primordia in the female floret mediates the suppression of stamen development, resulting in unisexual flowers in maize.
RESULTS
Isolation and Characterization of a Dwarf Mutant in Maize
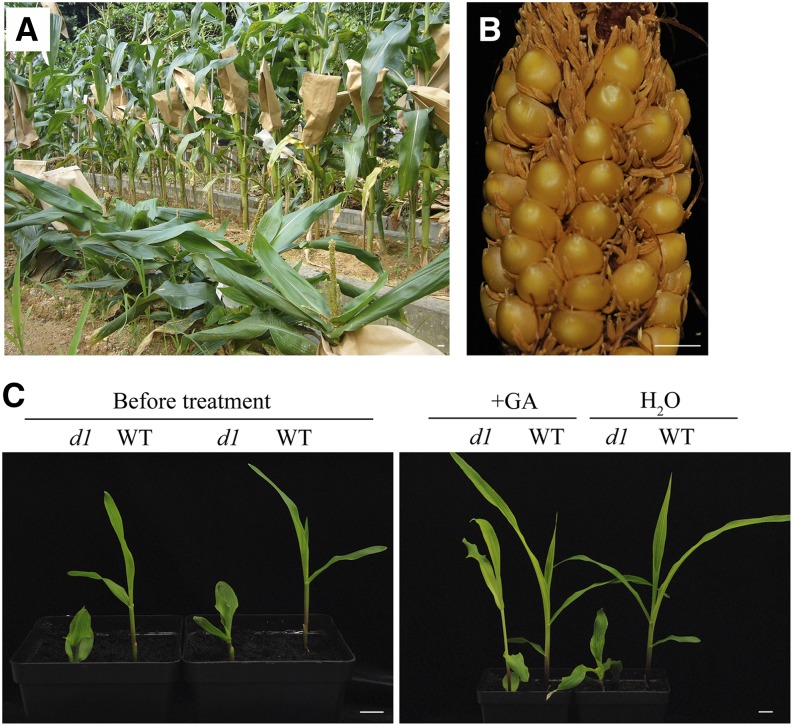
During a genetic analysis of a viviparous*3286 (vp*3286) mutant, we noticed that this line had an intriguingly high mutagenesis frequency. In one selfed progeny, it produced a dwarf mutant that exhibited a recessive nuclear mutation pattern. The mutant is approximately 30 cm tall at maturity with shortened internodes, and broad and dark-green leaves (Fig. 1A). Its tassel (male flowers) was erect and short with reduced branches. The anthers were fully developed, but shed poorly. The plant is andromonoecious, having bisexual flowers in the ear (female flowers in the wild type) and male flowers in the tassel (Fig. 1B). These phenotypes are reminiscent of typical GA-deficient or GA-insensitive mutants (Harberd and Freeling, 1989; Spray et al., 1996). To distinguish the two possibilities, we applied GA3 to the seedlings. The dwarfism was restored to the wild type by spraying 10 µm GA3 (Fig. 1C), indicating that the mutant is GA deficient. To determine which allele this mutant is, we crossed this mutant with other GA-deficient mutants. The cross with the classic d1 resulted in dwarf plants, indicating that the new mutant was allelic to d1. We named this new allele d1-3286.
Figure 1.
Phenotypes of the d1 allele and its response to GA treatment. A, Homozygous adult d1-3286 plants showed dwarfism with wide and compacted dark-green leaves (front row) in contrast with the wild type (back row). B, A homozygous ear of d1-3286 displayed andromonoecy and formation of anthers in the ear. C, The dwarf phenotype of the d1-3286 seedling before and after spray with 10 µm GA3 for 7 d. Bars = 1 cm.
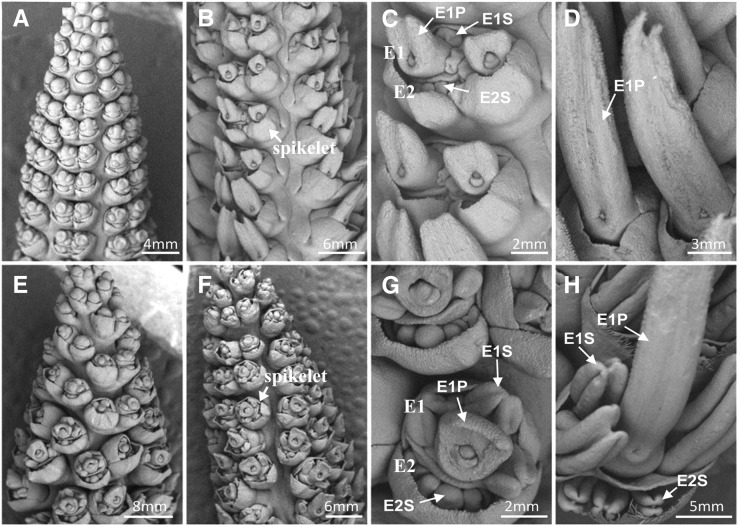
Mature maize plants are diclinous, having a male inflorescence (tassel) on the top and a female inflorescence (ear) in the leaf axil. To understand the formation of andromonoecy in the d1 mutant, we examined female flower development by scanning electron microscopy (SEM) (Fig. 2). At the early stage, the wild type (Fig. 2, A–D) and the d1 mutant (Fig. 2, E–H) form two florets within one female spikelet, named E1 and E2 (Fig. 2). The early E1 and E2 florets are bisexual, each containing one central pistil primordium surrounded by three stamen primordia. At the later stage, the E2 floret and the stamen of E1 abort in the wild type while the pistil primordium of E1 extends into a long silk that serves for reception of pollen, resulting in a unisexual female flower (Fig. 2, A–D). However, in the d1 mutant, the development of stamen primordia in E1 and E2 is not suppressed at the later stage, resulting in formation of anthers along with E1 elongated silks (Fig. 2, E–H). Relief of suppression on anther growth and development results in andromonoecy. This result as well as the observed andromonoecy in GA-deficient mutant anther ear1 (Bensen et al., 1995) and GA-insensitive mutant d8 (Peng et al., 1997) indicates that suppression of anther development in the female flowers in maize is mediated by GA.
Figure 2.
Female and male organ development in ear florets in the wild type and in the d1-3286 allele. The ears (female flowers) of the wild type (A–D) and the d1-3286 allele (E–H) were analyzed at different stages of flower development by SEM. E1, Primary ear floret; E1P, pistil of E1; E1S, stamen of E1; E2, secondary ear floret; E2S, stamen of E2.
Molecular Characterization of the d1 Alleles
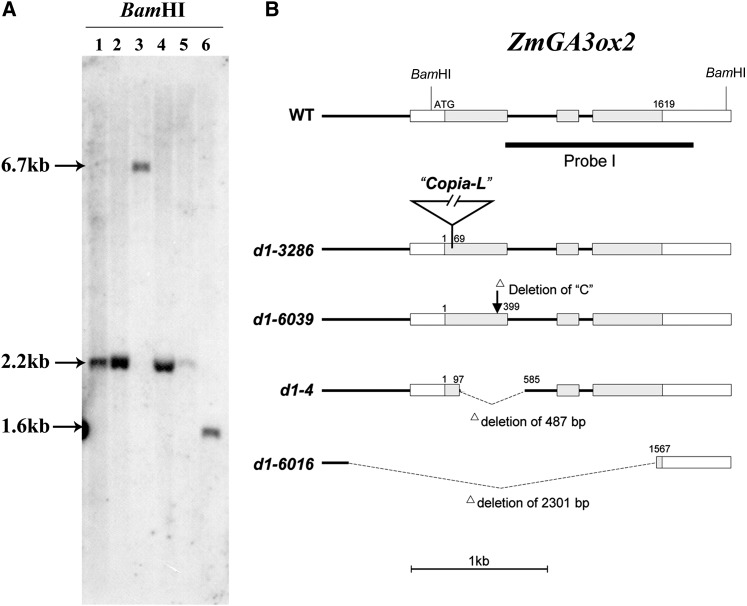
Previous studies suggested that the d1 mutant was blocked in the GA20 to GA5, GA5 to GA3, and GA20 to GA1 conversion (Fujioka et al., 1988; Spray et al., 1996), leading to the notion that D1 encodes either a GA3ox or a factor required for GA3ox expression. Searching of the maize draft genome (A Golden Path version 2.0) by using rice GA3oxs identified two homologs, named ZmGA3ox1 and ZmGA3ox2. In alignment with related GA3ox proteins from rice OsGA3ox1, OsGA3ox2 (Itoh et al., 2001), pea (Pisum sativum) PsGA3ox1 (Lester et al., 1997; Martin et al., 1997), Arabidopsis AtGA3ox1 (Chiang et al., 1995), barley (Hordeum vulgare) HvGA3ox2 (Spielmeyer et al., 2004), wheat TaGA3ox2 (Appleford et al., 2006), and Brachypodium distachyon BdGA3ox2 are roughly the same size with conserved residues throughout the entire protein sequences (Fig. 3). At the protein level, ZmGA3ox2 is more closely related to OsGA3ox2 (78% identity) than to ZmGA3ox1 (58%). At the nucleotide level, ZmGA3ox2 and ZmGA3ox1 are quite divergent, sharing no significant similarity. This divergence suggests that the two genes were separated long ago. In our experiments, DNA hybridization by using the ZmGA3ox2 as a probe did not hybridize to ZmGA3ox1 (Fig. 4A). ZmGA3ox2 is located on chromosome (Chr) 3S and ZmGA3ox1 on Chr 6L. Because the classic d1 mutation was mapped on Chr 3S (Neuffer and England, 1995), the ZmGA3ox2 gene remains a good candidate for the D1 gene. To test this notion, we analyzed four independent alleles of d1 (d1-3286, d1-6039, d1-4, and d1-6016). The d1-6039 allele was isolated from a targeted direct tagging experiment with d1-3286. The d1-4 allele was previously known as d4, but was proven later to be allelic to d1 (Stinard, 2009). The d1-6016 allele was also known as 302A in the Maize Genetics Cooperation Stock Center. DNA hybridization analysis by using the ZmGA3ox2 as a probe detected polymorphism among four d1 alleles and inbreds (W22 and B73; Fig. 4). With BamHI digestion, the inbred B73 and W22 were predicted to yield a 2.2-kb fragment. A similar size fragment was detected in d1-6039, B73, and W22, a 6.7-kb fragment in d1-3286, and a 1.6-kb fragment in d1-4 (Fig. 4A). Although the loading and fragment size detected in d1-6016 are similar to d1-6039, the signal in d1-6016 was much weaker than d1-6039. Molecular cloning and sequencing indicate that the d1-3286 allele contains an insertion of a 4.5-kb Copia-type element at position 69 (start from ATG) of the ZmGA3ox2 gene (Fig. 4B). The d1-6039 allele contains a deletion of C residue at position 399 in the first exon, causing a frame-shift that ends the translation at 163 amino acids. In addition, we also detected two short deletions of 15 and 7 bp in the 5′-untranslated region, six indels in the first intron, and several single nucleotide polymorphisms in the coding region. However, all of these changes did not change the protein sequence, suggesting that the d1-6039 allele is likely derived from a different maize background than B73. The d1-4 allele contains a deletion of 487 bp, covering 389 bp of the first exon and 98 bp of the first intron (Fig. 4B). The d1-6016 allele contains a deletion of 2,301 bp, covering 508 bp of upstream sequences and 1,793 bp of ZmGA3ox2 gene (Fig. 4B). The residual sequences of ZmGA3ox2 in d1-6016 contain a 291-bp fragment overlapping with the probe used in the DNA hybridization analysis. These 291 bp were also present in an approximately 2.2-kb BamHI fragment in d1-6016. This is the reason that a weak and similar wild-type size fragment was shown in the DNA hybridization result (Fig. 4B). Because all of these independent alleles contain mutations in the ZmGA3ox2 gene and cross of any two of these alleles gave rise to dwarf mutants, we conclude that the maize D1 locus encodes ZmGA3ox2, a putative GA3ox in the biosynthesis of GAs. The nature of mutation in each allele appears to abolish the ZmGA3ox2 function; hence, it is likely a null mutation. This is consistent with the severe dwarf phenotype.
Figure 3.
ClustalW sequence alignment and phylogenetic analysis of ZmGA3ox1 and ZmGA3ox2 with related GA3oxs. Amino acid alignment (A) and a phylogenetic tree (B) showing the relationships among maize ZmGA3ox2 (AFS50158), ZmGA3ox1 (AFS50160), Arabidopsis AtGA3ox1 (NP_173008), rice OsGA3ox1 (BAB62073) and OsGA3ox2 (BAB17075), pea PsGA3ox1 (AAB65829), barley HvGA3ox2 (AAT49061), B. distachyon BdGA3ox2 (XP_003569638), and wheat TaGA3ox2 (Q3I409). Mutations (deletions or insertions) in later characterized d1 alleles were marked (see Fig. 4). The highly conserved ferrous iron ligand binding residues for 2-oxoglutarate-dependent dioxygenases are marked as circles.
Figure 4.
Molecular characterization of d1 alleles. A, DNA hybridization analysis on the d1 alleles with ZmGA3ox2 probe I. 1, B73; 2, W22; 3, d1-3286; 4, d1-6039; 5, d1-6016; 6, d1-4. B, ZmGA3ox2 gene structure and different d1 alleles. The position of probe I is indicated. Exons are boxes. Shaded boxes are translated regions, and empty boxes are 5′- or 3′-untranslated regions. Introns and noncoding regions are shown as black lines. A Copia-like element (accession no. JX307642) was inserted at position 69 of the d1-3286 allele. One-base C was deleted at position 399 of the d1-6039 allele. A 487-bp fragment was deleted in the d1-4 allele. A 2,301-bp fragment was deleted in the d1-6016 allele.
Subcellular Localization of the D1 Protein
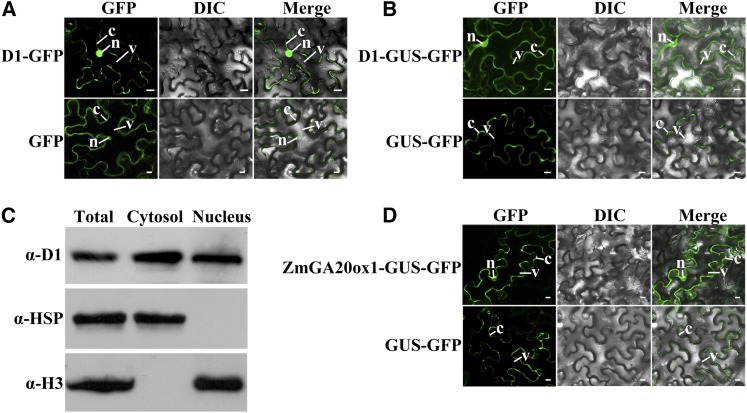
Because GA3ox catalyzes the final step in producing bioactive GAs, the enzyme localization determines the potential sites of bioactive GA production, given that the substrates are present. To determine the subcellular localization, D1 protein was fused with GFP and transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells. The tobacco epidermal cells contain a large vacuole in the middle and the cytosol is aligned along the cell wall. Strong GFP signals were found in both the cytosol and the nucleus by confocal laser microscopy (Fig. 5A). However, the control GFP protein also showed dual localization in the cytosol and the nucleus (Fig. 5A), raising the possibility that the nucleus localization of D1-GFP and GFP may be a result of passive diffusion if the size of the fusion protein is smaller than the nuclear pore. To address this possibility, D1 was fused with GUS-GFP to yield D1-GUS-GFP (approximately 132 kD), and tested with GUS-GFP (approximately 91 kD) as a negative control. The results show that D1-GUS-GFP is dual localized to the cytosol and the nucleus, whereas GUS-GFP is localized to the cytosol only (Fig. 5B), ruling out the possibility of passive diffusion. The nucleus localization of GFP (26.9 kD) may be subject to passive diffusion because it is smaller than the cutoff size (approximately 40 kD) of the nuclear pore (Marfori et al., 2011).
Figure 5.
Subcellular localization of D1 protein. A, B, and D, D1-GFP fusion and GFP proteins (A), D1-GUS-GFP and GUS-GFP fusion proteins (B), and ZmGA20ox1-GUS-GFP (NP_001241783) and GUS-GFP fusion proteins (D) were transiently expressed in tobacco leaf epidermal cells and were analyzed by fluorescent laser confocal microscope. C, Western-blot detection of D1 protein in nucleus and cytosol fractions of maize seedlings. Total, Total proteins from shoot and root. The protein amount was loaded identically for each antibody. α-D1, Anti-D1 (ZmGA3ox2) antibody; α-HSP82, anti-heat shock protein 82 antibody (cytosolic marker); C, cytosol; DIC, Differential interference contrast; N, nucleus; V, vacuole. Bars = 10 μm.
The dual localization of D1 in the nucleus and cytosol is surprising because GA3ox is widely believed to be in the cytosol (reviewed by Yamaguchi, 2008). To independently determine the subcellular localization of D1, we probed the D1 protein in subfractions of cytosol and nucleus by western-blot analysis. A polyclonal D1 antibody was raised by using His-D1 recombinant protein. The specificity of the antibody was tested by western-blot analysis on total proteins from the wild type and the d1-6016 mutant seedlings, which carry a deletion of the entire D1 gene. The antibody recognized a single band matching the expected size of D1 (41 kD) in the wild type and no band in the d1-6016 mutant (Supplemental Fig. S2), indicating that the D1 antibody is highly specific to the D1 protein. The wild-type seedling cells were fractionated into a nucleus and a cytosol fraction and the proteins were extracted. Western-blot analysis detected the presence of D1 protein in both the cytosol and the nucleus fraction (Fig. 5C). Antibodies against α-Heat Shock Protein82 (cytosol marker) and Histone3 (α-H3, nucleus marker) were used to monitor cross contamination. No cross contamination between the two fractions was detected. Thus, two independent approaches detected the consistent result that D1 is dual localized to the cytosol and the nucleus.
The substrates of GA3ox are produced via the function of GA20ox, which convert GA12 to GA9, and GA53 to GA20 (Hedden and Thomas, 2012). To test whether the upstream step is also localized in the nucleus and cytosol, we studied the subcellular localization of GA20ox. Maize ZmGA20ox1 complementary DNA (cDNA) was cloned and fused with GUS-GFP to create the ZmGA20ox1-GUS-GFP fusion. Using the same methodology, ZmGA20ox1-GUS-GFP was found to be dual localized to the cytosol and the nucleus (Fig. 5D). In conjunction with the D1 localization, we conclude that bioactive GAs can be synthesized in both the cytosol and the nucleus.
Determination of D1 Enzymatic Activity
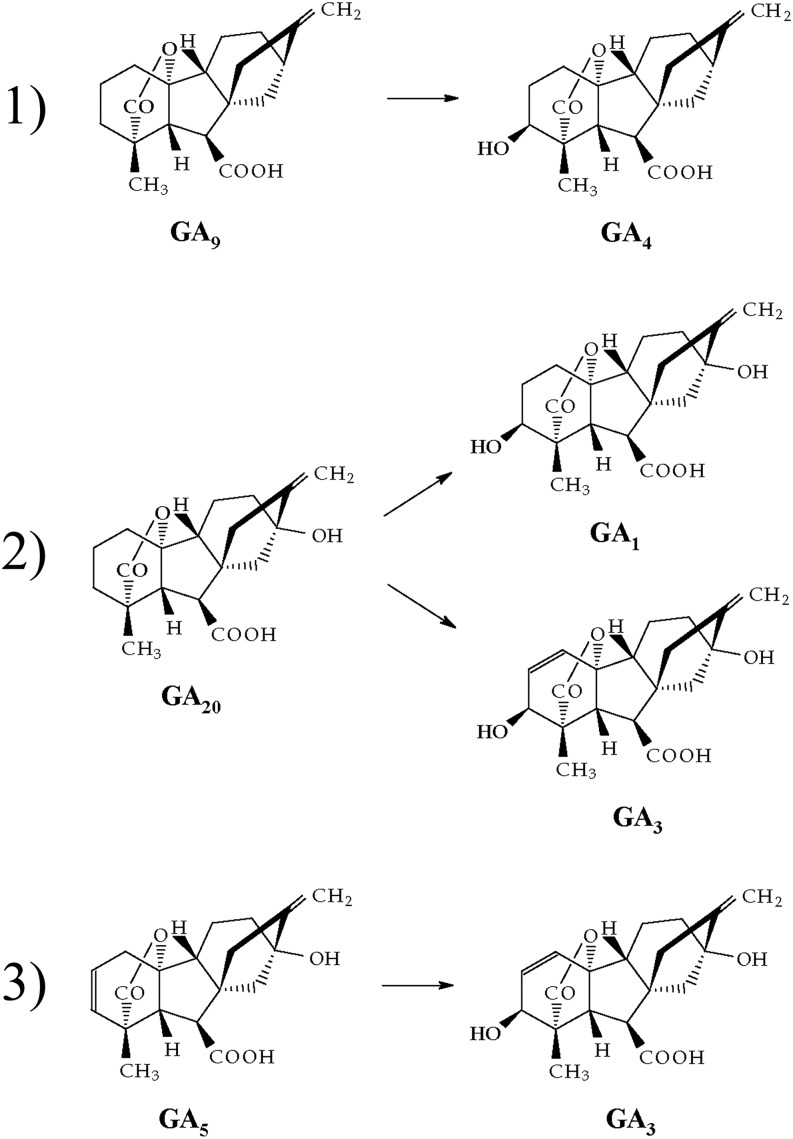
To determine the enzymatic function, the D1 protein was fused with GST protein in pGEX-2T vector and expressed in Escherichia coli. The fusion protein was purified and incubated with deuterium-labeled GA substrates 2H3-GA9, 2H2-GA20, and not deuterium-labeled 2H0-GA5 in the presence of cofactors 2-ketoglutarate, ascorbate, and FeSO4 (see “Materials and Methods”). The reaction products were analyzed by full-scan gas chromatography (GC)-mass spectrometry (MS) and identified with Kovats retention indices (KRIs). Recombinant GST-D1 protein is capable of catalyzing four reactions, converting GA9 to GA4, GA20 to GA1, GA20 to GA3, and GA5 to GA3 (Fig. 6; Table I). The MS results are provided in Supplemental Figure S3. In contrast with Arabidopsis in which the active GAs are GA1 and GA4 (Yamaguchi, 2006), maize, rice, and wheat (monocots) synthesize GA3 as an active form as well (Spray et al., 1996; Appleford et al., 2006). Although the biological function of GA3 remains to be elucidated (Magome et al., 2013), it is concluded that D1 (ZmGA3ox2) protein catalyzes the final step of biologically active GAs in maize. This conclusion is consistent with previous studies on physiological and biochemical features of the d1 mutant (Phinney, 1956; Fujioka et al., 1988; Spray et al., 1996).
Figure 6.
Recombinant D1 protein possesses four GA3ox activities in vitro.
Table I. Identification of the metabolites from GAs incubated with recombinant maize GA3ox2 by full-scan GC-MS and KRIs.
Characteristic ions are shown as mass-to-charge ratios, with the percentage of relative intensity of the base peak in parentheses.
| GA Substrate | GA Product | KRI | Characteristic Ions |
|---|---|---|---|
| 2H3-GA9 | 2H3-GA4 | 2,515 | 421 (100), 406 (16), 403 (42), 393 (51), 389 (57) |
| 2H2-GA20 | 2H2-GA1 | 2,674 | 508 (100), 493(9), 449(12), 418 (2), 392 (1) |
| 2H2-GA20 | 2H2-GA3 | 2,697 | 506 (100), 491(5), 475 (3), 447 (6), 433(4), 416 (1) |
| 2H0-GA5 | 2H0-GA3 | 2,699 | 504 (100), 489 (7), 473 (2), 445 (7), 431(3), 414 (2) |
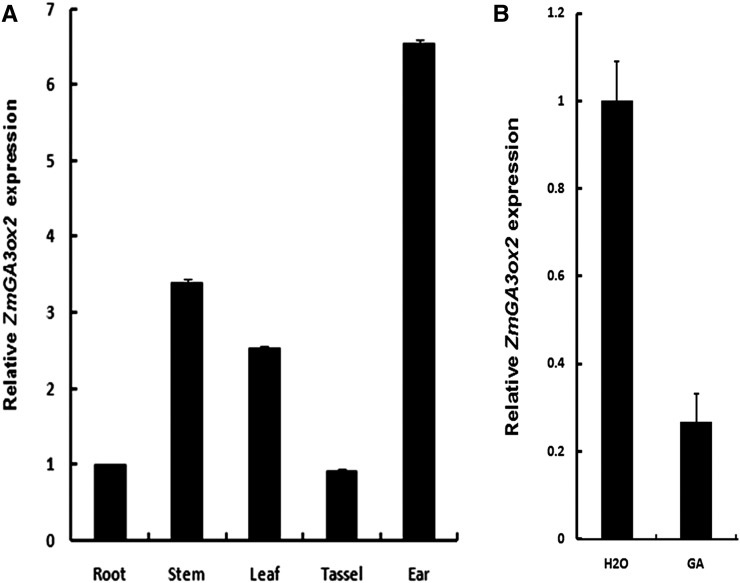
The expression pattern of ZmGA3ox2 in different organs was determined by quantitative reverse transcription (qRT)-PCR analysis. D1 (ZmGA3ox2) was found to be expressed in most tissues (e.g. roots, stems, leaves, tassels, and ears; Fig. 7A). By contrast, we could not detect the expression of ZmGA3ox1 in these tissues although the primers were approved working robustly on genomic DNA (data not shown). These results indicate that D1 is the predominant enzyme for the last step of bioactive GA production in most organs, whereas ZmGA3ox1 may be expressed in low-level cells or in highly specialized cells. In addition, we analyzed whether a negative feedback regulation exists on the expression of ZmGA3ox2. Application of 100 µm GA3 in whole seedlings strongly inhibited ZmGA3ox2 mRNA accumulation, suggesting that ZmGA3ox2 is subject to feedback regulation (Fig. 7B).
Figure 7.
Expression of ZmGA3ox2 in maize. qRT-PCR analysis of ZmGA3ox2 in different tissues (A) and in wild-type whole seedlings after treatment with GA3 (B). Actin1 was the reference gene. Error bars represent the se.
Sites of D1 Protein Expression during Maize Female and Male Floret Development
GA is believed to suppress the development of stamen primordia in the ear because GA-deficient or GA-insensitive mutants show andromonoecy in maize (Dellaporta and Calderon-Urrea, 1994). However, it is unclear whether GA plays this function in a cell-autonomous or nonautonomous fashion. Detection of GAs in specific cells is difficult because bioactive GAs exist at extremely low concentrations in plant tissues and can be readily transported. Because GA3ox predicts the potential sites of bioactive GA production, we determined the sites of D1 protein expression during maize female floret development by immunohistochemical analysis using the highly specific D1 antibody.
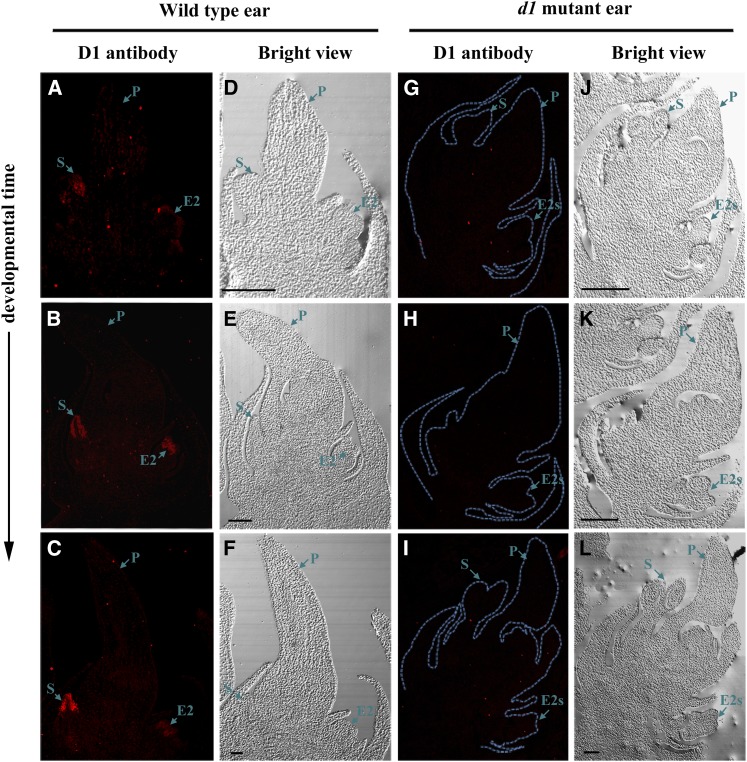
Wild-type and d1 ears (1–1.5 cm in length) were fixed in wax and sectioned. The D1 protein was detected with Alexa Fluor 594-labeled secondary antibody, which appeared red under confocal laser scanning microscopy (Fig. 8). The florets in the ears were mostly at bisexual stage. Red fluorescent signals were detected exclusively in the stamen regions of E1 and E2 florets in the wild-type spikelets (Fig. 8, A–C), but not in the d1 spikelets (Fig. 8, G–I). The signal became much stronger at late stages than in early stages (Fig. 8, A and C), probably because of the increased number of stamen cells. This result indicates that the D1 protein is specifically expressed in the stamen initials during female floret development. Thus, stamen cells in both E1 and E2 are the potential sites for bioactive GA production during maize sex determination. This result in conjunction with the SEM analysis suggests that GAs suppress stamen primordium development in female floret in a cell-autonomous manner.
Figure 8.
Immunohistochemical detection of D1 protein in developing maize female florets. Sections of wild-type and d1 ears (1–1.5 cm) were incubated with anti-D1 antibody and followed with secondary antibody labeled with Alexa Fluor 594, which produces red fluorescent signals under a fluorescent microscope. A to C, Wild-type female florets incubated with D1 antibody. D to F, Bright views of A to C. G to I, d1 Female florets incubated with D1 antibody. J to L, Bright views of G to I. E2, Secondary ear florets; E2s, stamen of E2 florets; P, pistil of E1 florets; S, stamen of E1 florets. Bars = 200 μm.
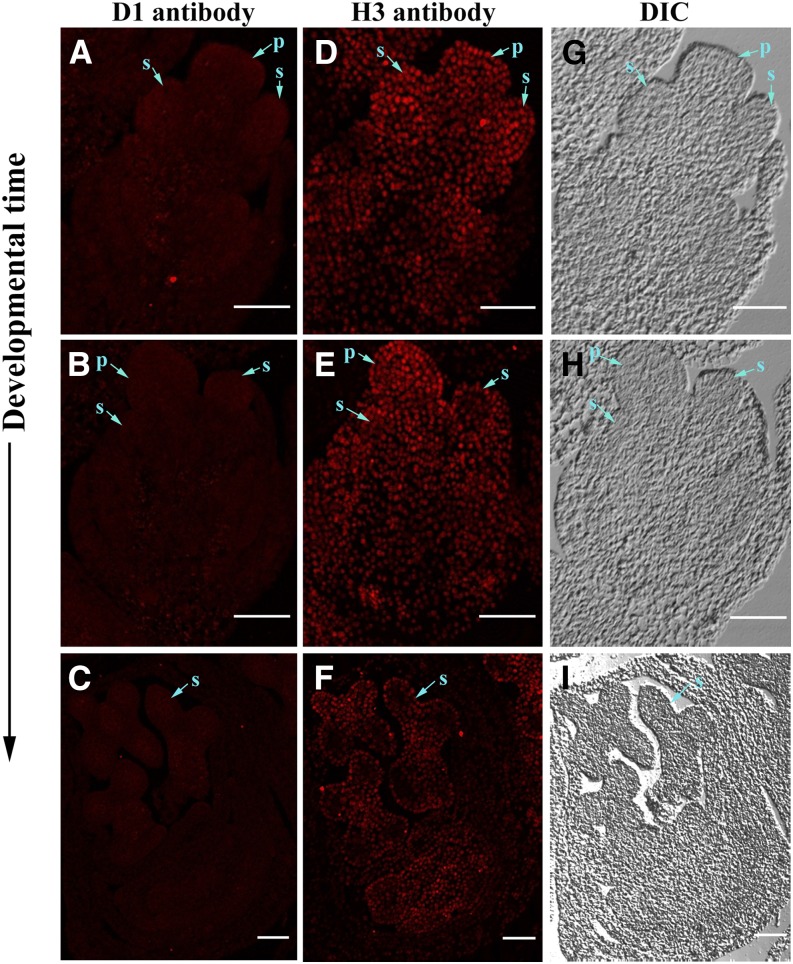
Similar to florets in the ear, tassel florets are bisexual at early stages. During sex determination, the stamen primordia develop into functional anthers while the pistil primordium in the tassel floret is aborted through programmed cell death (Calderon-Urrea and Dellaporta, 1999), resulting in only male florets in the tassel. The d1 mutant contains normal tassel florets. To test whether D1 plays a role in tassel floret sex determination, we performed immunohistochemical analysis on the wild-type tassel (Fig. 9). We did not detect a strong signal of D1 in tassel florets from the bisexual stage to the unisexual stage (Fig. 9, A–C). Detection of H3 displayed a strong signal, indicating that the experiment system is reliable (Fig. 9, D–F). These results suggest that D1 may be not expressed in the tassel floret or expressed at a low level that is beyond the limit of immunohistochemical detection.
Figure 9.
Immunohistochemical detection of D1 protein in developing maize male florets. Sections of wild-type tassels from 1 to 1.5 cm were incubated with anti-D1 antibody and anti-H3 antibody followed with secondary antibody labeled with Alexa Fluor 594, which produces red fluorescent signals under fluorescent microscope. A to C, Wild-type male florets incubated with D1 antibody. D to F, Wild-type male florets incubated with H3 antibody. G to I, Bright views of A to C and D to F. Developmental time arrow points to late stage. DIC, Differential interference contrast; P, pistil; S, stamen. Bars, 100 μm.
DISCUSSION
D1 Encodes a GA3ox Catalyzing the Final Step of GA Biosynthesis in Maize
We characterized a dwarf mutant in maize isolated during the study of vp*3286. Genetic analysis confirms that this mutant is an allele of d1. Biochemical characterization on d1 indicates that the mutation blocks three steps of GA biosynthesis (Fujioka et al., 1988; Spray et al., 1996). This raises two possibilities on the molecular identity of D1 gene: first, it encodes a GA3ox (ZmGA3ox); and second, it encodes proteins required for ZmGA3ox activity or expression.
We tested the first possibility by identifying mutations in the d1 alleles in candidate genes. The maize genome draft (A Golden Path version 2.0) contains two putative GA3oxs: ZmGA3ox1 on Chr 6L and ZmGA3ox2 on Chr 3S. Because the d1 allele was mapped to Chr 3S (Neuffer and England, 1995), ZmGA3ox2 stands as the most likely candidate. Analysis of four alleles of d1 including the d1-4 allele that was previously considered as d4 identified independent mutations in the ZmGA3ox2 gene in each allele (Fig. 4). In vitro enzymatic activity analysis confirmed that the recombinant ZmGA3ox2 protein catalyzes four reactions converting GA9 to GA4, GA20 to GA1, GA20 to GA3, and GA5 to GA3 (Fig. 6). This extends the previous analysis that the d1 mutation blocked three steps in GA biosynthesis (Spray et al., 1996). Together with the report that ZmGA3ox2 is a candidate gene for a major quantitative trait locus for plant height (qPH3.1; Teng et al., 2013), these results indicate that D1 encodes ZmGA3ox2, a GA3ox that catalyzes the final step of GA biosynthesis.
Although two GA3oxs exist in maize, we did not detect the expression of ZmGA3ox1 in the major tissues tested (Fig. 7), suggesting that ZmGA3ox1 plays a minor role in GA biosynthesis in major tissues. It was reported that ZmGA3ox1 was expressed in the immature tassel (Teng et al., 2013). Thus, it is possible that ZmGA3ox1 is expressed at a very low level or in limited cell types, such as in a few cells in immature tassels. By contrast, ZmGA3ox2 is highly expressed in all tissues tested, suggesting that ZmGA3ox2 plays a predominant role in GA biosynthesis through plant growth and development. A functional redundancy by ZmGA3ox1 is not evident in the d1 mutant because of the severe dwarfism of the d1 mutant. Interestingly, the rice genome contains two GA3ox genes. OsGA3ox2 is predominantly expressed and OsGA3ox1 is only found in unopened flowers in rice (Itoh et al., 2001). However, this pattern is different from Arabidopsis, in which four GA3ox genes have overlapping expressions (Mitchum et al., 2006; Hu et al., 2008). Addressing the specific role of GA3ox1 in maize tassels or rice immature flowers requires the analysis of corresponding loss-of-function mutants in either species.
Bioactive GA Potentially Can Be Synthesized in the Nucleus and the Cytosol
The compartment of bioactive GA synthesis inside the cell has not been resolved thus far. It is widely believed to be in the cytosol based on the evidence that GA3ox is a water-soluble enzyme and does not appear to contain any obvious targeting signals (Yamaguchi, 2008). Because GA3ox catalyzes the last step resulting in bioactive GA production, the site of GA3ox predicts at least the potential site of GA synthesis. By this logic, we examined the localization of GA3ox2 by two independent approaches. First, the D1-GFP fusion protein was found in both compartments although a Nuclear localization signal was not found in the protein sequence (Fig. 3). To rule out the possibility that nucleus localization may be resulted from nonspecific diffusion due to a smaller size of D1-GFP (approximately 68 kD), we tested D1-GUS-GFP (approximately 132 kD). This experiment showed the dual targeting to the nucleus and cytosol as well. Second, we raised a highly specific D1 antibody that reacted to a single band in the wild type that matches the D1 protein size and did not react to any band in the d1-6016 deletion mutant (Supplemental Fig. S2). The antibody detected presence of D1 in both the cytosol and the nucleus fraction (Fig. 5C). These results lead us to conclude that GA3ox is dual localized to the cytosol and the nucleus; thus, these two compartments are potential sites for GA biosynthesis.
Formation of bioactive GA in the nucleus required its precursor. ZmGA20ox1 catalyzes the penultimate reaction of bioactive GA synthesis leading to the formation of the substrate of D1. We found that ZmGA20ox1 was dual localized to the nucleus and cytosol as well (Fig. 5D), which suggests that the precursor of bioactive GA may be synthesized in the nucleus and cytosol. These results provide strong evidence that bioactive GA can be synthesized in the cytosol and nucleus.
Interestingly, this conclusion coincides with the subcellular localization of GA receptor GID1. The rice GID1, also a soluble protein, is primarily targeted to the nucleus, the proposed site of action. However, GID1 was also detected in the cytosol (Ueguchi-Tanaka et al., 2005). Consistently, the Arabidopsis GA receptor AtGID1 was also dual localized in the nucleus and cytosol (Willige et al., 2007). The similar dual localization of D1 and GID1 raises interesting questions regarding whether GA is synthesized and perceived in both compartments, and how GA synthesis is coordinated with the GA signal transduction components between the two compartments. Especially, the GA signaling repressor DELLA proteins are only found in the nucleus (Peng et al., 1997; Silverstone et al., 1998; Willige et al., 2007).
The Suppression of Stamen Primordia in Maize Female Florets Is Probably Mediated by Locally Synthesized GAs
GA promotes floral organ development in bisexual flower system such as Arabidopsis and rice (Koornneef and van der Veen, 1980; Aya et al., 2009). Although GA3ox genes are widely expressed in actively growing tissues (Kaneko et al., 2003), the sites of GA biosynthesis are not necessarily the locations of GA action. For instance, the development of Arabidopsis petals was defective in the GA-deficient mutant, but no GA3ox gene expression was found in the petal of wild-type flowers (Hu et al., 2008). In contrast with the promotion function in the bisexual flower system, GA has a suppression role in maize unisexual flower development. The presence of D1 protein in stamen initials in maize E1 and E2 florets (Fig. 8) is consistent with the andromonoecy phenomenon in which stamen growth and development are not suppressed in the E1 and E2 florets in the d1 mutant (Fig. 2, E–H). This result suggests that locally synthesis of bioactive GAs is required by the suppression of stamen cell development in E1 and E2 florets.
The influence of GA deficiency in maize male florets is not as severe as female florets, because the tassel of d1 mutant carries fertile male florets. At least two possibilities offer an explanation for this phenomenon: first, GA does not affect the stamen primordium development in male floret; or second, the level of GAs in the tassel is too low to induce the suppression of stamen primordia. A previous study reported that the amount of GA-like substances in the tassel was much lower than in the ear (Rood and Pharis, 1980). Consistently, no strong signal was detected by immunohistochemical analysis with D1 antibody in tassel florets (Fig. 9), indicating that the abundance of D1 in tassel florets is much lower than in the ear. Furthermore, exogenous GA converts male florets in the tassel to female florets (Nickerson, 1959). Considering the above evidence, we speculate that stamen development in tassel florets may be the result of deficiency in de novo GA biosynthesis, as indicated by the low expression of the D1 protein (ZmGA3ox2). In that case, the sex determination in maize is achieved by the regulation of bioactive GA synthesis.
The maize d8 mutant with impaired DELLA protein displays a similar andromonoecious phenotype as d1 (Dellaporta and Calderon-Urrea, 1994; Peng et al., 1999), indicating that GA regulates stamen primordium development through DELLA proteins. The sites of D1 expression in female florets are precisely the sites where the WEE1 gene is expressed (Kim et al., 2007). WEE1 is a negative regulator of cell cycle in maize. Thus, downstream components of DELLA proteins that are able to induce WEE1 gene expression need to be identified in order to understand the underlying mechanism of GA function in sex determination in maize.
An Active Copia-Like Element May Exist in the Progenitors of the d1-3286 Allele
Retrotransposable elements are abundant in many genomes. In maize, it accounts for more than 80% of the genome (Baucom et al., 2009; Schnable et al., 2009). The vast majority of retrotransposable elements are kept quiescent by epigenetic regulation (Slotkin and Martienssen, 2007), such that active retrotransposon elements are rarely found in nature (Picault et al., 2009). However, they can be reactivated by means of stresses and genomic shock (McClintock, 1984). In rice, tissue culture can activate Tos17 transposition, a type I or II transposable element (Hirochika, 2001). The isolation of d1-3286 suggested that the copia-like element inserted in the d1-3286 might be active. It was observed that the line vp*3286 that produced the d1-3286 allele had a higher mutagenic activity than similar lines derived from the active Mutator line. In addition to the d1-3286 mutation, a big embryo allele was isolated from the selfed vp*3286 lines (M. Suzuki and D.R. McCarty, unpublished data). Hence, an active retrotransposon such as the one identified in the d1-3286 allele would be a plausible explanation for the high mutagenic activity in the vp*3286 line.
MATERIALS AND METHODS
DNA Extraction and Southern-Blot Analysis
Genomic DNA extraction and Southern-blot analysis were carried out as previously reported (Tan et al., 2011).
Protein Sequence Alignment and Phylogenetic Analysis
Sequence data were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Homology searches in GenBank were done using the BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple alignments of protein sequences were performed by the ClustalW program, and the phylogenetic tree was created by MEGA 4.1 (Beta 3) software (Kumar et al., 2004; http://www.megasoftware.net) using the ClustalX and N-J plot programs (Saitou and Nei, 1987).
qRT-PCR Analysis
Total RNA was extracted from roots, stems, and leaves of 2-week-old seedlings, approximately 1.5-cm tassel and ear. Total RNA was isolated from whole seedlings treated with or without 100 μm GA3. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and treated with DNase I (NEB) prior to cDNA synthesis. cDNA was synthesized by using reverse transcriptase (SuperScript III; Invitrogen). qRT-PCR was performed by using primers specific for D1 (D1A-F6, CGCCCATCTCCTCCTTCTTCT; and qPCRR1, TCCATCACGTCACAGAAGCT) and normalized with Actin1 as a reference gene. The primers for the maize (Zea mays) Actin1 gene are ZmACT-RTF1 (ATGGTCAAGGCCGGTTTCG) and ZmACT-RTR1 (TCAGGATGCCTCTCTTGGCC). qRT-PCR was conducted with three technical replicates for each one of the three biological replicates of each tissue sample.
SEM Analysis
Fresh d1 and W22 ears (approximately 1.5–3 cm) were viewed on a Hitachi S-4700 scanning electron microscope at an accelerating voltage of 5 kV.
GA3ox Enzyme Assay
To demonstrate that D1 encodes active GA3oxs, the coding regions of D1 were cloned in expression vector pGEX-2T and transformed in the RIL strain of Escherichia coli to produce in-frame GST-D1 translational fusion protein. Cells were induced by 0.1 mm isopropylthio-β-galactoside at 28°C for 4 h. The same induction condition was used to produce GST protein. GST and GST-D1 recombinant proteins were purified by using Glutathione Sepharose 4B (GE) as described. The recombinant protein was used for GA3ox enzyme assays. GC-MS was performed with an automated mass spectrometer (JEOL) connected to a Hewlett-Packard 5890 series II gas chromatograph. The analytical conditions used were previously described (Itoh et al., 2001). [17,17-2H2]GA20 was purchased from Lewis Mander (Australian National University). [15,17,17-2H3]GA9 was synthesized from GA9-norketone and methyl-d3-triphenylphosphonium-bromide by Wittig reaction (Takahashi et al., 1986). All GAs used in this study were analyzed by full-scan GC-MS to show the absence of impurities.
Transient Expression of D1-GFP, D1-GUS-GFP, ZmGA20ox1-GUS-GFP, and GUS-GFP Fusion in Tobacco Epidermal Cells and Laser Confocal Microscopy Analysis
D1 and GUS cDNA were respectively cloned into pGWB5 vector and fused with a GFP protein in the C terminus by using Gateway technology (Invitrogen). D1-GUS, ZmGA20ox1, and GUS were fused through EcoRI and cloned into pGWB5 vector, respectively. Leaves of 3- to 5-week-old tobacco (Nicotiana benthamiana) plants were infiltrated with Agrobacterium tumefaciens EHA105 strains containing the pGWB5, pGWB5-D1, pGWB5-D1-GUS, pGWB5-ZmGA20ox1-GUS, and pGWB5-GUS constructs, respectively. Localization of fluorescent proteins was observed 36 h after infiltration by using a confocal laser scanning microscope (FV1000-IX81; Olympus). Vector pGWB5 without insertion was created by using Gateway technology (Invitrogen) with a self-ligated pENTR vector. Empty pGWB5 and pGWB5-GUS vectors were used as the negative control.
Genomic Library Construction and Screening
To clone the ZmGA3ox2 gene in the d1-3286 allele, a size-selected genomic library was constructed in λ-phage. The genomic DNA was digested with BamHI restriction enzyme, size fractionated, and ligated to a ZAP Express vector as previously reported (Tan et al., 1997). Approximately 2.5 × 105 plaques were screened with probe I of ZmGA3ox2 (Fig. 5A). Four clones were identified and excised into pBK-CMV plasmids according to the manufacturer’s instruction (Stratagene).
Purification of His-D1 Fusion Protein
To purified His-D1 recombinant protein, the coding region of D1 was cloned in the expression vector pET-30a(+) (Novagen). The plasmid was transformed in the BL21 strain of E. coli. Cells were induced by 1 mm isopropylthio-β-galactoside and cultured at 37°C overnight. Inclusion bodies were collected by centrifugation after sonication and dissolved in wash buffer (8 m urea, 0.5 m NaCl, and 10 mm Tris-HCl, pH 8.0). BD TALON Metal Affinity Resins (Clontech) were used in the purification. The purification procedure followed the manufacturer’s instructions with some modifications. Briefly, before binding with protein sample, resins were washed with H2O, 10 mm Tris-HCl, and wash buffer, respectively. The washing step was not stopped until the absorbance was stable and near 0. His-D1 fusion proteins were eluted from resins by incubation with elution buffer (500 mm imidazole, 8 m urea, 0.5 m NaCl, and 10 mm Tris-HCl, pH 8.0).
Fractionation of Nuclei and Cytosol and Western-Blot Analysis
Polyclonal antibody against D1 was raised in rabbit by using His-D1 fusion protein. To confirm the specificity of the D1 antibody, a western-blot analysis of the wild type versus the d1 mutant was carried out. Western-blot results indicated that the D1 antibody could not recognize any protein in the d1 mutant. This antibody was used in the following western-blot analysis and immunohistochemical analysis.
To isolate cytosolic protein, 3-d-old germinated seedlings were cut into pieces and incubated in cytosol isolation buffer (50 mm Tris-HCl, pH 7.6, 0.3 m Suc, 0.8% [v/v] Triton X-100, 15 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, and 1 mm dithiothreitol) on ice for 10 min. The supernatant was transferred to a new tube and then centrifuged at 12,000g for 10 min at 4°C. The supernatant was collected for western-blot analysis.
Isolation of intact nuclei from maize was as previously reported (Pandey et al., 2006). Three-day-old germinated maize seedlings were used. To prevent contamination from the supernatant, the nuclei pellet was washed three times by resuspension in wash buffer (0.1 m Tris, 0.8 m KCl, 0.1 m EDTA, 10 mm spermidine, and 10 mm spermine, pH 9.4) and pelleted by centrifugation at 1,800g at 4°C for 15 min. The integrity and purity of the nuclei were monitored by microscopic observation. Total protein extraction and western-blot detection were performed as previously described (Xu et al., 2006).
Immunohistochemical Analysis of D1 Protein in Developing Female and Male Florets
The maize ear and tassel (1–1.5 cm in length) were fixed in a formaldehyde-acetic acid-alcohol solution (45% [v/v] ethanol, 5% [v/v] acetic acid, and 1.9% [v/v] formaldehyde) and were then embedded in paraffin. After sectioning and deparaffinization, the samples were blocked in 3% (w/v) bovine serum albumin (BSA) in 1× phosphate-buffered saline for at least 2 h and then incubated with anti-D1 antibody or anti-H3 (1:100 dilution) at 4°C for overnight. After washing with 1% (w/v) BSA in 1× phosphate-buffered saline three times (each for 10 min), the samples were incubated with fluorescent secondary antibody (Alexa Fluor 594 goat anti-rabbit; Invitrogen) at room temperature for 1 h. The secondary antibody was removed and washed extensively with 1% BSA three times (each for 20 min). The fluorescent signals were observed and imaged under a confocal laser scanning microscope (Olympus FV1000-IX81).
Sequence data for ZmGA3ox2 genomic DNA and cDNA, ZmGA3ox1 genomic DNA, alleles d1-4, d1-6039, d1-3286, and d1-6016 can be found in the GenBank/EMBL database under accession numbers JX307637, JX307638, JX307639, JX307640, JX307641, JX307642, and KC867701, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The GA biosynthetic pathway in plants.
Supplemental Figure S2. Specificity of the D1 antibody.
Supplemental Figure S3. MS analysis of the in vitro activity of recombinant GST-ZmGA3ox2 fusion protein.
Supplementary Material
Acknowledgments
We thank the Maize Genetics Cooperation Stock Center for providing the d1 alleles, Philip Stinard (Maize Genetics Cooperation Stock Center) and Marty Sachs (University of Illinois) for the d1-4 allele, and Dr. Tsuyoshi Nakagawa (Shimane University) for the pGWB vectors.
Glossary
- SEM
scanning electron microscopy
- Chr
chromosome
- cDNA
complementary DNA
- GC
gas chromatography
- MS
mass spectrometry
- KRI
Kovats retention index
- qRT
quantitative reverse transcription
- BSA
bovine serum albumin
Footnotes
This work was supported by the National Natural Science Foundation of China (grant for project no. 31170298 to B.-C.T.) and the Research Grants Council of the Hong Kong Special Administrative Region (grants for project nos. 473512 and 473611 to B.-C.T.).
The online version of this article contains Web-only data.
References
- Appleford NE, Evans DJ, Lenton JR, Gaskin P, Croker SJ, Devos KM, Phillips AL, Hedden P (2006) Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 223: 568–582 [DOI] [PubMed] [Google Scholar]
- Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Østergaard L (2010) Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Urrea A, Dellaporta SL (1999) Cell death and cell protection genes determine the fate of pistils in maize. Development 126: 435–441 [DOI] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Calderon-Urrea A (1994) The sex determination process in maize. Science 266: 1501–1505 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, Phinney BO, Takahashi N (1988) Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol 88: 1367–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, Freeling M (1989) Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 121: 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sullivan JA, Mould RM, Gray JC, Peacock WJ, Dennis ES (2001) A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J 28: 201–208 [DOI] [PubMed] [Google Scholar]
- Hirochika H. (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4: 118–122 [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai WC, Hanada A, Alonso JM, Ecker JR, et al. (2008) Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20: 320–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender Rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M (2001) Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Kim JC, Laparra H, Calderón-Urrea A, Mottinger JP, Moreno MA, Dellaporta SL (2007) Cell cycle arrest of stamen initials in maize sex determination. Genetics 177: 2547–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel’s stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA 110: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Bodén M, Kobe B (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813: 1562–1577 [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1997) Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94: 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Neuffer MG, England D (1995) Induced mutations with confirmed locations. Maize Genet Coop Newsl 69: 43–46 [Google Scholar]
- Nickerson NH. (1959) Sustained treatment with gibberellic acid of five different kinds of maize. Ann Mo Bot Gard 46: 19–37 [Google Scholar]
- Pandey A, Choudhary MK, Bhushan D, Chattopadhyay A, Chakraborty S, Datta A, Chakraborty N (2006) The nuclear proteome of chickpea (Cicer arietinum L.) reveals predicted and unexpected proteins. J Proteome Res 5: 3301–3311 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Phinney BO. (1956) Growth response of single-gene dwarf mutants in maize to gibberellic acid. Proc Natl Acad Sci USA 42: 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N, Chaparro C, Piegu B, Stenger W, Formey D, Llauro C, Descombin J, Sabot F, Lasserre E, Meynard D, et al. (2009) Identification of an active LTR retrotransposon in rice. Plant J 58: 754–765 [DOI] [PubMed] [Google Scholar]
- Rood SB, Pharis RP (1980) Changes of endogenous gibberellin-like substances with sex reversal of the apical inflorescence of corn. Plant Physiol 66: 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis M, Robertson M, Ali S, Lenton JR, Chandler PM (2004) Isolation of gibberellin metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor Appl Genet 109: 847–855 [DOI] [PubMed] [Google Scholar]
- Sponsel VM, Hedden P (2010) Gibberellin Biosynthesis and Inactivation. In Plant Hormones: Biosynthesis, Signal Transduction, Action! Davies Peter J. eds, Springer, New York, 63-94. [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J (1996) The dwarf-1 (dt) Mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. Proc Natl Acad Sci USA 93: 10515–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard PS. (2009) d4 is allelic to d1. Maize Genet Coop Newsl 83: 51 [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Kamiya Y, Takahashi N, Graebe JE (1986) Metabolism of gibberellins in a cell-free system from immature seeds of Phaseolus vulgaris L. Planta 168: 190–199 [DOI] [PubMed] [Google Scholar]
- Tan BC, Chen Z, Shen Y, Zhang Y, Lai J, Sun SS (2011) Identification of an active new mutator transposable element in maize. G3 (Bethesda) 1: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z (2013) ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J 73: 405–416 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WH, Wang YS, Liu GZ, Chen X, Tinjuangjun P, Pi LY, Song WY (2006) The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor-like kinase. Plant J 45: 740–751 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2006) Gibberellin biosynthesis in Arabidopsis. Phytochem Rev 5: 39–47 [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.