Abstract
Background
Adolescent drinking is an important public health concern, one that is influenced by both genetic and environmental factors. The functional variant rs1229984 in alcohol dehydrogenase 1B (ADH1B) has been associated at a genome-wide level with alcohol use disorders in diverse adult populations. However, few data are available regarding whether this variant influences early drinking behaviors and whether social context moderates this effect. This study examines the interplay between rs1229984 and peer drinking in the development of adolescent drinking milestones.
Methods
1,550 European and African American individuals who had a full drink of alcohol before age 18 were selected from a longitudinal study of youth as part of the Collaborative Study on the Genetics of Alcoholism (COGA). Cox proportional hazards regression, with GxE product terms in the final models, was used to study two primary outcomes during adolescence: age of first intoxication and age of first DSM-5 alcohol use disorder symptom.
Results
The minor A allele of rs1229984 was associated with a protective effect for first intoxication (HR=0.56, 95% CI 0.41–0.76) and first DSM-5 symptom (HR=0.45, 95% CI 0.26–0.77) in the final models. Reporting that most or all best friends drink was associated with a hazardous effect for first intoxication (HR=1.81, 95% CI 1.62–2.01) and first DSM-5 symptom (HR=2.17, 95% 1.88–2.50) in the final models. Furthermore, there was a significant GxE interaction for first intoxication (p=.002) and first DSM-5 symptom (p=.01). Among individuals reporting none or few best friends drinking, the ADH1B variant had a protective effect for adolescent drinking milestones, but for those reporting most or all best friends drinking, this effect was greatly reduced.
Conclusions
Our results suggest that the risk factor of best friends drinking attenuates the protective effect of a well-established ADH1B variant for two adolescent drinking behaviors. These findings illustrate the interplay between genetic and environmental factors in the development of drinking milestones during adolescence.
Keywords: Gene-Environment Interaction, Adolescent, Alcohol Dehydrogenase, Peer drinking
INTRODUCTION
By age 17, most U.S. adolescents (54%–78%) have consumed alcohol, and a significant proportion (15%) meet the criteria for alcohol abuse (Merikangas et al., 2010; NSDUH, 2012; Swendsen et al., 2012). Patterns of alcohol use that begin in adolescence are important determinants for the development of alcohol use disorders during adulthood (Grant et al., 2006; Pitkanen et al., 2005). Therefore, understanding factors that contribute to early drinking behaviors is critical for disease prevention.
For decades, twin studies have recognized that both genetic and environmental factors influence individual risk for alcoholism (Heath et al., 1997; Kendler et al., 1994; Pickens et al., 1991; Prescott and Kendler, 1999). Recently, large-scale genetic studies have provided strong evidence for the contribution of specific genetic variants to alcohol use disorders in adults (Rietschel and Treutlein, 2013; Wang et al., 2012). An important next step in the translation of genetic findings identified in adults is to test whether these genetic variants also affect adolescent drinking behaviors and whether environmental risk factors moderate this role.
Among the most biologically well-understood genetic variants associated with alcohol use disorders is the polymorphism rs1229984 in the enzyme alcohol dehydrogenase 1B (ADH1B). The minor A allele (in the coding strand) of rs1229984 causes an amino acid change at position 48 by replacing arginine with histidine, which increases the activity of the ADH1B enzyme that oxidizes ethanol to acetaldehyde (Edenberg and Foroud, 2013; Hurley and Edenberg, 2012). After consuming alcohol, elevated ADH1B activity has been hypothesized to transiently increase the level of acetaldehyde, leading to unpleasant effects that limit further drinking. Meta-analysis of this variant in Asian populations, where the rs1229984 A allele is common (allele frequency=0.7 in 1000 Genomes)(Abecasis et al., 2012), has demonstrated strong effects on the risk of developing alcohol-related disorders (OR 0.45: p=7×10−42) (Li et al., 2011). Recently, this polymorphism was shown to have a similar effect on risk of alcohol dependence in European and African Americans (African and European OR 0.34: p=6.6×10−10 (Bierut et al., 2012); European: p=1.17×10−31(Gelernter et al., 2014)), where the rs1229984 A allele is less common (European American frequency=0.05; African American frequency=0.02 in Exome Variant Server)(http://evs.gs.washington.edu/EVS/).
Other studies suggest that social environments that encourage drinking may diminish the protective genetic effects of alcohol metabolizing variants (Hasin et al., 2002; Higuchi et al., 1994; Irons et al., 2007; Irons et al., 2012). However, to our knowledge, no study has explored the interplay of the ADH1B rs1229984 variant and the important social context of peer drinking during the critical developmental period of adolescence when alcohol use is initiated and drinking patterns are established. Peer drinking has long been recognized as a strong risk factor for adolescent drinking problems (Curran et al., 1997; Reifman et al., 1998), and recently, twin studies have provided evidence that peer drinking modifies heritable variation in adolescent alcohol involvement (Agrawal et al., 2010; Dick et al., 2007; Guo et al., 2009; Harden et al., 2008).
This study tests the interaction between a genome-wide significant functional ADH1B variant and the risk environment of peer drinking in the development of two adolescent drinking milestones: first intoxication and first DSM-5 alcohol use disorder symptom. Examining hypothesis-driven gene-by-environment (GxE) interactions using robust genetic and environmental risks during developmental transitions provides an important approach for untangling the complex etiology of alcohol use disorder.
MATERIALS AND METHODS
COGA Sample Description
Study participants were enrolled in the Collaborative Study on the Genetics of Alcoholism (COGA), a large, multi-center, family study designed to identify genes that contribute to alcohol use disorders in high-risk (defined as recruited through alcohol dependent probands) and community comparison families (Begleiter et al., 1995). Since 2005, the adolescent and young adult study in COGA has used a longitudinal design to examine the development of alcohol use disorders in young participants from these families. Individuals aged 12 to 22 were recruited from six sites across the US and interviewed every two years. Institutional review boards at all sites approved the study design. Adult participants provided informed consent, parents provided consent for all children younger than 18, and children provided assent.
Assessment of Phenotypes
Interview assessment was performed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) to gather reliable and valid information on alcohol use behaviors (Bucholz et al., 1995; Bucholz et al., 1994; Hesselbrock et al., 1999). Participants 18 years and older were assessed with the Phase IV SSAGA, and those less than 18 years were assessed with an age appropriate adolescent version called the Phase IV C-SSAGA (Kuperman et al., 2001).
Drinking Milestones
Two adolescent drinking milestones were used as primary outcomes among adolescent ever-drinkers: age of first intoxication, a common and clinically relevant variable, and first DSM-5 alcohol use disorder symptom, a heritable characteristic associated with future alcohol-related problems (Rhee et al., 2003; Young et al., 2006). These outcomes commonly occur during adolescence and therefore coincide with the environment of adolescent peer drinking. Age of first intoxication was derived from responses to the question “How old were you the first time you got drunk, that is, your speech was slurred or you were unsteady on your feet?” Age of first DSM-5 symptom was developed from examining the youngest age that individuals first experienced one of the 11 symptoms of alcohol use disorder. Given the longitudinal design of this study with multiple assessments over time, the earliest interview in which the participant endorsed first intoxication or first DSM-5 symptom was selected to assign the age of onset.
Peer Drinking
The environment of adolescent peer drinking was derived from participant responses to questions addressing the proportion of best friends who drink. With the longitudinal design of the study, 88% (1366/1550) of participants received at least one adult SSAGA assessment at age 18 years or older. Assignment of the level of peer drinking in these participants was determined from the first adult SSAGA interview with the question “When you were 12-17, how many of your best friends used alcohol?” and the 4 possible answers of none, few, most, or all. For participants who had not reached age 18 at the last assessment, peer drinking was evaluated with the maximum value from all C-SSAGA answers to the question “How many of your best friends use alcohol?” For the primary analyses, peer drinking was dichotomized into low peer drinking (few or no best friends drink) and high peer drinking (most or all best friends drink) as done in previous studies (Kuperman et al., 2013). The four level peer drinking variable (none, few, most, or all best friends) was also investigated in secondary analyses to assess a possible dose response, but interaction effects are not presented because of the small number of individuals in some groups.
To assess the concordance of the retrospective SSAGA interview peer drinking responses for ages 12-17 with current peer drinking reported in C-SSAGA assessments, we compared the first adult SSAGA response and the maximum value from all C-SSAGA assessments among individuals with at least one adult and one child questionnaire. For the 996 participants with both adult and child interviews, 73% of peer drinking assignments had the same dichotomous variable (none/few vs most/all best friends). This concordance demonstrates that our retrospective approach of using the first SSAGA interview when available is a reasonable strategy to assess peer drinking across adolescence. It also shows that for the 12% of participants without a single adult SSAGA assessment, using the maximum value from CSSAGA assessments reasonably estimates the proportion of best friends drinking from ages 12-17.
Genotyping
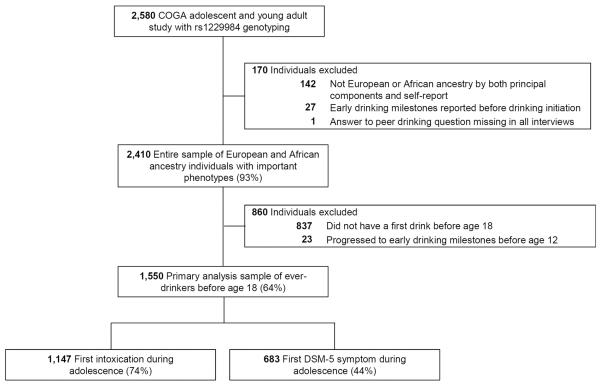
Blood samples were obtained for genetic analysis. The ADH1B rs1229984 variant was genotyped with Sequenom MassArray technology (Sequenom, San Diego, CA, USA) following standard procedures. Several quality control measures were employed. Genetic variants had a genotyping rate of greater than 99% and were in Hardy-Weinberg equilibrium in both the European and African American groups. The program PEDCHECK (O'Connell and Weeks, 1998) was used to examine Mendelian inheritance, and only individuals with no Mendelian inconsistencies were included in the rs1229984 genotyped sample (N=2580, Figure 1).
Fig. 1.
European and African American adolescent ever-drinkers with ADH1B rs1229984 genotyping were drawn from the Collaborative Study on the Genetics of Alcoholism (COGA) for the primary analyses of two early drinking milestones.
A set of 64 ancestry informative markers was genotyped as part of a 96 SNP Biorepository Panel by the Rutgers University Cell and DNA Repository. These markers were used in SNPrelate, a function in R, to assign ancestry groups. HapMap populations were included as reference groups. There was high concordance (97%) between self-reported and genetically determined ethnicity for European and African American individuals, and only concordant individuals were used in the analyses.
Sample Selection
In the COGA adolescent and young adult study, 2,580 individuals with a first interview age of 12 to 22 were genotyped for the ADH1B rs1229984 variant, and participants for the analyses were drawn from this group (Figure 1). Focusing on European and African American subjects and excluding individuals with missing or unreliable data left 2,410 individuals (entire sample described in Table 1). The samples used for the primary analyses of first intoxication and first DSM-5 symptom consisted of 1,550 ever-drinkers before age 18 (also described in Table 1). Ever-drinkers were targeted because the ADH1B variant is only expected to exhibit a protective effect in response to alcohol consumption. Because the peer drinking variable examined the age-range of 12-17, the primary analyses focused on events that occurred during this time.
Table 1.
Characteristics of samples used in analyses
| Characteristic | Entire sample (N=2,410) | Ever-drinkers before age 18 (N=1,550) |
|---|---|---|
| Ancestry, N (%) | ||
| European | 1,648 (68.4) | 1,130 (72.9) |
| African | 762 (31.6) | 420 (27.1) |
| Sex, N (%) | ||
| Males | 1,182 (49.1) | 784 (50.6) |
| Females | 1,228 (51.0) | 766 (49.4) |
| Age at first interview, years | ||
| Mean ± sd | 16.3 ± 3.2 | 16.7 ± 3.0 |
| Range | 12–22 | 12–22 |
| No. of interviews | ||
| Mean ± sd | 3.2 ± 1.1 | 3.2 ± 1.1 |
| Range | (1–5) | (1–5) |
| Family status, N (%) | ||
| From high-risk families | 2,096 (87.0) | 1,384 (89.3) |
| From comparison families | 314 (13.0) | 166 (10.7) |
| No. of extended families | 781 | 645 |
| No. of nuclear families | ||
| Only full-siblings | 1,629 | 1,151 |
| Including half-siblings | 1,438 | 1,044 |
| No. of individuals per extended family, median (range) | 2 (1–24) | 2 (1–17) |
| Drinking milestones reached before age 18, N (%) | ||
| First drink | 1,573 (65.3) | 1,550 (100.0) |
| First intoxication | 1,170 (48.6) | 1,147 (74.0) |
| First DSM-5 symptom | 702 (29.1) | 683 (44.1) |
| Among those who exhibit a first intoxication before age 18 | ||
| Mean age ± sd | 15.3 (1.5) | 15.4 (1.4) |
| Age range | 8–17 | 12–17 |
| Among those who exhibit a first DSM-5 symptom before age 18 | ||
| Mean age ± sd | 15.6 (1.3) | 15.6 (1.2) |
| Age range | 10–17 | 12–17 |
| rs1229984, N (%) | ||
| GG | 2,270 (94.2) | 1,452 (93.7) |
| GA | 137 (5.7) | 96 (6.2) |
| AA | 3 (0.1) | 2 (0.1) |
| Reported proportion of best friends who use alcohol between ages 12–17, N (%) | ||
| None | 746 (31.0) | 239 (15.4) |
| Few | 981 (40.7) | 708 (45.7) |
| Most | 513 (21.3) | 453 (29.2) |
| All | 170 (7.1) | 150 (9.7) |
Data Analysis
Data were analyzed using the Statistical Analysis System (SAS 9.3, Cary, NC, USA). Cox-Proportional Hazards Regression (SAS PROC PHREG) was used to model drinking milestones and all individuals who did not experience an event in adolescence were censored at their age of last interview or 18. Participants with rs1229984 GA genotype (N=96) and AA genotype (N=2) were collapsed into one group for comparison with the GG genotype participants (N=1,452), as done in previous studies (Bierut et al., 2012). Models were checked for violations of the proportional hazards assumption and Schoenfeld residuals were examined. The option COVSANDWICH (AGGREGATE) was used to statistically adjust for the non-independence of correlated familial data in all analyses, as done in previous studies (Kuperman et al., 2013).
Models in Primary Analyses
Main effects of the ADH1B variant and peer drinking were examined in univariate and multivariate models of age of first intoxication and first DSM-5 symptom in the sample of adolescent ever-drinkers (N=1,550, called univariate model set and multivariate model set, Table 2). All models presented in the tables employed STRATA statements for gender and ethnicity to adjust for differences in baseline hazards in these groups. The interplay between the ADH1B variant and peer drinking was assessed by adding product interaction terms to models of drinking milestones (called interaction model set, Table 2). This final proportional hazards model was λ(t)=λ(t)exp(β1*(rs1229984) + β2*(peer_drinking) + β3*(rs1229984*peer_drinking)). The possibility of a gene-environment correlation between ADH1B rs1229984 and peer drinking was also assessed because genetic factors influence selection of peers who drink (Fowler et al., 2007) and inadequate control of this correlation could produce false interactions. Using logistic regression, the outcome peer drinking was modeled with the variables of the ADH1B variant, gender, and ethnicity.
Table 2.
Cox proportional hazards regression models of adolescent drinking milestones
| Drinking milestones in ever-drinkers before age 18 (N=1,550) |
||||
|---|---|---|---|---|
| Models of first intoxication | Models of first DSM-5 symptom | |||
|
|
||||
| Hazard ratio (95% CI) | χ2 p value | Hazard ratio (95% CI) | χ2 p value | |
| Univariate model set | ||||
| rs1229984a | 0.72 (0.56−0.91) | .006 | 0.69 (0.50−0.94) | .02 |
| peer drinkingb | 1.89 (1.70−2.10) | <.0001 | 2.27 (1.98−2.60) | <.0001 |
| Multivariate model set | ||||
| rs 1229984 | 0.76 (0.61−0.96) | .02 | 0.73 (0.54−0.97) | .03 |
| peer drinking | 1.88 (1.69−2.09) | <.0001 | 2.26 (1.97−2.60) | <.0001 |
| Interaction model set | ||||
| rs 1229984 | 0.56 (0.41−0.76) | .0002 | 0.45 (0.26−0.77) | .004 |
| peer drinking | 1.81 (1.62−2.01) | <.0001 | 2.17 (1.88−2.50) | <.0001 |
| rs1229984*peer drinking | 2.10 (1.32–3.32) | .002 | 2.29 (1.21–4.30) | .01 |
| Examination of GxE term in interaction model set | ||||
| None/few best friends drink (GA/AA vs GG) | 0.56 (0.41–0.76) | .0002 | 0.45 (0.26–0.77) | .004 |
| Most/all best friends drink (GA/AA vs GG) | 1.16 (0.82–1.65) | .39 | 1.03 (0.73–1.45) | .87 |
Reference ADH1B rs1229984 genotype GG was compared to GA/AA;
Reference peer drinking status none/few best friends drink was compared to most/all best friends drink; All models adjusted for gender and ethnicity.
Secondary Analyses
Secondary analyses were performed to test the robustness of our primary findings. First, association of the ADH1B rs1229984 variant with the milestone of age of drinking initiation was examined in the entire sample, which included adolescent never-drinkers (N=2,410). Second, analyses stratified by ancestry were performed to examine the main and interaction effects within the subpopulations of European and African Americans.
RESULTS
Participant Characteristics
Demographic, behavioral, and genotypic characteristics of the study samples are presented in Table 1. The sample of ever-drinkers before age 18 used in the primary analyses consisted of 1,550 individuals from 1,151 nuclear families (defined by full-siblings) and 645 extended families. The mean first interview age was 17, 49% were female, and the majority came from high-risk families (89%) and were European American (73%). Before age 18, 74% had a first intoxication and 44% experienced a first DSM-5 symptom of alcohol use disorder. From ages 12 to 17, 39% reported that most or all of their best friends drank alcohol. Consistent with the expected population frequencies of the ADH1B variant, 6% carried at least one copy of the protective A allele (8% in European Americans and 3% in African Americans).
Effect of Peer Drinking
Most/all best friends drinking compared to none/few best friends drinking between ages 12-17 was associated with a main hazardous effect in univariate and multivariate models of early drinking behaviors (Table 2). In the final interaction model set with GxE product terms, self-reported peer drinking had a robust effect on first intoxication (Hazards ratio (HR)=1.81, 95% CI 1.62–2.01) and first DSM-5 symptom (HR=2.17, 95% CI 1.88–2.50). In secondary analyses examining all four responses for best friends drinking (none, few, most, all), an increase in the number of best friends drinking was similarly related to the first intoxication (multivariate model set with none as the reference; few HR=1.72, 95% CI 1.44–2.05; most HR=2.65, 95% CI 2.20–3.18; all HR=3.69, 95% CI 2.93–4.64) and first DSM-5 symptom (multivariate model set with none as the reference; few HR=2.43, 95% CI 1.77–3.33; most HR=4.29, 95% CI 3.12–5.92; all HR=5.84, 95% CI 4.16–8.21). These results indicate a “dosage effect” where the reported proportion of best friends drinking was positively associated with higher risk for developing adolescent drinking milestones.
Effect of ADH1B rs1229984 Variant
During adolescence, presence of the ADH1B variant (GA/AA genotypes) was associated with a protective main effect among ever-drinkers for first intoxication and first DSM-5 symptom in univariate and multivariate models (Table 2). In the final interaction model set with GxE product terms, the effect of the ADH1B variant was strong for both first intoxication (HR=0.56, 95% CI 0.41–0.76) and first DSM-5 symptom (HR=0.45, 95% CI 0.26–0.77). In secondary analyses of the entire sample that included never-drinkers, presence of the variant exhibited no effect on drinking initiation (HR in univariate model=1.12, 95% CI 0.92–1.36), consistent with the mechanism of the variant of only exhibiting an effect in response to alcohol consumption.
Interaction between ADH1B rs1229984 and Peer Drinking
The interaction between the ADH1B variant and peer drinking was tested by adding GxE product term to models of drinking milestones in adolescent drinkers (N=1,550), which illustrated a significant statistical interaction for first intoxication (p=.002) and first DSM-5 symptom (p=.01) (Table 2). Among individuals who reported none/few best friends drinking, the ADH1B GA/AA genotypes had a strong protective effect for first intoxication (HR=0.56, 95% CI 0.41–0.76) and first DSM-5 symptom (HR=0.45, 95% CI 0.26–0.77). In individuals who reported most/all best friends drinking, however, this protective effect was not observed for either first intoxication (HR=1.16, 95% CI 0.82–1.65) or first DSM-5 symptom (HR=1.03, 95% CI 0.73–1.45), as illustrated by the point estimates close to 1. Figure 2 more clearly illustrates this GxE interaction by presenting the survival estimates.
Fig. 2.
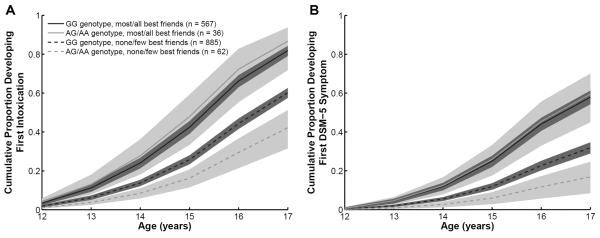
Cox proportional hazards regression survival estimates of (A) first intoxication and (B) first DSM-5 alcohol use disorder symptom in adolescent ever-drinkers (N=1,550) with the variables of ADH1B genotype, best friends drinking, and GXE interaction term.
Association between ADH1B Variant and Peer Drinking
No evidence of a gene-environment correlation between the ADH1B variant and peer drinking was observed. Specifically, the independent variable of the ADH1B rs1229984 variant was not significant in the logistic regression model of perceived peer drinking controlling for sex and ethnicity as covariates (most/all vs none/few best friends drink, Odds Ratio=1.19, 95% CI 0.78–1.83).
Assessment of Robustness of Results
The proportional hazards assumption was satisfied in first DSM-5 symptom models. Violations were noted in a subset of first intoxication analyses. Examination of Scholenfeld residuals indicated that the group of 17 year olds was driving this violation, perhaps reflecting important transitions at this age. Censoring at age 17 instead of 18 satisfied the proportional hazards assumption without substantially altering the parameter estimates, supporting our conclusions.
Ancestry-stratified analyses demonstrated consistent main and interaction effects in the European American subpopulation (N=1,130). In the interaction model set for European American individuals, peer drinking had a hazardous effect on first intoxication (HR=1.87, 95% 1.66–2.11) and first DSM-5 symptom (HR=2.23, 95% CI 1.89–2.63); rs1229984 had a protective effect on first intoxication (HR=0.60, 95% CI 0.44–0.82) and first DSM-5 symptom (HR=0.47, 95% CI 0.27–0.82); and interaction terms were significant (p<.02). The ADH1B GA/AA genotypes were protective among individuals reporting none/few best friends drinking, but not among those reporting most/all best friends drinking, corroborating our findings in the overall sample.
Stratified analyses of African Americans (N=420) provided trending evidence of main effects. In the interaction model set with GxE product terms, peer drinking had a hazardous effect on first intoxication (HR=1.62, 95% 1.27–2.08) and first DSM-5 symptom (HR=1.98, 95% CI 1.50–2.61); rs1229984 had a trending protective effect on first intoxication (HR=0.32, 95% CI 0.08–1.27) and first DSM-5 symptom (HR=0.35, 95% CI 0.05–2.28); and interaction terms were insignificant (p>.7). The limited sample size of African Americans combined with the low frequency of the rs1229984 minor allele limits power to detect interactions in this analysis. Nonetheless, the robust effect of peer drinking in both ancestry groups and the well-established role of rs1229984 across ancestry groups lends support for our conclusions drawn from the combined sample.
DISCUSSION
Alcohol use behaviors established during adolescence are important contributing factors for the later progression to alcohol dependence (Grant et al., 2006; Pitkanen et al., 2005). These data provide an example of the important interplay of genetic and environmental risks in the development of drinking milestones during this critical period of adolescence. Using a longitudinal sample of European and African American adolescent drinkers, we demonstrate that the ADH1B rs1229984 minor A allele is associated with a protective effect for early drinking behaviors, and in the environmental high-risk context of most or all best friends drinking, this genetic protection is negated.
The observation that the ADH1B variant is associated with a decreased risk of first intoxication and first DSM-5 symptom during adolescence (Table 2) extends previous findings that this variant protects against alcohol-related health problems in adulthood (Bierut et al., 2012; Gelernter et al., 2014; Li et al., 2011). Despite having an early role in the trajectory of drinking behaviors, the ADH1B variant was not associated with drinking initiation, consistent with the hypothesized mechanism of action that requires alcohol exposure (Edenberg and Foroud, 2013; Hurley and Edenberg, 2012). This specific example of a genetic variant that influences early drinking milestones, but not initiation, builds on twin and adoption study findings that genetic factors contribute to the development of adolescent alcohol-related problems, and environmental factors more strongly drive drinking initiation (Hopfer et al., 2003; Lynskey et al., 2010).
Beyond demonstrating an early protective role of the ADH1B GA/AA genotypes in the development of these drinking behaviors, the results illustrate that reporting most or all best friends drinking was associated with attenuation of this genetic protection (Figure 2). The observation that social context modifies the effect of an ADH1B variant extends previous studies on alcohol metabolizing variants. Higuchi et al. (1994) found that the proportion of alcohol dependent adults in Japan with one copy of a protective aldehyde dehydrogenase 2 (ALDH2) variant increased between 1979 and 1992, following the increased cultural pressure to drink alcohol. Similarly, Irons et al. (2007) reported that the high-risk environment of sibling substance use was associated with a diminished effect of this ALDH2 variant in East Asian adolescent adoptees, and more recently, this group demonstrated that high parental alcohol use and misuse reduced the effect of the ALDH2 protective allele (Irons et al., 2012). For the ADH1B rs1229984 variant, Hasin et al. (2002) observed a weaker protective role in certain groups, which was hypothesized to reflect differences in environmental exposure to heavy drinking. Our findings expand on these earlier observations by demonstrating that the critical high-risk social context of adolescent peer drinking is associated with the loss of the protective genetic effect of the ADH1B variant in European and African Americans.
Previous studies of metabolizing variants have focused on Asian populations where the ADH1B rs1229984 A allele is common, and only recently was this variant associated with alcoholism at a genome-wide level in an European and African American sample (p=6.6×10−10) (Bierut et al., 2012). A recent GWAS of alcohol dependence further supports a strong effect of this variant in European Americans (p=1.17×10−31) (Gelernter et al., 2014). To our knowledge, this study is the first to examine the effect of the ADH1B rs1229984 variant on adolescent drinking behaviors and incorporate environmental moderation in European and African Americans.
One challenge of studying the influence of the ADH1B rs1229984 variant in populations of European and African ancestry is the low frequency of the protective A allele. Although over 1,500 adolescent drinkers were examined in this analysis, only 98 (6%) carried an A allele (of which 36 reported most/all best friends drinking). Nonetheless, the influence of this variant and the GxE interaction was persistently strong in models of first intoxication and first DSM-5 symptom (Table 2). Secondary ancestry-stratified analyses also demonstrated consistent main and interaction effects in the European American subpopulation (N=1,130) and provided trending evidence of main effects in the African American subpopulation (N=420), where power was limited. These analyses, combined with previous studies supporting the protective role of rs1229984 across ancestry groups as well as the moderating effect of social environments, support our conclusion that this variant is associated with a protective effect for early drinking behaviors in European and African Americans, but this genetic protection may be eliminated by adolescent peer drinking.
The findings reported here have several limitations. First, studying a specific genetic variant provides limited information on the general genetic underpinnings of complex diseases such as alcohol use disorder (Dick and Kendler, 2012). Nevertheless, examination of specific robust variants provides important insight into underlying biological mechanisms that are not assessed by traditional studies of latent genetic influences. Second, other genetic variants may influence associations between ADH1B rs1229984 and drinking behaviors (Meyers et al., 2013; Toth et al., 2011). Third, self-reported peer drinking was viewed as an environmental risk factor in this study, but research suggests that genetic factors contribute to peer alcohol involvement (Fowler et al., 2007). Gene-environment correlations can arise when an individual's heritable behavior evokes an environmental response (evocative rGE) or when an individual possesses a heritable propensity to select an environment (active rGE). In this study, the ADH1B rs1229984 variant was not associated with self-reported peer drinking, supporting our interpretation that peer drinking acts as an environmental modifier, but other gene-environment correlations may still contribute to the observed effects. Fourth, the temporal ordering of peer drinking and the onset of drinking behaviors could not be assessed in this study (Table 1). It is possible that other risk factors correlated with peer drinking, such as parental monitoring or genetic risk for anti-social behavior, may account for the observed associations. Fifth, peer drinking was assessed by respondent report and may not reflect the actual proportion of best friends drinking. Finally, the majority of participants were from high-risk families, which may limit the generalizability of the findings. It is possible that only individuals at high-risk for alcohol use disorders lose the protective effect of the ADH1B rs1229984 variant under environments that encourage drinking. Replication of these findings in independent samples is a critical next step.
Despite these limitations, this study has several strengths. First, the analysis focused on a genetic variant with strong statistical and biological evidence for alcohol-related measures, which addresses common criticisms of GxE studies (Duncan and Keller, 2011; Joober et al., 2007; Risch et al., 2009). Second, focusing on a youth population and employing a longitudinal study design reduced recall bias, enabling more accurate assessment of drinking behaviors during the critical period of adolescence. Third, the robust environment of respondent report of best friends drinking from ages 12–17 coincided with the timing of the primary outcomes under study. This analysis focused on drinking behaviors that are common in adolescence and therefore are more likely to be directly influenced by peer drinking during this period. Finally, studying adolescent drinking milestones facilitated the characterization of the unfolding of genetic and environmental risks across development. Recent studies further support the discovery potential of examining genetic variants during important behavioral transitions in at-risk youth (Belsky et al., 2013; Dick et al., 2013). Future research on alcohol use disorders may benefit from similar hypothesis-driven study designs that examine well-established genes and environments during critical developmental periods.
From a public health perspective, this study provides a genetic argument in support of early social interventions to decrease affiliation with peer drinkers. Specifically, these findings support the use of a screening tool for practitioners to identify at-risk youth, developed by the National Institute on Alcohol Abuse and Alcoholism and the American Academy of Pediatrics, in which the first question addresses friends' drinking (NIAAA, 2011). Under the high-risk environment of best friends drinking, all adolescents were at increased risk for early drinking problems, and particularly, those at lower genetic risk experienced the greatest added risk. This study serves as a model of how understanding the interplay between genes and environments may increase etiological knowledge of alcohol use disorders and potentially inform interventions that aim to disrupt progression to alcoholism.
Acknowledgements
This work was supported by U10AA008401 from the National Institutes of Health. EO was supported by T32GM07200, UL1TR000448, and TL1TR000449. AA was supported by K02DA032573 and R21AA021235. DMD was supported by K02AA018755.
AA has previously received peer-reviewed funding from ABMRF/Foundation for Alcohol Research, which receives support from brewers. VMH is a member of the scientific advisory board of D&A Pharma, Paris, France. JCW, AG, and LJB are listed as inventors on Issued U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
Footnotes
Disclosures: The other authors declare no conflict of interest.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Balasubramanian S, Smith EK, Madden PA, Bucholz KK, Heath AC, Lynskey MT. Peer substance involvement modifies genetic influences on regular substance involvement in young women. Addiction (Abingdon, England) 2010;105:1844–1853. doi: 10.1111/j.1360-0443.2010.02993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T, Schuckit M, Edenberg H, Rice J. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Baker TB, Biddle AK, Evans JP, Harrington H, Houts R, Meier M, Sugden K, Williams B, Poulton R, Caspi A. Polygenic risk and the developmental progression to heavy, persistent smoking and nicotine dependence: evidence from a 4-decade longitudinal study. JAMA psychiatry. 2013;70:534–542. doi: 10.1001/jamapsychiatry.2013.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Molecular psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of studies on alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Hesselbrock VM, Shayka JJ, Nurnberger JI, Jr, Schuckit MA, Schmidt I, Reich T. Reliability of individual diagnostic criterion items for psychoactive substance dependence and the impact on diagnosis. Journal of studies on alcohol. 1995;56:500–505. doi: 10.15288/jsa.1995.56.500. [DOI] [PubMed] [Google Scholar]
- Curran PJ, Stice E, Chassin L. The relation between adolescent alcohol use and peer alcohol use: a longitudinal random coefficients model. Journal of consulting and clinical psychology. 1997;65:130–140. doi: 10.1037//0022-006x.65.1.130. [DOI] [PubMed] [Google Scholar]
- Dick DM, Cho SB, Latendresse SJ, Aliev F, Nurnberger JI, Jr, Edenberg HJ, Schuckit M, Hesselbrock VM, Porjesz B, Bucholz K, Wang JC, Goate A, Kramer JR, Kuperman S. Genetic influences on alcohol use across stages of development: GABRA2 and longitudinal trajectories of drunkenness from adolescence to young adulthood. Addiction biology. 2013 doi: 10.1111/adb.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Kendler KS. The impact of gene-environment interaction on alcohol use disorders. Alcohol research : current reviews. 2012;34:318–324. [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Changing environmental influences on substance use across development. Twin research and human genetics : the official journal of the International Society for Twin Studies. 2007;10:315–326. doi: 10.1375/twin.10.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. The American journal of psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. Genetics and alcoholism. Nature reviews. Gastroenterology & hepatology. 2013;10:487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [Accessed May, 2014]. Available at: URL: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- Fowler T, Shelton K, Lifford K, Rice F, McBride A, Nikolov I, Neale MC, Harold G, Thapar A, van den Bree MB. Genetic and environmental influences on the relationship between peer alcohol use and own alcohol use in adolescents. Addiction (Abingdon, England) 2007;102:894–903. doi: 10.1111/j.1360-0443.2007.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JD, Scherrer JF, Lynskey MT, Lyons MJ, Eisen SA, Tsuang MT, True WR, Bucholz KK. Adolescent alcohol use is a risk factor for adult alcohol and drug dependence: evidence from a twin design. Psychological medicine. 2006;36:109–118. doi: 10.1017/S0033291705006045. [DOI] [PubMed] [Google Scholar]
- Guo G, Elder GH, Cai T, Hamilton N. Gene-environment interactions: Peers' alcohol use moderates genetic contribution to adolescent drinking behavior. Social Science Research. 2009;38:213–224. doi: 10.1016/j.ssresearch.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Hill JE, Turkheimer E, Emery RE. Gene-environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behavior genetics. 2008;38:339–347. doi: 10.1007/s10519-008-9202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr L, Li TK. Alcohol and ADH2 in Israel: Ashkenazis, Sephardics, and recent Russian immigrants. The American journal of psychiatry. 2002;159:1432–1434. doi: 10.1176/appi.ajp.159.8.1432. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychological medicine. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction (Abingdon, England) 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet. 1994;343:741–742. doi: 10.1016/s0140-6736(94)91629-2. [DOI] [PubMed] [Google Scholar]
- Hopfer CJ, Crowley TJ, Hewitt JK. Review of twin and adoption studies of adolescent substance use. Journal of the American Academy of Child and Adolescent Psychiatry. 2003;42:710–719. doi: 10.1097/01.CHI.0000046848.56865.54. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ. Genes encoding enzymes involved in ethanol metabolism. Alcohol research : current reviews. 2012;34:339–344. [PMC free article] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, McGue M. Developmental trajectory and environmental moderation of the effect of ALDH2 polymorphism on alcohol use. Alcoholism, clinical and experimental research. 2012;36:1882–1891. doi: 10.1111/j.1530-0277.2012.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons DE, McGue M, Iacono WG, Oetting WS. Mendelian randomization: a novel test of the gateway hypothesis and models of gene-environment interplay. Development and psychopathology. 2007;19:1181–1195. doi: 10.1017/S0954579407000612. [DOI] [PubMed] [Google Scholar]
- Joober R, Sengupta S, Schmitz N. Promoting measured genes and measured environments: on the importance of careful statistical analyses and biological relevance. Archives of general psychiatry. 2007;64:377–378. doi: 10.1001/archpsyc.64.3.377. author reply 378–379. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. The American journal of psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kuperman S, Chan G, Kramer JR, Wetherill L, Bucholz KK, Dick D, Hesselbrock V, Porjesz B, Rangaswamy M, Schuckit M. A model to determine the likely age of an adolescent's first drink of alcohol. Pediatrics. 2013;131:242–248. doi: 10.1542/peds.2012-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman S, Schlosser SS, Kramer JR, Bucholz K, Hesselbrock V, Reich T, Reich W. Risk domains associated with an adolescent alcohol dependence diagnosis. Addiction (Abingdon, England) 2001;96:629–636. doi: 10.1046/j.1360-0443.2001.96462911.x. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biological psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey MT, Agrawal A, Heath AC. Genetically informative research on adolescent substance use: methods, findings, and challenges. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:1202–1214. doi: 10.1016/j.jaac.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, He JP, Burstein M, Swanson SA, Avenevoli S, Cui L, Benjet C, Georgiades K, Swendsen J. Lifetime prevalence of mental disorders in U.S. adolescents: results from the National Comorbidity Survey Replication--Adolescent Supplement (NCS-A) Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:980–989. doi: 10.1016/j.jaac.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers JL, Shmulewitz D, Aharonovich E, Waxman R, Frisch A, Weizman A, Spivak B, Edenberg HJ, Gelernter J, Hasin DS. Alcohol-Metabolizing Genes and Alcohol Phenotypes in an Israeli Household Sample. Alcoholism, clinical and experimental research. Alcoholism, clinical and experimental research. 2013;37:1872–1881. doi: 10.1111/acer.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAAA . National Institute on Alcohol Abuse and Alcoholism. Alcohol Screening and Brief Intervention for Youth: A Practitioner's Guide; National Institute on Alcohol Abuse and Alcoholism Publications Distribution Center; Rockville, MD: 2011. [Google Scholar]
- NSDUH [Accessed February 28, 2014];Results from the 2012 National Survey on Drug Use and Health: Detailed Tables. Available at: http://www.samhsa.gov/data/NSDUH/2012SummNatFindDetTables/Index.aspx.
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American journal of human genetics. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Archives of general psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Pitkanen T, Lyyra AL, Pulkkinen L. Age of onset of drinking and the use of alcohol in adulthood: a follow-up study from age 8–42 for females and males. Addiction (Abingdon, England) 2005;100:652–661. doi: 10.1111/j.1360-0443.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. The American journal of psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Reifman A, Barnes GM, Dintcheff BA, Farrell MP, Uhteg L. Parental and peer influences on the onset of heavier drinking among adolescents. Journal of studies on alcohol. 1998;59:311–317. doi: 10.15288/jsa.1998.59.311. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Archives of general psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J. The genetics of alcohol dependence. Annals of the New York Academy of Sciences. 2013;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA : the journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swendsen J, Burstein M, Case B, Conway KP, Dierker L, He J, Merikangas KR. Use and abuse of alcohol and illicit drugs in US adolescents: results of the National Comorbidity Survey-Adolescent Supplement. Archives of general psychiatry. 2012;69:390–398. doi: 10.1001/archgenpsychiatry.2011.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Fiatal S, Petrovski B, McKee M, Adany R. Combined effect of ADH1B RS1229984, RS2066702 and ADH1C RS1693482/ RS698 alleles on alcoholism and chronic liver diseases. Disease markers. 2011;31:267–277. doi: 10.3233/DMA-2011-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kapoor M, Goate AM. The genetics of substance dependence. Annual review of genomics and human genetics. 2012;13:241–261. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Rhee SH, Stallings MC, Corley RP, Hewitt JK. Genetic and environmental vulnerabilities underlying adolescent substance use and problem use: general or specific? Behavior genetics. 2006;36:603–615. doi: 10.1007/s10519-006-9066-7. [DOI] [PubMed] [Google Scholar]