Abstract
Understanding the mechanisms underlying ErbB3 over-expression in breast cancer will facilitate the rational design of therapies to disrupt ErbB2-ErbB3 oncogenic function. While ErbB3 over-expression is frequently observed in breast cancer, the factors mediating its aberrant expression are poorly understood. In particular, the ErbB3 gene is not significantly amplified, raising the question as to how ErbB3 over-expression is achieved. In this study we demonstrate that the ZNF217 transcription factor, amplified at 20q13 in ~20% of breast tumors, regulates ErbB3 expression. Analysis of a panel of human breast cancer cell lines (n = 50) and primary human breast tumors (n=15) demonstrated a strong positive correlation between ZNF217 and ErbB3 expression. Ectopic expression of ZNF217 in human mammary epithelial cells induced ErbB3 expression while ZNF217 silencing in breast cancer cells resulted in decreased ErbB3 expression. While ZNF217 has previously been linked with transcriptional repression due to its close association with CtBP1/2 repressor complexes, our results demonstrate that ZNF217 also activates gene expression. We demonstrate that ZNF217 recruitment to the ErbB3 promoter is CtBP1/2-independent and that ZNF217 and CtBP1/2 play opposite roles in regulating ErbB3 expression. In addition, we identify ErbB3 as one of the mechanisms by which ZNF217 augments PI-3K/Akt signaling.
Keywords: ZNF217, ErbB3, CtBP2, 20q13, breast cancer
Introduction
Over-expression of members of the ErbB family of receptor tyrosine kinases is frequently observed in cancer and correlates with poor patient prognosis and therapeutic resistance. Aberrant activation of ErbB3 has been linked to decreased survival in ovarian, prostate, breast, pancreatic and lung cancer (Sithanandam and Anderson, 2008) and has been functionally implicated in resistance to the targeted therapies Tamoxifen (Liu et al., 2007, Folgiero et al, 2008), Trastuzumab (Wang et al., 2008) and Gefitinib (Sergina et al, 2007, Engelman et al., 2007). Consequently, ErbB3 is evolving into an attractive therapeutic target in its own right. ErbB3 is unique in that it has an impaired tyrosine kinase activity (Guy et al., 1994) and in this respect, cannot be directly targeted although recent findings may revise this model (Shi et al., 2010). Various strategies for interfering with ErbB3 function have been examined, including siRNA (Sithanandam and Anderson, 2008) and inhibition of ADAM-mediated ligand cleavage (Zhou et al., 2006). A more thorough understanding of the endogenous mechanisms regulating ErbB3 expression will aid in the design of strategies to interfere with ErbB3 function.
In breast cancer, ErbB3 plays an essential role in ErbB2-driven breast cancer, with the ErbB2-ErbB3 heterodimer functioning as an “oncogenic unit”. In fact, ErbB3 is required for ErbB2-dependent breast tumor cell proliferation (Holbro et al., 2003) and strongly enhances ErbB2-dependent metastasis (Xue et al., 2006). While ErbB2 gene amplification is observed in approximately 20% of breast cancers, ErbB3 gene amplification is not a common event and mechanisms driving ErbB3 over-expression are not well understood. Interestingly, very little is known about the transcriptional regulation of ErbB3. Here, we report that the ZNF217 transcription factor, found amplified at 20q13, enhances ErbB3 transcription and contributes to ErbB3 protein expression in breast cancer cells.
20q13 amplification is associated with high histological grade, aneuploidy, high S-phase fraction, and with short disease-free survival of patients with node negative breast cancer (Tanner et al., 1995). The mapping of a commonly amplified region at 20q13 led to the positional cloning of the gene encoding ZNF217 (Collins et al., 1998). The ZNF217 gene is amplified in a variety of tumor types, such as breast (Kallioniemi et al.,1994), pancreas (Solinas-Toldo et al., 1996), ovarian (Iwabuchi et al., 1995), and colon (Schlegel et al., 1995). Retroviral transduction of normal human mammary epithelial cells (HMECs) and ovarian surface epithelial cells (OSE) with ZNF217 can facilitate immortalization (Nonet et al., 2001, Li et al., 2007), supporting a role for ZNF217 in carcinogenesis.
Protein motif analysis indicates that the ZNF217 gene encodes a transcription factor with eight C2H2 Kruppel-like DNA-binding motifs and a proline-rich transactivation domain at the C-terminus (Collins et al., 1998). ZNF217 has been reported to physically interact with C-terminal Binding Protein (CtBP) (Quinlan et al., 2006), a known co-repressor, and biochemically purify with histone deacetylases, histone methyltransferases, and other proteins associated with transcriptional repressor complexes (Shi Y et al., 2003). Since ZNF217 is thought to be a DNA-binding protein, it has been proposed that ZNF217 functions in gene repression by recruiting CtBP and an associated repressor complex to DNA (Chinnadurai, 2007). CtBP family members CtBP1 and CtBP2 are found together in repressor complexes suggesting redundant transcriptional regulatory roles (Hildebrand and Soriano, 2002). Various studies using mouse fibroblast cells null for CtBP1/2 have revealed both redundant and unique transcriptional regulatory roles for CtBP1 and CtBP2 during development, although mechanisms underlying the individual roles are still unclear (Chinnadurai, 2009).
Over-expression of ZNF217 may provide a selective advantage to tumor cells by interfering with pathways associated with normal regulation of growth, death, or differentiation. Previously published work has demonstrated that over-expressed ZNF217 attenuates apoptotic events triggered by transiently induced telomere dysfunction or doxorubicin-induced DNA damage (Huang et al., 2005). This work connected elevated levels of ZNF217 with increased phosphorylation of the serine/threonine kinase AKT, a known mediator of apoptosis resistance. This finding prompted a search to identify the ZNF217 target genes upstream of the PI3K/AKT pathway and our current studies make the novel finding that ZNF217 contributes to ErbB3 over-expression in breast cancer.
RESULTS
ZNF217 and ErbB3 expression correlate in breast cancer cells and primary tumors
To identify novel transcriptional targets of ZNF217, expression profiling was used to generate a list of genes displaying altered expression upon RNAi-mediated silencing of ZNF217 (Krig et al., 2007). Data analysis identified the ErbB3 receptor tyrosine kinase as a candidate transcriptional target of ZNF217. We therefore examined ZNF217 and ErbB3 transcript levels in a panel of 50 breast cancer cell lines (Chin et al., 2006). The breast cancer cell lines are listed along with the corresponding log2 signals for ZNF217 and ErbB3 RNA levels in supplementary Table 1. These values are graphed in Figure 1. Regression models were fitted to assess the strength of the apparent association, using the R statistical package (http://www.r-project.org/). On average, we found that a two-fold difference in ErbB3 expression between cell lines was associated with a 26% higher level of ZNF217 RNA (95% confidence interval, 15% to 38% increase). This linear relationship on the log2 scale was statistically significant (P<0.001), with a correlation coefficient of 0.69, accounting for 47% of the variation in ZNF217 (r2 =0.47) (Figure 1). This positive correlation suggested that, despite its reported association with transcriptional repressor proteins (Shi Y et al., 2003; Shi Y. J. et al., 2005), ZNF217, when bound to the ErbB3 promoter, activates, rather than represses, transcriptional activity.
Figure 1. ErbB3 and ZNF217 mRNA expression in 50 breast cancer cell lines.
Expression data for 50 breast cancer cell lines were determined using Affymetrix U133A arrays. A single probe set represents ErbB3 expression (202454_s_at) and ZNF217 expression (203739_at) in this dataset (Chin et al., 2006). The breast cancer cell lines are listed with the corresponding relative log2 ratio for ZNF217 and ErbB3 in Supplementary Table 1. The correlation coefficient (determined by Pearson's method of correlation analysis) between ErbB3 and ZNF217 was 0.69 (R2=0.47), indicating a significant linear association between the expression of these two genes.
To determine if the correlation between transcript levels observed in the breast cancer cell lines was reflected at the protein level in primary human breast tumors, we next screened 15 breast tumor lysates. Densitometry of immunoblots probed with antibodies to ErbB3, ZNF217, and actin (control) revealed a strong association between ZNF217 and ErbB3 protein levels (r2=0.7) (Figures 2A&B). A doubling of ErbB3 protein level across these samples was associated with a 75% higher level of ZNF217 (95% confidence interval, 41% to 132% increase, P<0.001).
Figure 2. ErbB3 and ZNF217 protein expression in human and murine specimens.
(A) An immunoblot of 15 human primary breast tumor lysates probed with antibodies to ZNF217, ErbB3 and actin (loading control). (B) The intensities of the ZNF217 and ErbB3 signals relative to actin were quantified by densitometry and plotted. (C) Immunoblot of lysates from wild-type FvB mouse mammary tissue versus matched normal and tumor tissues from NDL 2-5 mice probed with antibodies to ErbB3, ZNF217 and actin.
In another test of the link between ZNF217 and ErbB3 levels, we analyzed tissue lysates from the NDL murine mammary tumor model in which ErbB2 expression is driven by the MMTV promoter/enhancer (Siegelet al., 1999). In this model, transgenic over-expression of activated ErbB2 (NDL2-5 or Neu deletion mutant) in mouse mammary epithelium yields focal adenocarcinomas that evolve after long latency and metastasize to lungs with high frequency (Ursini-Siegel et al., 2007). Previous studies have shown that endogenous ErbB3 protein levels are elevated 10- to 15-fold in NDL tumor tissue, underscoring the link between ErbB2 and ErbB3 in mammary tumorigenesis (Siegel et al., 1999). We examined the expression of ErbB3 and ZNF217 in normal mammary glands from non-transgenic age-matched FvB littermates, in normal tissue from two independent NDL2-5 transgenic mice, and in tumor tissues from these mice (Figure 2C). Endogenous ErbB3 protein expression was significantly elevated in tumor tissues, as previously reported (Siegel et al., 1999), as was ZNF217 protein.
Over-expression of ZNF217 up-regulates endogenous ErbB3 expression in HMECs
Previous studies (Nonetet al., 2001) found that expression of retrovirally-introduced ZNF217 in HMECs could lead to immortalization. We analyzed total ZNF217 and ErbB3 protein levels in HMECs before immortalization (p9), after retroviral infection (p10), and after immortalization (p28), and found that ErbB3 levels corresponded to those of ZNF217 (Figure 3A). To model ZNF217 gene amplification and directly assess its role in regulating ErbB3 expression, we infected HMECs (which express modest endogenous ZNF217 levels) with either control adenovirus or one expressing ZNF217. RNA and protein lysates were collected at several time-points post-infection. ZNF217 protein levels accumulated with time, as expected. At the 48 hour time-point where ZNF217 expression was the most robust, expression of ErbB3 mRNA and protein were also significantly increased (Figures 3B & C). Moreover, the results indicated that infection with the ZNF217 adenovirus led to a detectable increase in ErbB3 transcripts as early as 16 hours post-infection. These results indicate that ZNF217 overexpression is sufficient to up-regulate ErbB3.
Figure 3. Exogenous ZNF217 gene expression enhances endogenous ErbB3 expression in HMECs.
(A) Protein expression levels of ZNF217 and ErbB3 in HMEC 184 before (passage 9) and after infection (passage 10) with ZNF217-encoding retroviruses, and after immortalization (passage 28). (B) Immunoblot of ZNF217 and ErbB3 protein expression in HMECs infected with control or ZNF217- encoding adenoviruses (C) Relative levels of ErbB3 mRNA in HMECs infected with control or ZNF217-encoding adenoviruses, determined at indicated time-points by Taqman real-time PCR analyses. Actin was used as an internal control. Note that error bars are present but very small.
Silencing ZNF217 reduces ErbB3 expression
To further implicate ZNF217 in ErbB3 regulation, we depleted ZNF217 with ZNF217-specific siRNA in two different human breast cancer cell lines in which it is robustly expressed (Figure 4A, MCF7, left panel; ZR75-1, right panel). Depletion of ZNF217 led to significant decreases in endogenous ErbB3 protein (Figure 4A) and transcript (Figure 4B; MCF7 cells), demonstrating that ZNF217 is required for robust ErbB3 expression. Since ZNF217 has been previously characterized as a transcriptional repressor, we next examined its impact on the activity of the ErbB3 promoter. To accomplish this, we performed a dual luciferase reporter assay. MCF7 cells were co-transfected with an ErbB3 promoter-driven luciferase reporter along with control or ZNF217 expression vectors and assayed for luciferase activity. Although MCF7 cells express high levels of endogenous ZNF217, ectopic ZNF217 expression increased the relative luciferase activity by approximately three-fold compared to the control (Figure 4C, left panel). Conversely, ZNF217 depletion in ZR75-1 (Figure 4C, right panel) and MCF7 cells (not shown) led to a significant decrease in reporter activity. Taken together, these results support the conclusion that ZNF217 regulates ErbB3 expression through activation of the ErbB3 promoter.
Figure 4. Silencing ZNF217 reduces ErbB3 transcript and protein expression.
(A) Scrambled control or ZNF217 siRNAs were transiently transfected into MCF7 and ZR75-1 cells. Lysates were harvested at 72h post-transfection and subjected to immunoblotting with ZNF217, ErbB3 and actin antibodies. Densitometric analysis of ZNF217 and ErbB3 relative to actin from three independent experiments is shown for both cell lines. (B) RNA samples from MCF7 cells treated in triplicate for 72 h with scrambled control or ZNF217-specific siRNAs were subjected to Taqman real-time PCR to determine relative ZNF217 and ErbB3 mRNA levels. Actin mRNA levels were used as an internal standard. Note that error bars are present but very small. (C) MCF7 cells were co-transfected with control or ZNF217 expression construct and an ErbB3-promoter driven luciferase reporter construct. Lysates were harvested 24h post-transfection and assayed for firefly and renilla luciferase activity using Promega Dual Luciferase Kit. Relative luciferase activity is shown. (D) ZR75-1 cells were co-transfected with scrambled control or ZNF217 siRNA and an ErbB3-promoter driven luciferase reporter construct. Cells were lysed at 24h post-transfection and assayed for firefly luciferase and beta-galactosidase activity (as an internal control). Relative luciferase activity is shown for a representative experiment performed in triplicate. p values for all comparisons in (A) through (D) are < 0.01.
ZNF217 and CtBP2 bind to the ErbB3 proximal promoter
Earlier ChIP-chip array data indicated that ZNF217 and CtBP1/2 binding overlapped at the majority (~75%) of promoters where they were detected (Krig et al., 2007). However, ZNF217-CTBP complexes have been implicated in transcriptional repression (Cowger et al., 2006). To determine whether both proteins are bound to the ErbB3 promoter, which our data suggests is activated rather than repressed by ZNF217, we performed chromatin immunoprecipitation (ChIP) using specific antibodies to ZNF217 and CtBP2. MCF7 lysates immunoprecipitated with either ZNF217 or CtBP2 antibodies show enrichment at the promoter sequence −458 to −200 upstream of the transcription start site (TSS) identified as the ZNF217 binding sequence from ChIP-chip array experiments (Krig et al, 2007) (Figure 5A and 5B). ZNF217 and CtBP2 also show weak but detectable binding approximately 3kb upstream from the TSS.
Figure 5. ZNF217 and CtBP2 occupy the ErbB3 proximal promoter in MCF7 breast cancer cells.
(A) Cartoon illustrating the locations of the sequences amplified in the ChIP studies shown in B and C. (B) Far right: PCR-amplified DNA enriched by immunoprecipitation with antibodies to ZNF217 or CtBP2, or with IgG (negative control) and 1:50 dilutions of un-enriched lysate (Input). Left: Quantitative PCR on ZNF217 ChIP (far left) and CtBP2 ChIP (middle) at the erbB3 proximal promoter versus 3kb upstream. (C) PCR-amplified DNA enriched by immunoprecipitation with antibodies to ZNF217, CtBP1, CTBP2, coREST, PolII, indicated histone modification marks, IgG (negative control), or 1:50 dilution of input are shown for comparison at the proximal erbB3 promoter. Results for ChIP-qPCR with primers amplifying the proximal region of the ErbB3 promoter are shown below gel panels. (D). ChIP-qPCR was performed on DNA enriched by immunoprecipitation with antibodies to ZNF217, CtBP2, AcH3K9, or TrimeK4 from MCF7 cells treated with siRNA to ZNF217 or scrambled control. Relative enrichment between siZNF217 and scramble control is compared (also graphed as fold change in Supplementary Figure 1). For (B) through (D), all qPCR was performed in triplicate and graphed as averaged relative DNA over 1:50 input for each ChIP sample (relative enrichment). A representative experiment is shown.
Since the ZNF217-CtBP complex has thus far been associated with transcriptional repression, we next examined a series of histone “marks” to probe the chromatin state of the ErbB3 proximal promoter (−458 to −200) where both ZNF217 and CtBP1/2 reside. Chromatin immunoprecipitations were performed on MCF7 lysates with antibodies for histone activation and repression marks along with an antibody for RNA polymerase II (Pol II). The histone activation marks, AcH3K9 and TrimeK4, were clearly evident at the ErbB3 proximal promoter, along with a robust signal for Pol II (Figure 5C top panel). In contrast to the activation marks, histone methylation marks (TrimeK9 and TrimeK27) commonly associated with promoter repression (Martin and Zhang, 2005) were notably absent from the ErbB3 proximal promoter (Figure 5C lower panel), despite the presence of CTBP1 and its repressor partner, coREST (Shi Y et al., 2003; Cowger et al., 2007). The same DNA was used to analyze the status of the NRK gene, previously shown to be repressed by ZNF217 (Krig et al., 2007). We found that ZNF217, CtBP1/2 and coREST were bound to the proximal promoter of the NRK gene. In contrast to the ErbB3 promoter, TrimeK27 was clearly present at the NRK promoter along with modest levels of TrimeK9 (Supplementary Figure 1). Collectively, the presence of robust activation marks and the absence of any repressive marks at the ErbB3 promoter support the conclusion that ZNF217 occupancy at the ErbB3 promoter is associated with gene activation rather than repression.
To directly examine the link between ZNF217 and the activation marks, we depleted ZNF217 by siRNA in MCF7 cells and performed chromatin immunoprecipitation with antibodies to ZNF217, CtBP2, AcH3K9 and trimeK4. Depletion of ZNF217 reduced the ZNF217 occupancy of the ErbB3 promoter, as expected (Figure 5D, top panel). CtBP2 levels were reduced commensurate with ZNF217 depletion suggesting that CtBP2 is recruited to the ErbB3 promoter, at least in part, through interaction with ZNF217 (Figure 5D, top panel). In agreement with this, ZNF217 and CtBP are known to physically interact through the PXDLS protein motif (Qunilan et al., 2006). Interestingly, ZNF217 depletion dramatically reduced the TrimeK4 levels but had no significant effect on AcH3K9 levels (Figure 5D and Supplementary Figure 2).
CtBP function in ErbB3 regulation
Our data support the conclusion that ZNF217 activates rather than represses the ErbB3 promoter. However, ZNF217 is found at the ErbB3 proximal promoter along with CtBP1/2, a known ZNF217 interaction partner and transcriptional repressor. To more clearly delineate the individual roles of ZNF217 and CtBP1/2 in ErbB3 transcriptional regulation, we turned to a defined genetic model, CtBP1/2 null mouse embryo fibroblasts (MEFs, a gift from the Hildebrand lab). Lysates were collected from wild type MEFs as well as CtBP1/2 null MEFs and subjected to immunoblotting. As shown in Figure 6A, CtBP1/2 protein was not detectable in the null MEFs, as expected. Interestingly, ErbB3 protein was dramatically up-regulated in the null MEFs, strongly suggesting that CtBP1/2 represses ErbB3 transcription.
Figure 6. CtBP2 represses ErbB3 expression.
(A) Protein lysates from wildtype and CtBP1/2 null MEFs were immunoblotted for ErbB3, CtBP1, CtBP2, and actin. (B) Mouse primers for the comparable ErbB3 proximal promoter sequence were PCR-amplified on DNA from wildtype or CtBP1/2 null MEFs enriched by immunoprecipitation with antibodies to ZNF217, CtBP2, indicated histone modification marks or IgG (negative control). 1:50 dilutions of input also shown. ChIP-qPCR was performed in triplicate and is shown as relative enrichment of immunoprecipitated DNA for mek27 and ZNF217 (left lower graph) and AcH3K9 and trimeK4 (right lower graph). (C) CtBP1/2 null MEFs were transiently transfected with a CtBP2 expression construct or empty control vector and the levels of ErbB3, CtBP2, and actin were analyzed by immunoblotting. Densitometric analysis of ErbB3 from three independent experiments is shown in the right panel. p < 0.05. (D) CtBP1/2 null MEFs were transiently transfected with a CtBP2 expression construct and an ErbB3-promoter driven luciferase reporter construct. Lysates were collected at 24h and assayed for firefly luciferase and beta-galactosidase activity. Relative luciferase activity is shown for a representative experiment performed in triplicate. p < 0.01.
We next examined the presence of activation and repression marks at the ErbB3 proximal promoter (found on ch: 10 in the mouse genome) in wild type and CtBP1/2 null MEFs. As shown in Figure 6B, ZNF217 was enriched at the ErbB3 promoter in both cases, demonstrating that despite its tight association with CtBP1/2 (Cowger et al., 2007;Quinlan et al., 2006), ZNF217 is recruited to the ErbB3 promoter in a CtBP1/2-independent manner. In wild type cells, where ErbB3 expression was very modest, a mix of activation (AcH3K9, TrimeK4) and repression marks (TrimeK27) were present (Figure 6B top gel and ChIP-qPCR graphs below). However, in the CtBP1/2-null MEF cells where ErbB3 expression was significantly elevated, the activation marks were preserved (and enriched in the case of AcH3K9) while the repression marks (TrimeK9, TrimeK27) were nearly absent (Figure 6B lower gel and ChIP-qPCR graph below). Notably, the methylation of histone3 lysine 27 is dramatically reduced in CtBP-null mefs (Figure 6B and Supplementary Figure 3). To examine this further and directly implicate CtBP proteins in ErbB3 repression, a rescue experiment was performed. CtBP1/2 null MEFs were transiently transfected with either empty vector or a vector expressing CtBP2, and ErbB3 protein levels were examined by immunoblotting. A representative experiment is shown in Figure 6C (left panel) and quantified in the right panel. Ectopic expression of CtBP2 in CtBP1/2 null MEFs was clearly sufficient to suppress ErbB3 expression, defining a role for CtBP2 as an ErbB3 repressor. In agreement with this, ectopic expression of CtBP2 in null MEFs led to a significant decrease in ErbB3-promoter driven reporter activity (Figure 6D). These data demonstrate that despite their close association and overlapping promoter occupancy, ZNF217 and CtBP play opposing roles in the regulation of ErbB3 expression.
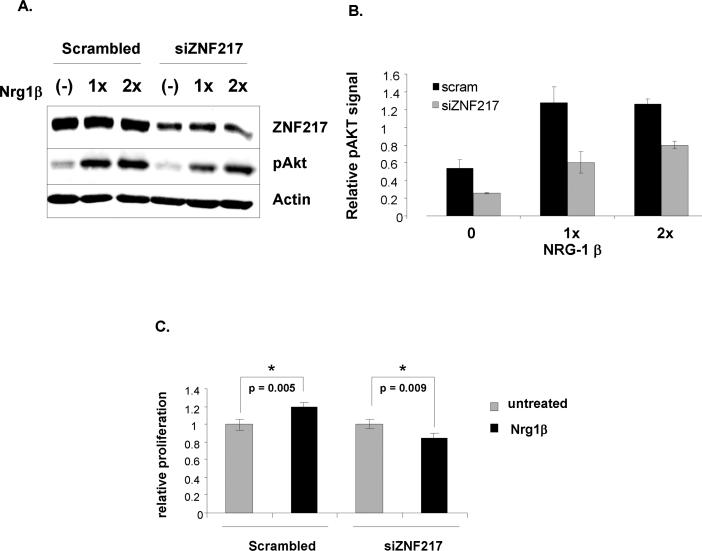
Depletion of ZNF217 limits downstream signaling through ErbB3
To examine the functional impact of ZNF217 modulation of ErbB3, ErbB3-mediated activation of the PI3K/Akt signaling cascade was examined in ZR75-1 breast cancer cells. ErbB3 is unique amongst the ErbB family in its propensity to potently activate the PI3K/Akt signaling cascade. To determine whether ZNF217 status impacts downstream signaling through ErbB3, ZR75-1 cells were treated with either scramble control or siZNF217 and then stimulated with two different concentrations of the ErbB3 activating ligand, Neuregulin-1β. Lysates were probed for ZNF217, phospho-Akt (S473) and actin control and a representative immunoblot is shown in Figure 7A with quantification of several experiments in Figure 7B. Depletion of ZNF217 decreased basal Akt phosphorylation and limited ligand-stimulated Akt phosphorylation such that at both Neuregulin 1β concentrations, Akt phosphorylation was significantly diminished. This attenuation of Neuregulin 1β-stimulated Akt phosphorylation is likely a direct consequence of the ErbB3 depletion observed following ZNF217 knockdown, although other means by which ZNF217 may influence ErbB3 signaling through the PI3K/Akt pathway cannot presently be ruled out. To examine whether this attenuation of signaling has consequences for tumor cell growth, an MTT assay was performed. ZR75-1 cells were treated with either scramble control or ZNF217 siRNA and then stimulated with a sub-saturating concentration of Neuregulin-1β (necessary to appreciate the differences between ZNF217-replete and -depleted cells). Although ZR75-1 cells have a high basal rate of proliferation, they demonstrated a modest but reproducible response to a low concentration of Neuregulin-1β(p = 0.005). Conversely, in ZNF217-depleted cells, the proliferative response to Neuregulin-1β was abolished and cells instead proliferated less in the presence of Neuregulin-1β that in its absence (p = 0.009). Similar results were observed in MCF7 cells (data not shown). These findings have important implications for future therapeutic approaches which aim at targeting ErbB3. Depletion of ZNF217 may be beneficial in this regard.
Figure 7. ZNF217 depletion suppresses ErbB3-mediated downstream signaling.
(A) ZR75-1 cells were transiently transfected with either scramble control or ZNF217-specific siRNA. 48 hours after transfection, cells were serum starved overnight and then treated with two different concentrations of Neuregulin1βfor twenty minutes. Lysates were prepared and blotted for ZNF217, p-AKT and actin (loading control). A representative western blot is shown. (B) Densitometric analysis of phospho Akt signal from three independent experiments. p < 0.01 for all comparisons between scramble and siZNF217. (C) Cell viability was determined in ZR75-1 cells transiently transfected with either scramble control or ZNF217-specific siRNA. 48 hours after transfection, cells were serum-starved overnight followed by treatment with Neuregulin1β for an additional 48h. Proliferation of cells at 96h post-transfection was measured by MTT assay.
DISCUSSION
In this study, we find that ZNF217 and ErbB3 expression strongly correlate in human breast cancer cell lines and murine and human tumor samples. In addition, we demonstrate that ZNF217 binds to the proximal ErbB3 promoter and activates this promoter in a reporter assay. Ectopic expression of ZNF217 is sufficient to augment ErbB3 expression in human mammary epithelial cells. Conversely, depletion of endogenous ZNF217 leads to significant decreases in ErbB3 expression, indicating that ZNF217 expression is required for the maintenance of robust ErbB3 expression in breast cancer cells. These gain and loss of function studies indicate that ZNF217 is upstream of ErbB3 expression. On the basis of these findings, we propose that cells bearing amplified ZNF217 genes may be selected for during breast cancer evolution, at least in part due to increases in ErbB3 levels and associated Akt signaling, which would contribute to a pro-survival phenotype of ZNF217-positive breast tumors. Our findings strongly suggest that ErbB3 is one link between ZNF217 and enhanced phospho-Akt levels (Huang et al, 2005).
While the ErbB2 gene at ch.17q12 is frequently found amplified, the mechanisms leading to over-expression of ErbB3 in breast cancer are not well understood. Low-level ErbB3 gene amplification has been reported (Sassen et al., 2008), but the frequency with which this contributes to ErbB3 over-expression in cancers is unknown. Presumably, over-expression of ErbB3 can occur through loss of regulatory controls at the transcriptional, translational or protein stability levels (Yen et al., 2006, Folgiero et al., 2007). Indeed, loss of the ubiquitin ligase Nrdp1 has been associated with ErbB3 overexpression (Yen et al., 2006). Our data demonstrate that ZNF217 contributes to the transcriptional dys-regulation of ErbB3 in breast cancer. While we favor the model that ZNF217 directly activates the ErbB3 promoter, due to its consistent presence at the promoter in various cell types, we cannot rule out the possibility that ZNF217 regulates ErbB3 transcription in an indirect manner. Nevertheless, our data indicate that ZNF217 is both necessary and sufficient for robust ErbB3 expression.
ErbB3 has been implicated in resistance to targeted therapies such as Tamoxifen (Liu et al., 2007) and Trastuzumab and has been linked to breast tumor cell invasion (Yen et al., 2006) and metastasis (Xue et al., 2006). ErbB3 plays a central role in ErbB2-driven breast tumor cell growth (Holbro et al., 2003) and has also been implicated in signaling by other receptor tyrosine kinases such as the Met receptor (Lee et al., 2007) and TGF beta type 1 receptors (Wang et al., 2008). Manipulation of ZNF217 levels and/or function in cancer cells may present a novel means for lowering ErbB3 expression and limiting its oncogenic activity and contributions to therapeutic resistance. The ErbB2-targeted antibodies Trastuzumab (Junttila et al., 2009) and Pertuzumab (Agus et al., 2002) disrupt ErbB2-ErbB3 heterodimers, limiting ErbB3 contribution to ErbB2 signaling, but leave ErbB3 free to interact with other receptors. Targeting ZNF217 in cancer cells may have benefits beyond its effects on ErbB3, since our previous work and the work of others have identified several genes subject to both transcriptional activation and repression by ZNF217 (Krig et al., 2007, Thillainadesan et al., 2008). Although more challenging than enzymes, transcription factors are emerging as viable therapeutic targets in cancer (Frank, 2009, Shakya et al., 2009). Approaches that disrupt protein-protein interactions necessary for transcription factor activity (Plowright et al., 2009) or utilize RNA interference to silence transcription factor expression (Heidenreich, 2009, Singh et al., 2008) are on the horizon.
ZNF217 and CtBP2 proteins have been demonstrated to interact (Quinlan et al., 2006) and their occupancy at promoters shows substantial overlap (Krig et al., 2007), leading to the model that one is involved in the recruitment of the other to target promoters (see model in Krig et al. 2007). Previous work (Krig et al., 2007), together with our present work, demonstrates that ZNF217 recruitment to the ErbB3 promoter is independent of CtBP. However, CtBP may depend, to some degree, on ZNF217 for its recruitment to the ErbB3 promoter in some cell lines, suggesting a complex relationship between the two. Overall, our data demonstrate that ZNF217 and CtBP regulate ErbB3 expression in an opposing manner. Given the limited understanding of ErbB3 over-expression in breast cancer, our findings have important implications for the future design of therapies targeting ErbB3.
MATERIALS AND METHODS
Cell culture
MCF7 cells and mouse embryo fibroblasts were cultured in DMEM (Gibco) supplemented with 10% fetal calf serum (Gibco) and 1% penicillin-streptomycin antibiotics (Cellgro). ZR75-1 cells were cultured in RPMI 1640 (Mediatech) with 10 mM Hepes (Gibco), 1 mM sodium pyruvate (Gibco), 14 mM glucose, 1 ug/mL insulin (Sigma), 10% fetal calf serum (Gibco) and 1% antibiotics (Cellgro). Finite life span HMEC 184 were obtained from reduction mammoplasty tissue and grown in a serum-free MCDB 170 medium (Clonetics Division of BioWhittaker)
Primary human tissue specimens
Frozen human breast tumors were provided by the UCD Cancer Center Specimen Repository and the NCI Cooperative Human Tissue Network. All samples were de-identified, and the study was approved by the Institutional Review Board of the UCD School of Medicine. Tissues were homogenized in ice-cold T-PER Tissue Protein Extraction Reagent (Pierce) with protease and phosphatase inhibitors, then centrifuged to remove insoluble debris.
Transgenic mice and tissues
FVB mice expressing the activated Neu transgene (NDL2-5, neu deletion mutant) (Siegel et al, 1999) were maintained at UCD animal facilities. Mammary tissues were collected and snap-frozen in liquid nitrogen. Tissue lysates were prepared as described above.
Recombinant ZNF217 adenoviral infections
To amplify the ZNF217-adenovirus or control Adeno-X virus (Clontech), HEK293 cells were infected and supernatants collected, centrifuged, and flash frozen until use. Post-stasis HMECs 184 at early passage were pre-treated with polybrene and infected with adenoviral supernatants. Extracts for protein or RNA analyses were collected at various time points. Extracts for protein were collected and quantitated using the Pierce BCA protein detection reagent. RNA was isolated using the RNeasy Kit (Qiagen). To check the integrity of the RNA, aliquots from RNA samples were prepared using the Agilent RNA 6000 Nano Kit and assayed on a Bioanalyzer (Agilent).
Luciferase reporter assays
MCF7 cells or MEFs were plated at 2 × 105 cells per well in 12-well plates and allowed to settle overnight. Cells were transiently transfected using Lipofectamine LTX Plus (Invitrogen) with ErbB3_pGL4 Luciferase Reporter construct, ZNF217 expression vector, and pRLO renilla luciferase vector (Promega) or beta-galactosidase pCMX reporter vector to normalize cell numbers and transfection efficiency. Lysates were harvested 24 or 48 hours after transfection with 1x Passive Lysis Buffer (Promega) and assayed for firefly and renilla luciferase activity using the Dual Luciferase Kit (Promega), or beta-galactosidase actvity was measured using CPRG (chlorophenolred-ß-D-galactopyranoside) at 550nm in a plate reader format.
Immunoblotting
Lysates were resolved by 8% SDS-PAGE and immunoblotted with various primary antibodies. See supplemental data for antibodies used.
Cell assays using RNAi
ZR75-1 or MCF7 cells were plated at a density of 1 × 105 per well in 24-well culture plates. Cells were transfected 24 hours after plating with 100 nM siRNA targeting ZNF217 (OnTarget plus SMARTpool, Dharmacon, cat # L-004987-01) or non-targeting pool (OnTarget plus non-targeting pool, Dharmacon, cat # D-001810-10) using DharmaFect 1 (Dharmacon). Medium was replaced after 24 h, and cells were left for an additional 48 h or as indicated. For cell signaling and growth assays following RNAi treatment, cells were serum starved in medium containing 0.1% FBS before Neuregulin1β treatments. To measure proliferation, cells were allowed to grow under Neuregulin1β treatment for 48h and assayed by MTT (3-(4,5-dimethylthiaszol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma). Crystals formed from the MTT were dissolved in acidic isopropanol and the absorption measured at 570 nm with a baseline subtraction at 655nm.
RNA isolation and real-time PCR
RNA was purified using a commercial kit (Qiagen; RNAeasy Kit). RNA (2.5ug) was converted to cDNA using the High Capacity cDNA Reverse Transcription Synthesis Kit (Applied Biosystems). Analyses were performed using TaqMan Gene Expression primers (Applied Biosystems) and probes that were labeled with FAM and Two Step RT qPCR Master Mix (EuroGentec). Further details are listed in Supplementary Methods.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed following the method previously described (O'Geen et al, 2006). For MCF7 ChIPs +/−siZNF217, cells were plated, transfected with 100nM siRNA for 48h and cross-linked with 1% formaldehyde at 96h. Quantitative real-time PCR was performed on a Bio-Rad DNA Engine Opticon real-time PCR system using SYBR® Green Master PCR Mix according to the manufacturer's instructions (BioRad). The relative fold enrichment of each target site was calculated as 2 to the power of the cycle threshold (cT) difference between input chromatin and ChIP samples. For MCF7 siRNA ChIP-qPCR, primers to an exon of the ZNF10 gene (unaffected by histone marks) were used to normalize differences in DNA between siRNA treatments. See supplemental Methods for antibodies and primers used.
Statistical analysis
Regression models were fitted in the R statistical Package (R Development Core Team.) R: A Language and Environment for Statistical Computing. Vienna, Austria; R Foundation for Statistical Computing, 2006). All experiments were repeated at least 3 times with representative experiments chosen. All numerical data are expressed as mean ± s.e.m. (standard error of the mean) from a representative experiment performed in triplicate. p values were determined with the Student's t test.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01 CA118384 (CS); NIH grant RO1 CA45250 (P.J.F); U54 CA112970 (P.Y., R.N.), and the Office of Energy Research, Office of Health and Biological Research, US Department of Energy under Contract No. DE-AC03-76SF00098 (PY) and NIH Center for Research Resources (NCRR) UL1 RR024146 (L.A.B.). JM was a recipient of a DOD Breast Cancer Research Program Pre-doctoral fellowship: W81XWH-06-1-0402. We would like to thank Dr. Nelly Auersperg (U. Vancouver) for the recombinant adenoviral ZNF217 construct and Jeremy Semeiks for his assistance with the adenoviral transduction experiments. We thank Dr. Jeffrey Hildebrand for the CtBP2-null mef-90 fibroblasts and Dr. Merlin Crossly for the CtBP2_pcDNA_3.1 expression construct. We thank Dr. Hongwu Chen for the pCmX vector, Dr. James Trimmer for the co-REST ascites and Dr. Martha Stampfer for the HMEC 184 cells. CTCS statistical support is made possible by Grant Number UL1 RR024146 from the Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. A special thank you to members of the Farnham lab for valuable discussions.
Footnotes
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Agus D, Akita R, Fox W, Lewis G, Higgins B, Pisacane P, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2002;2:127–137. doi: 10.1016/s1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- Bates NP, Hurst HC. An intron 1 enhancer element mediates oestrogen-induced suppression of ERBB2 expression. Oncogene. 1997;15:473–81. doi: 10.1038/sj.onc.1201368. [DOI] [PubMed] [Google Scholar]
- Berns EM, Foekens JA, van Staveren IL, van Putten WL, de Koning HY, Portengen H, et al. Oncogene amplification and prognosis in breast cancer: relationship with systemic treatment. Gene. 1995;159:11–18. doi: 10.1016/0378-1119(94)00534-y. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Chinnadurai D. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Chinnadurai D. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad SciU S A. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CoreTeam R-D . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–86. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Folgiero V, Bachelder RE, Bon G, Sacchi A, Falcioni R, Mercurio AM. The alpha6beta4 integrin can regulate ErbB-3 expression: implications for alpha6beta4 signaling and function. Cancer Res. 2007;67:1645–52. doi: 10.1158/0008-5472.CAN-06-2980. [DOI] [PubMed] [Google Scholar]
- Folgiero V, Avetrani P, Bon G, Di Carlo SE, Fabi A, Nistico C, et al. Induction of ErbB-3 expression by alpha6beta4 integrin contributes to tamoxifen resistance in ERbeta1-negative breast carcinomas. PLoS One. 2008;3:e1592. doi: 10.1371/journal.pone.0001592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. Targeting transcription factors for cancer therapy. Drugs. 2009;12:29–33. [PubMed] [Google Scholar]
- Funes M, Miller JK, Lai C, Carraway KLr, Sweeney C. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J Biol Chem. 2006;281:19310–19319. doi: 10.1074/jbc.M603225200. [DOI] [PubMed] [Google Scholar]
- Gaudray P, Szepetowski P, Escot C, Birnbaum D, Theillet C. DNA amplification at 11q13 in human cancer: from complexity to perplexity. Mutat Res. 1992;276:317–328. doi: 10.1016/0165-1110(92)90018-5. [DOI] [PubMed] [Google Scholar]
- Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway K., 3rd Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich O. Targeting oncogenes with siRNAs. Methods Mol Biol. 2009;487:221–242. doi: 10.1007/978-1-60327-547-7_11. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP1 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Beerli R, Maurer F, Koziczak M, Barbas Cr, Hynes N. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, et al. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Human Mol Genetics. 2005;14:3219–3225. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, et al. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res. 1995;55:6172–6180. [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis PG, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krig SR, Jin VX, Bieda MC, O'Geen H, Yaswen P, Green R, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Engelman J, Cantley L. Biochemistry. PI3K charges ahead. Science. 2007;317:239–242. doi: 10.1126/science.1146073. [DOI] [PubMed] [Google Scholar]
- Li P, Maines-Bandiera S, Kuo WL, Guan Y, Sun Y, Hills M, et al. Multiple roles of the candidate oncogene ZNF217 in ovarian epithelial neoplastic progression. Int J Cancer. 2007;120:1863–1873. doi: 10.1002/ijc.22300. [DOI] [PubMed] [Google Scholar]
- Liu B, Ordonez-Ercan D, Fan Z, Edgerton SM, Yang X, AD T. Downregulation of erbB3 abrogates erbB2-mediated tamoxifen resistance in breast cancer cells. Int J Cancer. 2007;120:1874–1882. doi: 10.1002/ijc.22423. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of lysine methylation. Nature Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–1254. [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J, Loboda A, Showe M, Showe L, McMahon S. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Plowright L, Harrington K, Pandha H, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer). Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, et al. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–8172. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revillion F, Pawlowski V, Lhotellier V, Louchez MM, Peyrat JP. mRNA expression of the type I growth factor receptors in the human breast cancer cells MCF-7: regulation by estradiol and tamoxifen. Anticancer Res. 2003;23:1455–60. [PubMed] [Google Scholar]
- Sassen A, Rochon J, Wild P, Hartmann A, Hofstaedter F, Schwarz S, et al. Cytogenetic analysis of HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients. Breast Cancer Res. 2008;10:R2. doi: 10.1186/bcr1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, et al. Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer Res. 1995;55:6002–6005. [PubMed] [Google Scholar]
- Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–41. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A, Cooksey R, Cox J, Wang V, McClain D, Tantin D. Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nature Cell Biology. 2009;11:320–327. doi: 10.1038/ncb1840. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci. 2010 doi: 10.1073/pnas.1002753107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Ryan ED, Cardiff RD, Muller WJ. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. Embo J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Boldin-Adamsky S, Thimmulappa R, Rath S, Ashush H, Coulter J, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413–48. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. 1996;56:3803–3807. [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, et al. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- Thillainadesan G, Isovic M, Loney E, Andrews J, Tini M, Torchia J. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28:6066–6077. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer. 2007;7:389–397. doi: 10.1038/nrc2127. [DOI] [PubMed] [Google Scholar]
- Wang SE, Xiang B, Guix M, Olivares MG, Parker J, Chung CH, et al. Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol Cell Biol. 2008;28:5605–5620. doi: 10.1128/MCB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Liang F, Mahmood R, Vuolo M, Wyckoff J, Qian H, et al. ErbB3-dependent motility and intravasation in breast cancer metastasis. Cancer Res. 2006;66:1418–1426. doi: 10.1158/0008-5472.CAN-05-0550. [DOI] [PubMed] [Google Scholar]
- Yen L, Cao Z, Wu X, Ingalla ER, Baron C, Young LJ, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Research. 2006;66:11279–11286. doi: 10.1158/0008-5472.CAN-06-2319. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CH, Huang Y, Oberley LW, Domann FE. A family of AP-2 proteins down-regulate manganese superoxide dismutase expression. J Biol Chem. 2001;276:14407–13. doi: 10.1074/jbc.M009708200. [DOI] [PubMed] [Google Scholar]
- Zhu CH, Domann FE. Dominant negative interference of transcription factor AP-2 causes inhibition of ErbB-3 expression and suppresses malignant cell growth. Breast Cancer Res Treat. 2002;71:47–57. doi: 10.1023/a:1013378113916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.