Abstract
Until recently, Staphylococcus aureus from clonal complex (CC)398 were mostly described as colonizing asymptomatic raised pigs and pig-farmers. Currently, the epidemiology of the CC398 lineage is becoming more complex. CC398 human-adapted isolates are increasingly being identified in bloodstream infections in humans living in animal-free environments. In addition, CC398 isolates are increasingly responsible for invasive infections in various animals. CC398 isolates that colonize asymptomatic pigs and the isolates that infect humans living in animal-free environments (human-adapted isolates) both lack several clinically important S. aureus–associated virulence factors but differ on the basis of their prophage content. Recent findings have provided insight into the influence of a φMR11-like helper prophage on the ability of CC398 isolates to infect humans. To assess the recent spread of the CC398 lineage to various animal species and to investigate the links between the φMR11-like prophage and the emergence of CC398 isolates infecting animals, we studied 277 isolates causing infections in unrelated animals. The prevalence of CC398 isolates increased significantly between 2007 and 2013 (p < 0.001); 31.8% of the animal isolates harbored the φMR11-like prophage. High-density DNA microarray experiments with 37 representative infected-animal isolates positive for φMR11-like DNA established that most infected-animal isolates carried many genetic elements related to antimicrobial resistance and virulence genes, and a φ3 prophage encoding immune-modulating proteins and associated with animal-to-human jumps. Our findings suggest recent clonal expansion and dissemination of a new subpopulation of CC398 isolates, responsible for invasive infections in various animals, with a considerable potential to colonize and infect humans, probably greater than that of human-adapted CC398 isolates, justifying active surveillance.
Keywords: bacteriophage, lysogeny, virulence factor, genome plasticity, genome content
Introduction
Staphylococcus aureus belonging to the clonal complex (CC) 398 has become a worldwide health threat (van Belkum et al., 2008). Until recently, CC398 isolates were mostly described as colonizing asymptomatic raised pigs and pig-farmers, and rarely infecting farmers and veterinarians (Huijsdens et al., 2006). Currently, the epidemiology of the CC398 lineage is becoming more complex. The lineage is now extending its territory as CC398 isolates are increasingly being identified in invasive infections in humans living in animal-free environments (Valentin-Domelier et al., 2011; Uhlemann et al., 2012; Verkade et al., 2012), in pet dogs, cats, and horses (Loncaric et al., 2014) and in animals of livestock species, including poultry, cattle, and rabbits (Argudín et al., 2013; Agnoletti et al., 2014).
CC398 isolates that colonize asymptomatic pigs (named below colonizing isolates) and the strains that infect humans living in animal-free environments (human-adapted isolates) both lack several clinically important S. aureus–associated virulence factors (Valentin-Domelier et al., 2011) but differ on the basis of their prophage content (McCarthy et al., 2011; Price et al., 2012). Phages play a critical role in bacterial diversity and evolution. They serve as a driving force in microbial evolution by transducing genes that supply their host with novel genetic information. This may enable host adaptation to new environments, and in some instances, confer virulence properties associated with pathogenesis in bacterial infections (Canchaya et al., 2004). Recent findings have provided insight into the influence of a helper prophage on the ability of CC398 isolates to infect humans. The sequence of this prophage named φMR11-like, shows high homology with that of φMR11, a phage isolated from a bacterium infecting the human bloodstream that is a candidate therapeutic phage (Matsuzaki et al., 2003). The φMR11-like prophage is specifically associated with the human-adapted CC398 isolates, and interacts with a human-associated β-converting φ3 prophage encoding immune-modulating proteins CHIP and SCIN such that virulence genes are expressed during stress situations and during lysogeny (van der Mee-Marquet et al., 2013).
The prophage content of CC398 isolates responsible for infections in animals has not been thoroughly investigated. Here, we report an analysis of a large collection of S. aureus isolates responsible for infections in various animal species. We determined the prevalence of CC398 isolates among these infected-animal isolates, and of isolates harboring the φMR11-like prophage which has been found to be associated with the human-adapted CC398 subpopulation. Then, using high-resolution whole-genome microarrays, we compared the mobile genetic element (MGE) content of infected-animal isolates harboring the φMR11-like prophage and of isolates belonging to the colonizing and human-adapted CC398 subpopulations.
Materials and methods
Bacterial isolates
We studied 292 isolates. The collection contains three subpopulations of isolates: (1) 277 epidemiologically independent isolates recovered from infected animals, between 2007 and 2013, in geographically distant locations in France (n = 263) and western Switzerland (n = 14); these isolates were obtained from 101 cows, 10 pigs, 15 rabbits, 29 chickens, 28 horses, 27 dogs, 20 cats, 40 sheep, three goats, and four primates; (2) eight colonizing CC398 isolates: four isolated from asymptomatic farmers and four from asymptomatic pigs; (3) four human-adapted CC398 isolates from epidemiologically unrelated French patients with bloodstream infection and not exposed to animals or farming (van der Mee-Marquet et al., 2013). The reference strains COL, Newman, and RF122 were also included for comparison purposes.
Epidemiological study
We used the disk diffusion method (Bio-Rad, France) to test the antibiotic susceptibility of isolates. The mecA gene was detected by PCR. All isolates were tested by PCR to identify those that are CC398 isolates (Stegger et al., 2011); positive isolates were assigned to CC398 by MLST (Robinson and Enright, 2004). CC398 isolates were spa-typed using a previously published procedure (Frenay et al., 1996). We used a previously developed PCR-based test that targets three MR11-like prophage sequences (van der Mee-Marquet et al., 2013) to search for MR11-like prophage in the genome of the isolates.
Analysis of MGE content by DNA microarray
A set of 37 isolates, representative of the diversity of the infected-animal isolates positive for φMR11-like DNA, was established for extensive MGE content analysis. The 37 isolates were chosen based on their antibiotic susceptibility profile, spa-type and host species origin. We used a microarray produced by in situ synthesis of 60 base-long oligonucleotide probes (Agilent, Palo Alto, CA), selected as described previously (Charbonnier et al., 2005). The array covers >96% of all open reading frames (ORFs), including those on plasmids, annotated in strains N315, Mu50, MW2, COL, NCTC8325, USA300, MRSA252, MSSA476, and Newman. Purified genomic DNA (gDNA) was extracted from each strain used for the design of the microarray (DNeasy; Qiagen), labeled with Cy5 dCTP with the Klenow fragment of DNA polymerase I (BioPrime, Invitrogen, Carlsbad, CA), and used for normalization (Talaat et al., 2002). Cy5-labeled pooled DNA (500 ng) and Cy3-labeled DNA from the test strain were mixed, diluted to 50 μl in Agilent hybridization buffer and hybridized with the microarray at a temperature of 60°C for 17 h in a dedicated hybridization oven (Robbins Scientific, Sunnyvale, CA). The slides were washed, dried under a nitrogen flow, and scanned (Agilent, Palo Alto, CA) with 100% photon multiplier tube power for both wavelengths. Fluorescence intensities were extracted with Feature Extraction software (version 9; Agilent). Local background-subtracted signals were corrected for unequal dye incorporation or unequal load of the labeled product. The algorithm consisted of a rank consistency filter and a curve fitted with the default LOWESS (locally weighted linear regression) method. Data were analyzed with GeneSpring, version 8.0 (Silicon Genetics, Redwood City, CA).
Statistical data
Chi-squared tests and Fisher's exact test (two-tailed) were used to test for statistical significance; a P < 0.05 was considered significant.
Ethics statement
Animal isolates
The 263 French isolates were collected from infected animals during the period 2007–2013 in diverse regions of the country via the Resapath network, which ensures the surveillance of antimicrobial resistance in diseased animals in France (www.resapath.anses.fr). Fourteen bovine isolates were recovered from cases of subclinical bovine mastitis in animals on Swiss farms. Isolates from Dutch pigs were obtained by non-invasive sampling during a field study. All studies were performed in accordance with the national guidelines and consequently did not require the approval of an ethics committee; the farmers gave informed consent and agreed to the collection of samples.
Human isolates
The French human isolates were obtained from clinical samples during annual surveillance studies that were run in accordance with the French Healthcare recommendations for the prevention of infection. Ethical approval of the monitoring programs was obtained from the appropriate national committee: the Réseau Alerte Investigation Surveillance des Infections Nosocomiales. The human isolates from the Netherlands were obtained during prospective surveillance, approved by the medical ethics committee of the Sint Elisabeth Hospitaal, Tilburg, the Netherlands (protocol number 0749). All participants provided written, informed consent.
Results
Epidemiological study
Sixty-nine of the 277 infected-animal isolates were identified as belonging to the CC398 lineage (24.9%) (Table 1). The prevalence of CC398 isolates was significantly higher during the period 2011–2013 (33.8%) than during the period 2007–2010 (3.6%) (p < 0.001). CC398 isolates were significantly more prevalent among isolates from rabbits and horses than those from the other host species (60.5% for rabbits and horses vs. 18.4% for the other species combined, p < 0.001).
Table 1.
Prevalence of CC398 isolates and of φMR11-like prophage elements in the genomes of the 277 animal isolates studied.
| Host species | CC398 isolates (%) | Isolates positive for φMR11-like prophage (%)a | |||
|---|---|---|---|---|---|
| All isolates | in CC398 | in other lineages | |||
| Cattle | 101 | 20 (20) | 15 (15) | 9/20 (45) | 6/81 (7) |
| Pig | 10 | 3 (30) | 2 (20) | 0/3 | 2/7 (29) |
| Rabbit | 15 | 12 (80) | 12 (80) | 12/12 (100) | 0/3 |
| Poultry | 29 | 5 (17) | 21 (72) | 2/5 (40) | 19/24 (79) |
| Horse | 28 | 14 (50) | 18 (64) | 9/14 (64) | 9/14 (64) |
| Dog | 27 | 8 (30) | 8 (30) | 2/8 (25) | 6/19 (32) |
| Cat | 20 | 5 (25) | 11 (55) | 0/5 | 11/15 (73) |
| Primate | 4 | 1 (25) | 1 (25) | 0/1 | 1/3 (33) |
| Sheep | 40 | 0 | 0 | – | 0/40 |
| Goat | 3 | 1 (33) | 0 | 0/1 | 0/2 |
| All | 277 | 69 (25) | 88 (32) | 34/69 (49) | 54/208 (26) |
| MRSA | |||||
| Cattle | 16 (16) | 16/16 (100) | 7/16 (44) | 7/16 (44) | – |
| Pig | 1 (10) | 1/1 | 0/1 | 0/1 | – |
| Rabbit | 2 (13) | 0/2 | 0/2 | – | 0/2 |
| Poultry | 1 (3) | 1/1 | 0/1 | 0/1 | – |
| Horse | 25 (89) | 14/25 (56) | 15/25 (60) | 9/14 (64) | 6/11 (54) |
| Dog | 18 (67) | 5/18 (28) | 6/18 (33) | 1/5 (20) | 5/13 (38) |
| Cat | 20 (100) | 5/20 (25) | 11/20 (55) | 0/5 | 11/15 (73) |
| Primate | 1 (25) | 1/1 | 0/1 | 0/1 | – |
| Sheep | 5 (12) | 0/5 | 0/5 | – | 0/5 |
| Goatb | 0 | – | – | – | – |
| All MRSA | 89 (32) | 43/89 (48) | 39/89 (44) | 17/43 (39) | 22/46 (48) |
| MSSA | |||||
| Cattle | 85 (84) | 4/85 (5) | 8/85 (9) | 2/4 (50) | 6/81 (7) |
| Pig | 9 (90) | 2/9 (22) | 2/9 (22) | 0/2 | 2/7 (29) |
| Rabbit | 13 (87) | 12/13 (92) | 12/13 (92) | 12/12 (100) | 0/1 |
| Poultry | 28 (97) | 4/28 (14) | 21/28 (75) | 2/4 (50) | 19/24 (79) |
| Horse | 3 (11) | 0/3 | 3/3 (100) | – | 3/3 (100) |
| Dog | 9 (33) | 3/9 (33) | 2/9 (22) | 1/3 (33) | 1/6 (17) |
| Catb | 0 | – | – | – | – |
| Primate | 3 (75) | 0/3 | 1/3 (33) | – | 1/3 (33) |
| Sheep | 30 (88) | 0/35 | 0/35 | – | 0/35 |
| Goat | 3 (100) | 1/33 (33) | 0/3 | 0/1 | 0/2 |
| All MSSA | 188 (68) | 26/188 (14) | 49/188 (26) | 17/26 (65) | 32/162 (20) |
isolates with at least one positive PCR test were considered positive.
no isolate.
Of the 277 infected-animal isolates, 89 were MRSA (32.1%), 106 were TetR (38.3%) and 55 were EryR (19.8%). The prevalence of resistance to methicillin was significantly higher among CC398 isolates than isolates of other lineages (62% among CC398 vs. 22.1% among non-CC398 isolates, p < 0.001). In the CC398 group, susceptibility to methicillin was significantly associated with isolates from infected rabbits (100% among rabbit CC398 vs. 24.6% among non-rabbit CC398 isolates, p < 0.001).
The spa-type t011 was the most common among the 69 CC398 isolates (44.9%), and there were eight other spa-types: t034, t571, t899, t1190, t108, t002, t097, and t2922. Four spa-types (t011, t034, t571, and t899) have previously been found in colonized-pig-associated CC398 isolates (1–3). The spa-type t1190 was only observed in isolates from rabbits, and t002 in isolates from poultry.
φMR11-like DNA was detected in 88 of the 277 isolates from infected animals (31.8%) and was significantly more frequent among strains isolated during the period 2011–2013 (40.5%) than during the period 2007–2010 (9.7%) (p < 0.001) (Table 1). The prevalence of φMR11-like prophage fragments was (i) higher among isolates from rabbits (80.0%), poultry (72.4%), and horses (64.3%) than other hosts, (ii) significantly higher among MRSA (43.8%) than MSSA (26.1%) isolates (p = 0.003), and (iii) significantly higher among CC398 isolates (49.3%) than isolates of other lineages (26.0%) (p < 0.001).
DNA microarray analysis of the MGE content of genomes in isolates positive for ΦMR11-like DNA from infected animals, colonizing CC398 isolates, and human-adapted CC398 isolates
We studied 37 isolates (Table 2) representative of the 88 infected-animal isolates positive for φMR11-like DNA. On the microarrays, 351 probes appeared to be discriminative for the CC398 lineage (van der Mee-Marquet et al., 2013) (Supplementary Files 1–8). The 351 probes can be classified into four groups: 26 probes related to human-associated β-converting φ3-prophage (Supplementary File 1); 24 probes related to φMR11-like-prophage (Supplementary File 2); 38 φL54a-related probes (Supplementary File 3); and 263 probes related to diverse elements of the accessory genome of S. aureus (Supplementary Files 4–8). This last group includes 70 probes related to diverse prophages (Supplementary File 4), 39 MGE-related probes (Supplementary File 5), 18 probes related to antimicrobial resistance-associated elements (Supplementary File 6), 48 probes related to putative virulence-associated elements (Supplementary File 7), and 88 probes related to other diverse genetic elements (Supplementary File 8).
Table 2.
Characteristics of the 37 animal isolates studied with high resolution microarrays.
| Host species | Year of isolation | Isolate | CC | Spa-type | Antibiotic resistance | ||
|---|---|---|---|---|---|---|---|
| MetR | TetR | EryR | |||||
| Cattle | 2011 | 11329 | 398 | t011 | + | + | |
| 2012 | 32348 | 398 | t899 | + | + | ||
| 2011 | bovSW | 398 | t899 | + | + | ||
| 2011 | 32086 | 398 | t011 | + | + | + | |
| 2011 | 11104 | 398 | t011 | + | + | ||
| 2010 | M52 | 151 | t529 | ||||
| 2007 | 20263 | new | t092 | ||||
| 2013 | 33612 | new | t529 | ||||
| Pig | 2013 | 34048 | new | t1130 | + | ||
| 2012 | 34050 | new | t318 | ||||
| Rabbit | 2013 | 34053 | 398 | t1190 | + | + | |
| 2012 | 34066 | 398 | t1190 | ||||
| Poultry | 2013 | 33972 | 398 | t011 | + | + | |
| 2013 | 33924 | 398 | t002 | + | |||
| 2012 | 33982 | 1495 | t002 | + | |||
| 2013 | 33928 | 1495 | t3478 | ||||
| 2012 | 33983 | 1495 | t002 | ||||
| 2013 | 33963 | new | t056 | + | |||
| Horse | 2010 | 26023 | 398 | t011 | + | + | |
| 2013 | 33877 | 1 | t1508 | ||||
| 2012 | 32718 | 8 | t394 | + | + | + | |
| 2012 | 32721 | 8 | t394 | + | + | + | |
| Dog | 2012 | 32978 | 398 | t011 | + | + | |
| 2013 | k1343 | 398 | t6605 | ||||
| 2011 | 27741 | 8 | t211 | + | |||
| 2011 | 27267 | 8 | t2054 | + | + | ||
| 2011 | 27095 | 8 | t121 | + | |||
| 2012 | 32313 | 8 | t008 | + | |||
| Cat | 2012 | 32811 | 5 | t002 | + | ||
| 2010 | 25175 | 5 | t002 | + | + | + | |
| 2012 | 32820 | 5 | t777 | + | |||
| 2012 | 32775 | 5 | t003 | + | + | ||
| 2010 | 26695 | 8 | t574 | + | + | ||
| 2010 | 26451 | 8 | t068 | + | + | ||
| 2012 | 32305 | 8 | t008 | + | |||
| 2011 | 27744 | 22 | t032 | + | |||
| Primate | 2012 | 33318 | new | t318 | + | ||
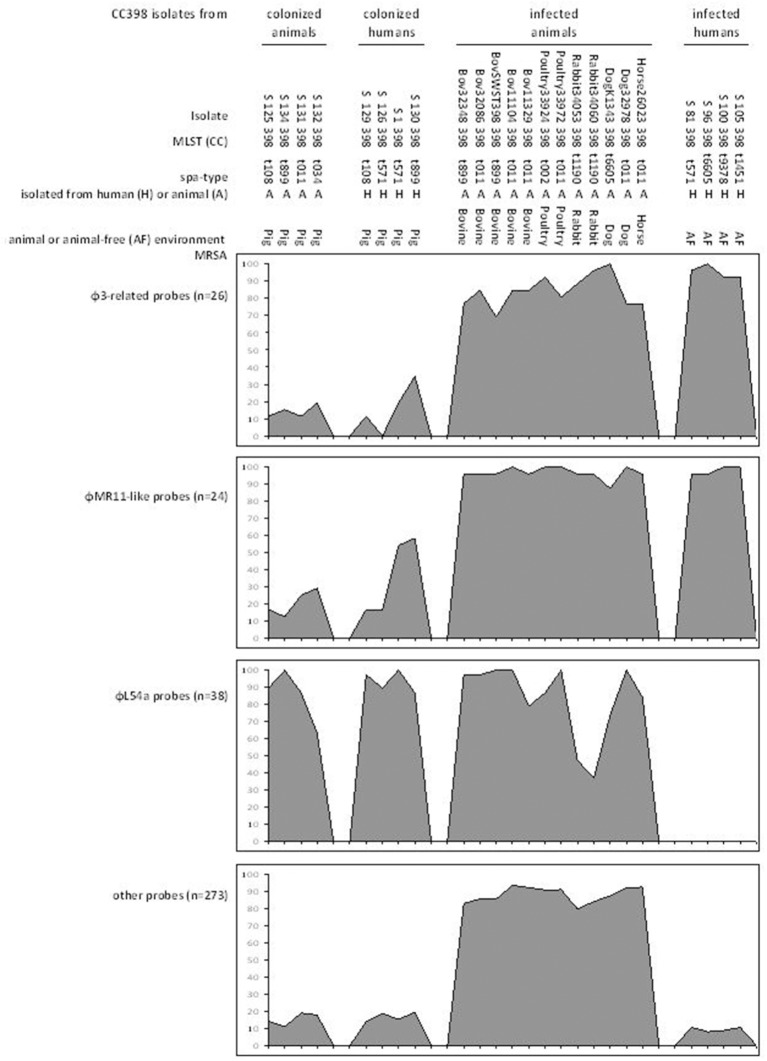
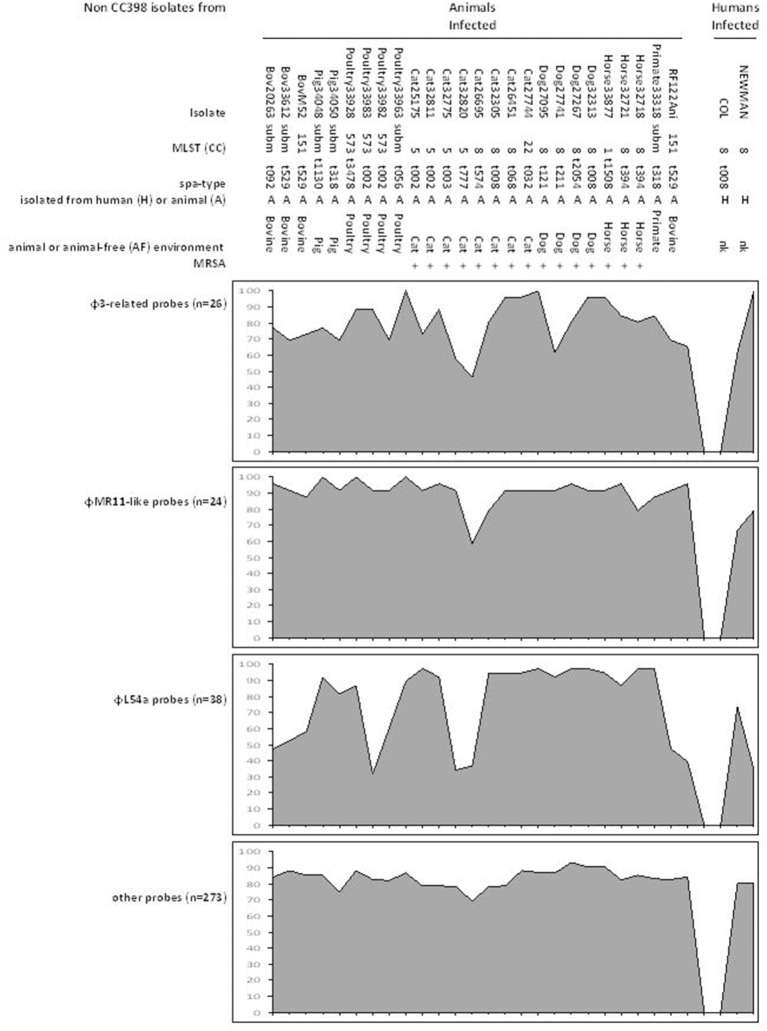
Figures 1, 2 show the prevalence of hybridization signals for each isolate and each group of probes. The three different subpopulations of isolates (infected-animal, colonizing CC398, and human-adapted CC398) each showed different and characteristic hybridization patterns, and can thereby be clearly distinguished. Colonizing isolates and human-adapted isolates gave few positive hybridization signals. The colonizing isolates mostly only hybridized with probes for the φL54a prophage whereas the human-adapted isolates mostly only hybridized with the probes related to the φ3- and φMR11-like prophages. Few of the isolates in these two groups hybridized with probes of the large group of 263 probes related to diverse elements (Supplementary Files 4–8 and Figures 1, 2). By contrast, the infected-animal isolates hybridized with the vast majority of the capture probes recognizing the φ3 and φMR11-like prophages and numerous MGEs containing putative virulence genes and antibiotic resistance determinants. Most of the 37 infected-animal isolates, especially the MRSA isolates, also hybridized with the φL54a-related probes (Figures 1, 2). The infected-animal isolates all gave similar hybridization patterns, whether or not they belonged to the CC398, suggesting that the infected-animal isolates all possess similar set of accessory genes, regardless of their lineage of origin.
Figure 1.
Prevalence of hybridization of the four groups of probes with the genomes of the tested isolates belonging to CC398. The different subpopulations of isolates showed different hybridization patterns. The colonizing isolates mostly only hybridized with probes for the φL54a prophage whereas the human-adapted isolates mostly only hybridized with the probes related to the φ3- and φMR11-like prophages. By contrast, the infected-animal CC398 isolates hybridized with the vast majority of the capture probes.
Figure 2.
Prevalence of hybridization of the four groups of probes with the genomes of the tested isolates belonging to other lineages. The infected-animal isolates and the strains COL and NEWMAN responsible for invasive infections in humans hybridized with the vast majority of the capture probes.
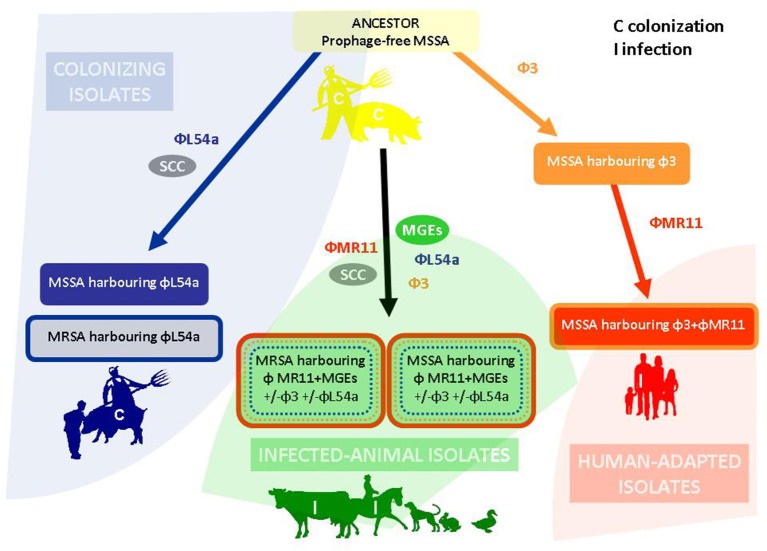
Our analyses reveal genetic elements that the three CC398 subpopulations studied have acquired by horizontal genetic transfer. We propose a model in which the ancestral prophage-free colonizing CC398 MSSA isolates diverged into three subpopulations (Figure 3).
Figure 3.
Schematic representation of the diversification within the CC398 lineage. Representatives of the ancestral prophage-free CC398 population were identified in the form of pig-borne colonizing isolates (upper part) (Hernandez et al., 2013; van der Mee-Marquet et al., 2013). The acquisition by such prophage-free isolates of the φ3 prophage and the φL54a prophage resulted in the ancestral MSSA isolates: human isolates carrying the φ3-prophage and pig-borne colonizing isolates carrying the φL54a prophage, respectively (Corvaglia et al., 2013; Hernandez et al., 2013; van der Mee-Marquet et al., 2013). The acquisition of the φMR11-like prophage by the human isolates resulted in human-adapted MSSA carrying φ3- and φMR11-like-prophages (Corvaglia et al., 2013; van der Mee-Marquet et al., 2013); the acquisition by the pig-borne colonizing isolates of the SCCmec cassette resulted in the pig-borne colonizing MRSA isolates (Hernandez et al., 2013; van der Mee-Marquet et al., 2013). The acquisition of MGEs, including the φMR11-like prophage, which contains genes contributing to bacterial virulence, resulted in the emergence of MSSA and MRSA isolates responsible for infections in both livestock and pet species (lower part).
Discussion
We studied a large set of S. aureus isolates representative of infections in animals. Our findings confirm that S. aureus CC398 isolates initially described as colonizers of asymptomatic pigs are now entering new territories, and infecting numerous animal species (van Belkum et al., 2008). Animal CC398 isolates, formerly exclusively associated with pigs and pig-farmers, have spread into other niches; and this event is evolutionarily recent. We show the chronology of this epidemiological evolution of the CC398 lineage: these isolates were able to infect only a limited range of hosts until 2007 but since 2010 have spread to numerous livestock and pet species. Note that our study only included a collection of selected strains and is therefore not representative of true incidence. Our findings need to be confirmed by, for example, prospective surveillance of the incidence of CC398 infections in these various animal species and in various geographical areas.
Population genetic studies have identified S. aureus clones with distinct genetic backgrounds as causative agents of infections in various host species. For example, sequence type (ST) 97 and ST151 appear to be ruminant-specific (Spoor et al., 2013) and ST5 is frequently isolated from poultry (Lowder et al., 2009), whereas ST5, ST8, ST22, ST30, and ST45 are mainly associated with human samples (Oliveira et al., 2002). By contrast, and consistent with recent reports (Argudín et al., 2013; Agnoletti et al., 2014; Loncaric et al., 2014), our work documents the capacity of strains from the CC398 lineage to colonize and adapt to numerous host species, including livestock, pets, and humans. For example, we show that a clone known to colonize pigs, MRSA-t011, is now frequently detected in cattle, poultry, horse, and dog. The CC398 isolates from infected animals nevertheless included two sub-clones that appeared host-specific: a rabbit-specific strain, MSSA-t1190 (Agnoletti et al., 2014) and a poultry-specific strain, showing the t002 spa-type usually detected in the avian-adapted ST5 lineage (Lowder et al., 2009). These observations suggest an appreciable genome plasticity of the CC398 lineage facilitating evolution leading to extended host diversity.
Our microarrays containing probes corresponding to numerous sequenced strains from diverse origins, cover a wide diversity of MGEs. Analyses with these microarrays indicated that among CC398 isolates, those colonizing asymptomatic pigs have the smallest pool of accessory genes. By contrast, infected-animal isolates had larger numbers of MGEs in their genomes. Many MGEs carry virulence genes and antimicrobial resistance determinants, consistent with our suggestion that these MGEs contribute to strain pathogenicity. These MGE were also observed in the genomes of strains COL and NEWMAN responsible for invasive infections in humans, and in the genome of strain RF122 responsible for infections in cows. This strongly suggests that these isolates have a considerable potential for virulence. In addition, the degree of conservation of MGEs among CC398 isolates suggests a recent clonal expansion and dissemination of this subpopulation of CC398 from infected animals.
Unlike colonizing isolates, both human-adapted and one third of the infected-animal CC398 isolates hybridized with φMR11-like related probes. As there is substantial similarity between the sequences of the φMR11-like prophage and Avβ, a prophage carrying avian niche-specific genes, our findings are consistent with evidence of an Avβ prophage in CC398 isolates from infected turkeys (Argudín et al., 2013). Thus, it appears that the φMR11-like prophage is associated with human-adapted CC398 isolates and also with isolates infecting various animal species. The carriage of a φMR11-like prophage was associated with the carriage of several SaPIs-like MGEs encoding putative virulence genes. The simultaneous presence of a helper phage and other MGEs, especially SaPIs, may result in significantly increased bacterial virulence (Ram et al., 2012). Therefore, our findings suggest that the interaction of the φMR11-like prophage and the MGEs encoding putative virulence genes may significantly alter bacterial pathogenicity. Using an animal model, we are currently investigating whether the φMR11-like prophage promotes virulence in CC398 infected-animal isolates.
There is evidence that acquisition of φ3 prophage facilitated animal-to-human jumps by CC97 isolates (Goerke et al., 2006; Spoor et al., 2013), leading to the spread of this pathogen. Most of the infected-animal isolates we studied carry a human-associated β-converting φ3 prophage encoding immune-modulating proteins. Therefore, it is likely that these CC398 infected-animal isolates are capable of adapting to, colonizing and finally infecting humans. Frequent contacts between humans and animals contribute to the transmission and spread of S. aureus between host species. Thus, our work suggests that livestock and pets colonized and/or infected with this third subpopulation of CC398 strains represent a major reservoir, favoring the emergence of CC398 S. aureus with the capacity for pandemic spread in humans.
Conclusion
Our data support a model of rapid adaptation of the S. aureus CC398 lineage to diverse hosts. Originating from this lineage that formerly only colonized animals and humans in contact with them, two subpopulations emerged in parallel following the acquisition of MGEs. The common acquisition of the φMR11-like prophage by these two CC398 subpopulations coincide with epidemiological changes, particularly the evolution from colonizing hosts toward becoming agents responsible for severe invasive infections. Importantly, unlike human-adapted CC398 isolates that lack several clinically important S. aureus–associated virulence factors (Valentin-Domelier et al., 2011), the infected-animal isolates are similar to virulent S. aureus strains, harboring numerous genetic elements related to microbial resistance and virulence genes, and the β-converting φ3 prophage associated with animal-to-human jumps. This observation strongly suggests that these infected-animal isolates have a considerable potential for virulence, probably greater than that of human-adapted CC398 isolates. Given the specific features of the genomic content of the infected-animal CC398 isolates described here, our findings indicate that there is a need for prospective surveillance of both humans and animals to monitor the evolution of the CC398 lineage.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Gregory Resch and Prof. Philippe Moreillon (Department of Fundamental Microbiology, University of Lausanne, Lausanne, Switzerland) for providing data and comments related to mastitis isolates from Switzerland. This work was supported by grants 31003A_153474/1 from the Swiss National Science Foundation (to Patrice François).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00652/abstract
References
- Agnoletti F., Mazzolini E., Bacchin C., Bano L., Berto G., Rigoli R., et al. (2014). First reporting of methicillin-resistant Staphylococcus aureus (MRSA) ST398 in an industrial rabbit holding and in farm-related people. Vet. Microbiol. 170, 172–177. 10.1016/j.vetmic.2014.01.035 [DOI] [PubMed] [Google Scholar]
- Argudín M. A., Cariou N., Salandre O., Le Guennec J., Nemeghaire S., Butaye P. (2013). Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from diseased turkeys. Avian Pathol. 42, 572–580. 10.1080/03079457.2013.854308 [DOI] [PubMed] [Google Scholar]
- Canchaya C., Fournous G., Brüssow H. (2004). The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53, 9–18. 10.1111/j.1365-2958.2004.04113.x [DOI] [PubMed] [Google Scholar]
- Charbonnier Y., Gettler B., Francois P., Bento M., Renzoni A., Vaudaux P., et al. (2005). A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 6:95. 10.1186/1471-2164-6-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvaglia A. R., François P., Bertrand X., Quentin R., Hernandez D., van der Mee-Marquet N. (2013). Whole-genome sequences of two Staphylococcus aureus ST398 strains of human origin, S94 and S100. Genome Announc. 1:e00691-13. 10.1128/genomeA.00691-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenay H. M., Bunschoten A. E., Schouls L. M., van Leeuwen W. J., Vandenbroucke-Grauls C. M., Verhoef J., et al. (1996). Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15, 60–64. 10.1007/BF01586186 [DOI] [PubMed] [Google Scholar]
- Goerke C., Wirtz C., Flückiger U., Wolz C. (2006). Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 61, 1673–1685. 10.1111/j.1365-2958.2006.05354.x [DOI] [PubMed] [Google Scholar]
- Hernandez D., van der Mee-Marquet N., Kluytmans J., Donnio P.-Y., Quentin R., Corvaglia A. R., et al. (2013). Whole-genome sequences of Staphylococcus aureus ST398 strains of animal origin. Genome Announc. 1:e00689-13. 10.1128/genomeA.00689-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijsdens X. W., van Dijke B. J., Spalburg E., van Santen-Verheuvel M. G., Heck M. E., Pluister G. N., et al. (2006). Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. 10.1186/1476-0711-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncaric I., Künzel F., Licka T., Simhofer H., Spergser J., Rosengarten R. (2014). Identification and characterization of methicillin-resistant Staphylococcus aureus from Austrian companion animals and horses. Vet. Microbiol. 168, 381–387. 10.1016/j.vetmic.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Lowder B. V., Guinane C. M., Ben Zakour N. L., Weinert L. A., Conway-Morris A., Cartwright R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. 10.1073/pnas.0909285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki S., Yasuda M., Nishikawa H., Kuroda M., Ujihara T., Shuin Y., et al. (2003). Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phiMR11. J. Infect. Dis. 187, 213–624. 10.1086/374001 [DOI] [PubMed] [Google Scholar]
- McCarthy A. J., Witney A. A., Gould K. A., Moodley A., Guardabassi L., Voss A., et al. (2011). The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biol. Evol. 3, 1164–1174. 10.1093/gbe/evr092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. C., Tomasz A., De Lencastre H. (2002). Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2, 180–189. 10.1016/S1473-3099(02)00227-X [DOI] [PubMed] [Google Scholar]
- Price L. B., Stegger M., Hasman H., Aziz M., Larsen J., Andersen P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3:e00305-11. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G., Chen J., Kumer K., Ross H. F., Ubeda C., Damle P. K., et al. (2012). Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc. Natl. Acad. Sci. U.S.A. 109, 16300–16305. 10.1073/pnas.1204615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Enright M. C. (2004). Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 10, 92–97. 10.1111/j.1469-0691.2004.00768.x [DOI] [PubMed] [Google Scholar]
- Spoor L. E., McAdam P. R., Weinert L. A., Rambaut A., Hasman H., Aarestrup F. M., et al. (2013). Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio 4:e00356-13. 10.1128/mBio.00356-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegger M., Lindsay J. A., Moodley A., Skov R., Broens E. M., Guardabassi L. (2011). Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J. Clin. Microbiol. 49, 732–734. 10.1128/JCM.01970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat A. M., Howard S. T., Hale W., Lyons R., Garner H., Johnston S. A. (2002). Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30, e104. 10.1093/nar/gnf103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann A. C., Porcella S. F., Trivedi S., Sullivan S. B., Hafer C., Kennedy A. D., et al. (2012). Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. MBio 3:e00027-12. 10.1128/mBio.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Domelier A. S., Girard M., Bertrand X., Violette J., François P., Donnio P.-Y., et al. (2011). Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: an emerging human-adapted subclone? PLoS ONE 6:e28369. 10.1371/journal.pone.0028369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Melles D. C., Peeters J. K., van Leeuwen W. B., van Duijkeren E., Huijsdens X. W., et al. (2008). Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerging Infect. Dis. 14, 479–483. 10.3201/eid1403.070760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Mee-Marquet N., Corvaglia A. R., Valentin A. S., Hernandez D., Bertrand X., Girard M, et al. (2013). Analysis of prophages harboured by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect. Genet. Evol. 18, 299–308. 10.1016/j.meegid.2013.06.009 [DOI] [PubMed] [Google Scholar]
- Verkade E., Bergmans A. M., Budding A. E., van Belkum A., Savelkoul P., Buiting A. G., et al. (2012). Recent emergence of Staphylococcus aureus clonal complex 398 in human blood culture. PLoS ONE 7:e41855. 10.1371/journal.pone.0041855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.