Abstract
The title compound, C33H35NO6 [systematic name: (Z)-3-(4-{(E)-[(E)-1a,5-dimethyl-9-oxo-2,3,7,7a-tetrahydrooxireno[2′,3′:9,10]cyclodeca[1,2-b]furan-8(1aH,6H,9H,10aH,10bH)-ylidene]methyl}phenyl)-2-(3,4,5-trimethoxyphenyl)acrylonitrile methanol hemisolvate], C33H35NO6·0.5CH3OH, was prepared by the reaction of (Z)-3-(4-iodophenyl)-2-(3,4,5-trimethoxyphenyl)acrylonitrile with parthenolide [systematic name: (E)-1a,5-dimethyl-8-methylene-2,3,6,7,7a,8,10a,10b-octahydrooxireno[2′,3′:9,10]cyclodeca[1,2-b]furan-9(1aH)-one] under Heck reaction conditions. The molecule is built up from fused ten-, five- (lactone) and three-membered (epoxide) rings with a {4-[(Z)-2-cyano-2-(3,4,5-trimethoxyphenyl)ethenyl]phenyl}methylidene group as a substituent. The 4-[(Z)-2-cyano-2-(3,4,5-trimethoxyphenyl)ethenyl]phenyl group on the parthenolide exocyclic double bond is oriented in a trans position to the lactone ring to form the E isomer. The dihedral angle between the benzene ring of the phenyl moiety and the lactone ring mean plane is 21.93 (4)°.
Keywords: crystal structure, parthenolide derivatives, Heck synthesis, biological activity
Related literature
For the biological activity of parthenolide, see: Hall et al. (1979 ▶). For the biological activity of parthenolide derivatives similar to the title compound, see: Hanson et al. (1970 ▶); Hehner et al. (1998 ▶); Kupchan et al. (1971 ▶); Neelakantan et al. (2009 ▶); Oka et al., 2007 ▶); Ralstin et al. (2006 ▶); Sun et al. (2006 ▶); Penthala et al. (2013b
▶). For the synthesis and crystal structures of similar molecules, see: Han et al. (2009 ▶); Penthala et al. (2013a
▶). For details of the experimental procedure, see: Hope (1994 ▶); Parkin & Hope (1998 ▶);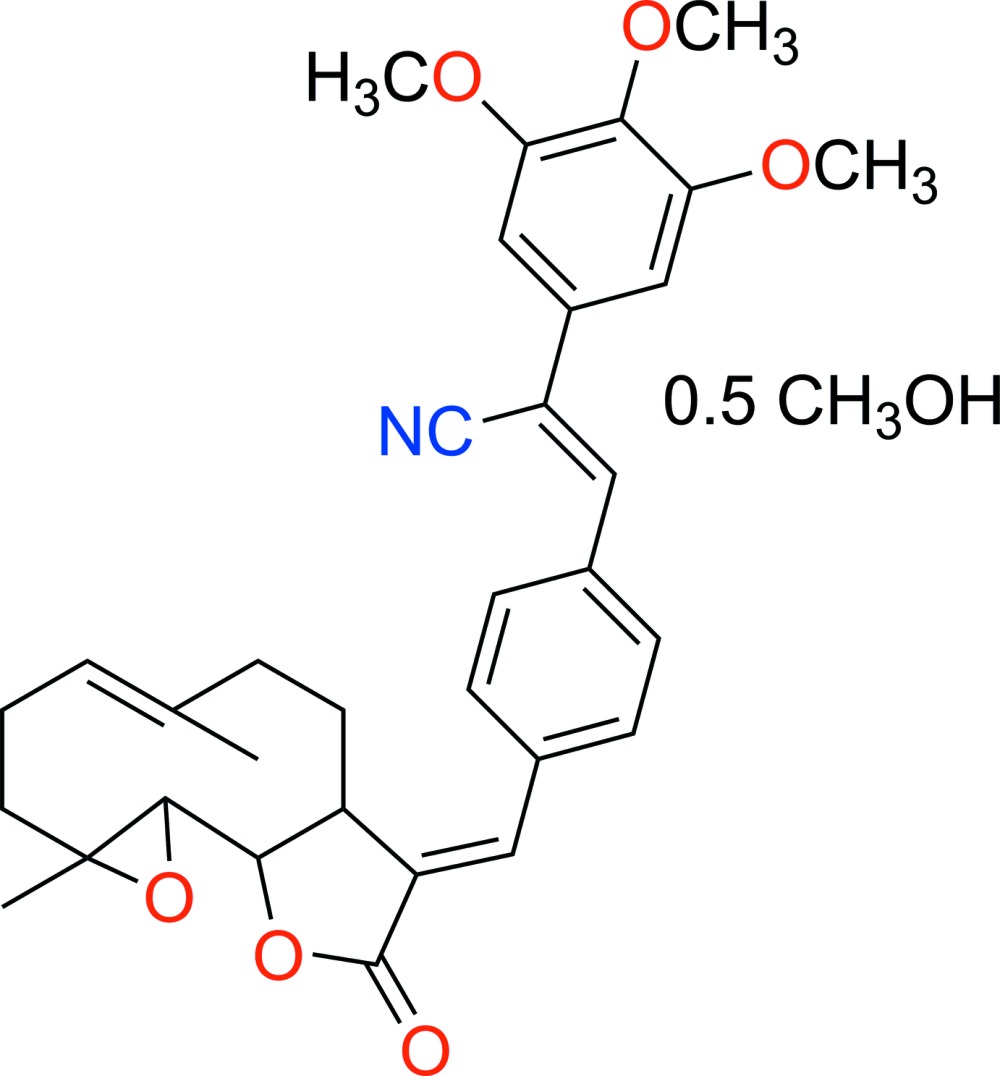
Experimental
Crystal data
C33H35NO6·0.5CH4O
M r = 557.64
Orthorhombic,
a = 9.3347 (2) Å
b = 16.2442 (3) Å
c = 19.2580 (4) Å
V = 2920.18 (10) Å3
Z = 4
Cu Kα radiation
μ = 0.71 mm−1
T = 90 K
0.18 × 0.15 × 0.10 mm
Data collection
Bruker X8 Proteum diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2008b ▶) T min = 0.836, T max = 0.963
40379 measured reflections
5349 independent reflections
5303 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.024
wR(F 2) = 0.065
S = 1.03
5349 reflections
387 parameters
H-atom parameters constrained
Δρmax = 0.14 e Å−3
Δρmin = −0.13 e Å−3
Absolute structure: Flack x determined using 2283 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013 ▶)
Absolute structure parameter: 0.02 (2)
Data collection: APEX2 (Bruker, 2006 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008a ▶); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2008a ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008a ▶); software used to prepare material for publication: SHELXL97 (Sheldrick, 2008a ▶), CIFFIX (Parkin, 2013 ▶), PLATON (Spek, 2009 ▶) and local program (Parkin, 2000 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814019333/sj5404sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814019333/sj5404Isup2.hkl
. DOI: 10.1107/S1600536814019333/sj5404fig1.tif
A view of the molecule with displacement ellipsoids drawn at the 50% probability level.
CCDC reference: 1021449
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by NIH/NCI (grant No. CA158275).
supplementary crystallographic information
S1. Comment
Parthenolide (PTL) and its analogs belong to the class of sesquiterpene lactones. These compounds are currently being used in the development of anti-cancer agents for the treatment of hematological tumours (Sun et al., 2006; Hehner et al., 1998; Ralstin et al. 2006; Oka et al., 2007; Kupchan et al., 1971 and Hanson et al., 1970). Recently, we have reported the crystal structure of (E)-13-(4-aminophenyl)parthenolide, a Heck reaction derivative of parthenolide (Penthala et al. 2013a), and we have also reported on Z-2-(3,4,5-trimethoxyphenyl)acrylonitrile analogs (Penthala et al. 2013b) as anti-cancer agents. As part of a program for the development of parthenolide analogs as anti-leukemic agents (Neelakantan et al. 2009), and small molecule analogs as anti-cancer agents, our research group is focusing on the synthesis of E-olefinic analogues of PTL which can be obtained from the reaction of parthenolide with iodoaromatic reagents utilizing Heck chemistry (Han et al. 2009). The title compound was obtained from the reaction of parthenolide with (Z)-3-(4-iodophenyl) -2-(3,4,5- trimethoxyphenylacrylonitrile under Heck reaction conditions. To obtain detailed information on the structure of the title compound and to establish the geometry of the exocyclic C13—C14 double bond, a single-crystal X-ray structure determination has been carried out.
Recrystallization of the title compound from methanol afforded light yellow coloured crystals that were suitable for X-ray analysis. The X-ray studies revealed that the title compound was identified as the E-isomer (conformation about the exocyclic methylidene C═C bond; the conformation about the C═C bond in the ten-membered ring is also E). The molecule is built up from fused ten-, five- (lactone) and three-membered (epoxide) rings with a (Z)-3-(4-phenyl)-2-(3,4,5- trimethoxyphenyl)acrylonitrile group as a substituent. The dihedral angle between the benzene ring of the phenyl moiety and the lactone ring mean plane is 21.93 (4) Å.
S2. Experimental
A mixture of parthenolide (1.0 mmol), diisopropylethylamine (3.0 mmol), and (Z)-3-(4-iodophenyl)-2-(3,4,5-trimethoxyphenyl acrylonitrile (1.1 mmol) in toluene (1 ml) was treated with palladium (II) ferrocene (0.01 mmol) and then stirred at 353 K for 24 h. The reaction mixture was cooled to room temperature, water (8 ml) was added, and the mixture was extracted with ethyl acetate (10 mlx3). The separated organics were dried over anhydrous Na2SO4 and concentrated under reduced pressure. The obtained crude residue was purified using silica flash chromatography (7:3, hexanes/EtOAc) to afford the title compound, which was recrystallized from methanol as light yellow coloured crystals suitable for X-ray analysis (87% yield; M·P.: 478–480 K); 1H NMR (400 MHz, CDCl3): δ 7.95 (d, J = 8.4 Hz, 2H), 7.67 (d, J = 3.6 Hz, 1H), 7.52 (d, J = 8.4 Hz, 2H), 7.46 (s, 1H), 6.88 (s, 2H), 5.29 (d, J = 11.2 Hz, 1H), 3.94 (s, 6H, 2xOCH3), 3.90 (s, 3H, OCH3), 3.3 (m, 1H), 2.85 (d, J = 8.4 Hz, 1H), 2.41–2.46 (m, 1H), 2.10–2.27 (m, 5H), 1.69 (s, 3H, CH3), 1.46–1.55 (m, 2H), 1.32 (s, 3H, CH3), 1.27–1.30 (m, 1H) p.p.m.. 13C NMR (100 MHz, CDCl3): δ 17.60,17.70, 24.54, 30.58, 36.33, 42.13, 47.16, 55.60, 61.27, 61.94, 66.71, 83.30, 103.75, 113.21, 118.04, 125.48, 129.53, 129.95, 130.56, 130.97, 134.88, 134.96, 135.69, 137.06, 139.77, 140.34, 153.89, 170.85 p.p.m..
S3. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 1. H atoms were found in difference Fourier maps, but subsequently included in the refinement using riding models, with constrained distances set to 0.95%A (Csp2H), 0.98Å (RCH3), 0.99Å (R2CH2), 1.00Å (R3CH) and 0.84Å (OH). Uiso(H) parameters were set to values of either 1.2Ueq or 1.5Ueq (RCH3 and OH only) of the attached atom.
The partial occupancy methanol molecule refined to an occupancy of about one half. For the final rounds of refinement its occupancy was fixed at exactly 0.5 for the sake of simplicity. This is reasonable because other crystals from the same batch would almost certainly have had varying amounts of solvent incorporated, due to unpredictable rates of solvent loss dependent on such things as crystal handling. The position of this half-occupancy methanol is consistent with an O—H···π weak hydrogen bonding interaction in which the distance between atom O1M and the centroid of the trimethoxyphenyl ring (C24-C29) is 3.212 (3)Å.
Figures
Fig. 1.
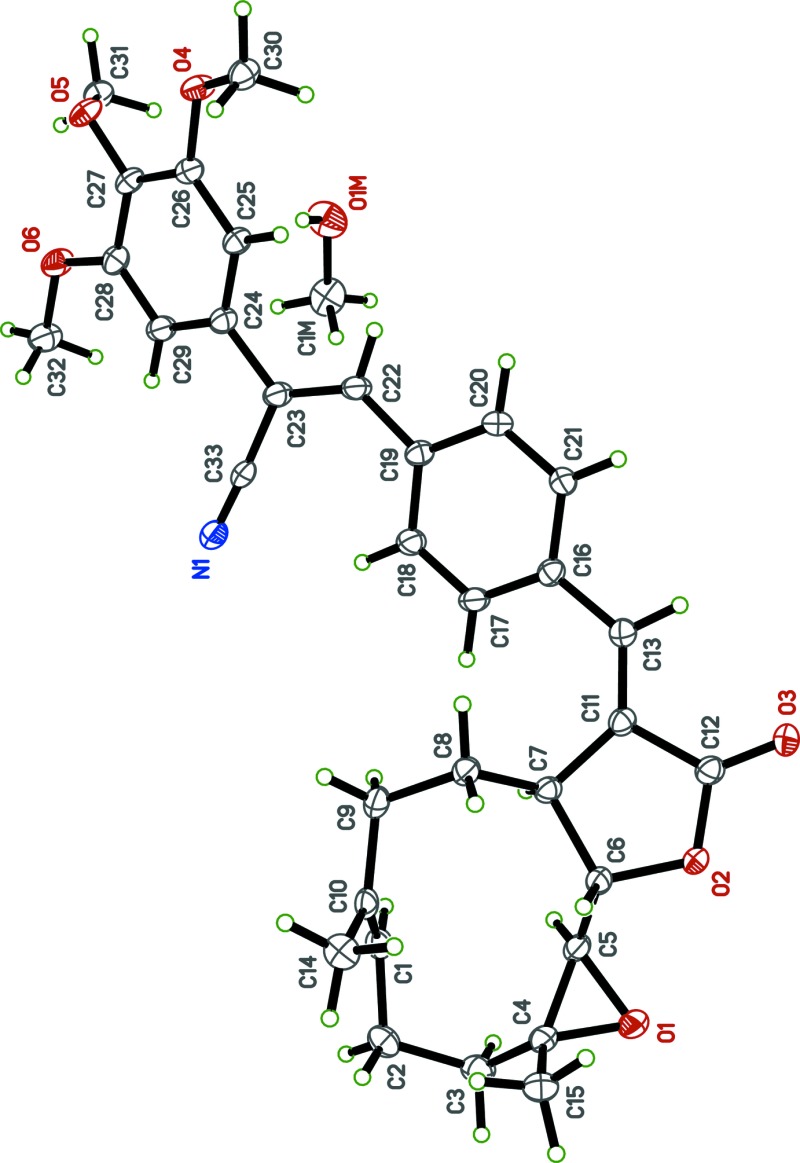
A view of the molecule with displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C33H35NO6·0.5CH4O | Dx = 1.268 Mg m−3 |
| Mr = 557.64 | Cu Kα radiation, λ = 1.54178 Å |
| Orthorhombic, P212121 | Cell parameters from 9693 reflections |
| a = 9.3347 (2) Å | θ = 3.6–68.4° |
| b = 16.2442 (3) Å | µ = 0.71 mm−1 |
| c = 19.2580 (4) Å | T = 90 K |
| V = 2920.18 (10) Å3 | Irregular cut wedge, pale yellow |
| Z = 4 | 0.18 × 0.15 × 0.10 mm |
| F(000) = 1188 |
Data collection
| Bruker X8 Proteum diffractometer | 5349 independent reflections |
| Radiation source: fine-focus rotating anode | 5303 reflections with I > 2σ(I) |
| Detector resolution: 5.6 pixels mm-1 | Rint = 0.036 |
| φ and ω scans | θmax = 68.4°, θmin = 3.6° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2008b) | h = −11→11 |
| Tmin = 0.836, Tmax = 0.963 | k = −13→19 |
| 40379 measured reflections | l = −20→23 |
Refinement
| Refinement on F2 | Hydrogen site location: difference Fourier map |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.024 | w = 1/[σ2(Fo2) + (0.0363P)2 + 0.5907P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.065 | (Δ/σ)max < 0.001 |
| S = 1.03 | Δρmax = 0.14 e Å−3 |
| 5349 reflections | Δρmin = −0.13 e Å−3 |
| 387 parameters | Extinction correction: SHELXL2014 (Sheldrick, 2008a), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.00092 (14) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack x determined using 2283 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| Secondary atom site location: difference Fourier map | Absolute structure parameter: 0.02 (2) |
Special details
| Experimental. The crystal was mounted with polyisobutene oil on the tip of a fine glass fibre, which was fastened in a copper mounting pin with electrical solder. It was placed directly into the cold gas stream of a liquid nitrogen based cryostat, according to published methods (Hope, 1994; Parkin & Hope, 1998).Diffraction data were collected with the crystal at 90 K, which is standard practice in this laboratory for the majority of flash-cooled crystals. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement progress was checked using PLATON (Spek, 2009) and by an R-tensor (Parkin, 2000). The final model was further checked with the IUCr utility checkCIF. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.48351 (13) | 0.18048 (7) | 0.54248 (6) | 0.0242 (3) | |
| O2 | 0.35142 (13) | 0.32648 (7) | 0.48438 (5) | 0.0210 (2) | |
| O3 | 0.34451 (13) | 0.45934 (7) | 0.45520 (6) | 0.0237 (3) | |
| O4 | 0.02191 (12) | 0.41510 (7) | −0.22367 (6) | 0.0225 (3) | |
| O5 | 0.14060 (12) | 0.30869 (7) | −0.31036 (5) | 0.0219 (2) | |
| O6 | 0.30389 (14) | 0.18270 (7) | −0.26366 (6) | 0.0261 (3) | |
| N1 | 0.35753 (19) | 0.14312 (9) | 0.01607 (8) | 0.0311 (4) | |
| C1 | 0.35744 (17) | 0.04178 (10) | 0.37932 (8) | 0.0203 (3) | |
| H1 | 0.4318 | 0.0590 | 0.3489 | 0.024* | |
| C2 | 0.40090 (19) | −0.01340 (10) | 0.43865 (9) | 0.0231 (3) | |
| H2A | 0.4617 | −0.0586 | 0.4208 | 0.028* | |
| H2B | 0.3143 | −0.0381 | 0.4598 | 0.028* | |
| C3 | 0.48435 (19) | 0.03537 (10) | 0.49458 (9) | 0.0249 (4) | |
| H3A | 0.5000 | 0.0001 | 0.5358 | 0.030* | |
| H3B | 0.5792 | 0.0516 | 0.4761 | 0.030* | |
| C4 | 0.40199 (18) | 0.11124 (10) | 0.51545 (8) | 0.0213 (3) | |
| C5 | 0.42653 (17) | 0.18504 (10) | 0.47296 (8) | 0.0187 (3) | |
| H5 | 0.5005 | 0.1772 | 0.4360 | 0.022* | |
| C6 | 0.31309 (17) | 0.24713 (9) | 0.45417 (8) | 0.0180 (3) | |
| H6 | 0.2182 | 0.2288 | 0.4727 | 0.022* | |
| C7 | 0.30239 (17) | 0.26039 (9) | 0.37424 (8) | 0.0170 (3) | |
| H7 | 0.3909 | 0.2376 | 0.3522 | 0.020* | |
| C8 | 0.17085 (17) | 0.21982 (10) | 0.33974 (8) | 0.0197 (3) | |
| H8A | 0.0932 | 0.2152 | 0.3745 | 0.024* | |
| H8B | 0.1362 | 0.2559 | 0.3019 | 0.024* | |
| C9 | 0.20236 (18) | 0.13380 (10) | 0.30982 (8) | 0.0211 (3) | |
| H9A | 0.1211 | 0.1168 | 0.2801 | 0.025* | |
| H9B | 0.2887 | 0.1370 | 0.2801 | 0.025* | |
| C10 | 0.22602 (17) | 0.06913 (9) | 0.36481 (8) | 0.0189 (3) | |
| C11 | 0.30617 (16) | 0.35309 (9) | 0.36819 (8) | 0.0175 (3) | |
| C12 | 0.33264 (17) | 0.38809 (9) | 0.43821 (8) | 0.0191 (3) | |
| C13 | 0.29827 (17) | 0.40414 (9) | 0.31380 (8) | 0.0188 (3) | |
| H13 | 0.2959 | 0.4610 | 0.3254 | 0.023* | |
| C14 | 0.09048 (18) | 0.04115 (11) | 0.39995 (9) | 0.0253 (4) | |
| H14A | 0.1121 | −0.0048 | 0.4313 | 0.038* | |
| H14B | 0.0213 | 0.0231 | 0.3648 | 0.038* | |
| H14C | 0.0498 | 0.0869 | 0.4267 | 0.038* | |
| C15 | 0.2653 (2) | 0.09726 (11) | 0.55515 (9) | 0.0265 (4) | |
| H15A | 0.2874 | 0.0715 | 0.5999 | 0.040* | |
| H15B | 0.2021 | 0.0610 | 0.5283 | 0.040* | |
| H15C | 0.2175 | 0.1501 | 0.5631 | 0.040* | |
| C16 | 0.29273 (16) | 0.38648 (9) | 0.23919 (8) | 0.0175 (3) | |
| C17 | 0.33360 (19) | 0.31137 (10) | 0.20977 (8) | 0.0217 (3) | |
| H17 | 0.3699 | 0.2690 | 0.2389 | 0.026* | |
| C18 | 0.32232 (19) | 0.29741 (10) | 0.13921 (8) | 0.0226 (3) | |
| H18 | 0.3514 | 0.2459 | 0.1207 | 0.027* | |
| C19 | 0.26861 (17) | 0.35823 (10) | 0.09458 (8) | 0.0182 (3) | |
| C20 | 0.23537 (17) | 0.43490 (10) | 0.12358 (8) | 0.0189 (3) | |
| H20 | 0.2037 | 0.4782 | 0.0942 | 0.023* | |
| C21 | 0.24764 (17) | 0.44897 (9) | 0.19415 (8) | 0.0191 (3) | |
| H21 | 0.2252 | 0.5018 | 0.2123 | 0.023* | |
| C22 | 0.24427 (18) | 0.34952 (10) | 0.01995 (8) | 0.0202 (3) | |
| H22 | 0.2131 | 0.3986 | −0.0022 | 0.024* | |
| C23 | 0.25803 (18) | 0.28451 (10) | −0.02315 (8) | 0.0201 (3) | |
| C24 | 0.22173 (17) | 0.28790 (10) | −0.09850 (8) | 0.0201 (3) | |
| C25 | 0.13268 (17) | 0.35015 (10) | −0.12349 (8) | 0.0194 (3) | |
| H25 | 0.0903 | 0.3882 | −0.0922 | 0.023* | |
| C26 | 0.10621 (17) | 0.35628 (10) | −0.19439 (8) | 0.0188 (3) | |
| C27 | 0.16914 (17) | 0.30063 (10) | −0.24085 (8) | 0.0189 (3) | |
| C28 | 0.25283 (18) | 0.23611 (10) | −0.21509 (8) | 0.0213 (3) | |
| C29 | 0.27962 (18) | 0.22993 (10) | −0.14397 (8) | 0.0229 (3) | |
| H29 | 0.3372 | 0.1863 | −0.1266 | 0.027* | |
| C30 | −0.07334 (18) | 0.45919 (11) | −0.17877 (8) | 0.0228 (3) | |
| H30A | −0.1350 | 0.4201 | −0.1541 | 0.034* | |
| H30B | −0.1327 | 0.4966 | −0.2064 | 0.034* | |
| H30C | −0.0175 | 0.4910 | −0.1451 | 0.034* | |
| C31 | 0.26452 (18) | 0.32355 (10) | −0.35293 (8) | 0.0219 (3) | |
| H31A | 0.3352 | 0.3552 | −0.3265 | 0.033* | |
| H31B | 0.2362 | 0.3547 | −0.3943 | 0.033* | |
| H31C | 0.3065 | 0.2709 | −0.3671 | 0.033* | |
| C32 | 0.3951 (2) | 0.11810 (11) | −0.24020 (9) | 0.0294 (4) | |
| H32A | 0.4758 | 0.1416 | −0.2143 | 0.044* | |
| H32B | 0.4314 | 0.0873 | −0.2802 | 0.044* | |
| H32C | 0.3409 | 0.0810 | −0.2099 | 0.044* | |
| C33 | 0.31313 (19) | 0.20649 (10) | 0.00030 (8) | 0.0219 (3) | |
| O1M | 0.4544 (3) | 0.42249 (17) | −0.16592 (16) | 0.0371 (6) | 0.5 |
| H1M | 0.3705 | 0.4054 | −0.1596 | 0.056* | 0.5 |
| C1M | 0.5516 (4) | 0.3704 (3) | −0.1313 (2) | 0.0338 (8) | 0.5 |
| H1M1 | 0.6414 | 0.4001 | −0.1230 | 0.051* | 0.5 |
| H1M2 | 0.5705 | 0.3218 | −0.1600 | 0.051* | 0.5 |
| H1M3 | 0.5104 | 0.3532 | −0.0868 | 0.051* | 0.5 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0286 (6) | 0.0255 (6) | 0.0187 (5) | −0.0042 (5) | −0.0075 (5) | 0.0020 (5) |
| O2 | 0.0323 (6) | 0.0182 (5) | 0.0125 (5) | −0.0012 (5) | −0.0003 (5) | −0.0012 (4) |
| O3 | 0.0345 (7) | 0.0187 (5) | 0.0181 (5) | −0.0004 (5) | −0.0002 (5) | −0.0042 (4) |
| O4 | 0.0247 (6) | 0.0275 (6) | 0.0153 (5) | 0.0082 (5) | −0.0013 (5) | 0.0017 (5) |
| O5 | 0.0190 (6) | 0.0349 (6) | 0.0118 (5) | 0.0016 (5) | −0.0008 (4) | 0.0001 (5) |
| O6 | 0.0331 (7) | 0.0291 (6) | 0.0161 (5) | 0.0110 (5) | −0.0011 (5) | −0.0044 (5) |
| N1 | 0.0477 (10) | 0.0227 (7) | 0.0228 (7) | 0.0045 (7) | −0.0131 (7) | −0.0036 (6) |
| C1 | 0.0219 (8) | 0.0184 (7) | 0.0205 (8) | −0.0015 (6) | 0.0034 (6) | −0.0030 (6) |
| C2 | 0.0208 (8) | 0.0193 (7) | 0.0293 (9) | 0.0014 (6) | 0.0023 (7) | 0.0014 (7) |
| C3 | 0.0251 (8) | 0.0239 (8) | 0.0258 (8) | 0.0009 (7) | −0.0036 (7) | 0.0060 (7) |
| C4 | 0.0245 (8) | 0.0228 (8) | 0.0165 (7) | −0.0033 (7) | −0.0049 (7) | 0.0017 (6) |
| C5 | 0.0200 (8) | 0.0223 (8) | 0.0137 (7) | −0.0030 (6) | −0.0017 (6) | −0.0007 (6) |
| C6 | 0.0217 (8) | 0.0178 (7) | 0.0145 (7) | −0.0029 (6) | 0.0008 (6) | −0.0011 (6) |
| C7 | 0.0189 (7) | 0.0183 (7) | 0.0137 (7) | 0.0018 (6) | 0.0008 (6) | −0.0001 (6) |
| C8 | 0.0226 (8) | 0.0192 (7) | 0.0172 (7) | 0.0022 (6) | −0.0032 (6) | −0.0011 (6) |
| C9 | 0.0252 (8) | 0.0208 (8) | 0.0174 (7) | −0.0003 (6) | −0.0019 (6) | −0.0049 (6) |
| C10 | 0.0232 (8) | 0.0165 (7) | 0.0169 (7) | −0.0015 (6) | 0.0018 (6) | −0.0057 (6) |
| C11 | 0.0179 (7) | 0.0192 (7) | 0.0154 (7) | 0.0020 (6) | 0.0005 (6) | −0.0017 (6) |
| C12 | 0.0209 (8) | 0.0205 (8) | 0.0158 (7) | 0.0009 (6) | 0.0022 (6) | 0.0000 (6) |
| C13 | 0.0206 (7) | 0.0175 (7) | 0.0183 (7) | 0.0008 (6) | −0.0012 (6) | −0.0012 (6) |
| C14 | 0.0209 (8) | 0.0256 (8) | 0.0295 (9) | 0.0005 (7) | 0.0010 (7) | 0.0009 (7) |
| C15 | 0.0329 (9) | 0.0265 (8) | 0.0201 (8) | −0.0039 (7) | 0.0032 (7) | 0.0039 (7) |
| C16 | 0.0160 (7) | 0.0200 (7) | 0.0165 (7) | −0.0013 (6) | −0.0008 (6) | 0.0002 (6) |
| C17 | 0.0293 (9) | 0.0201 (7) | 0.0158 (7) | 0.0052 (7) | −0.0011 (6) | 0.0029 (6) |
| C18 | 0.0316 (9) | 0.0192 (8) | 0.0172 (7) | 0.0065 (7) | −0.0005 (7) | −0.0003 (6) |
| C19 | 0.0194 (7) | 0.0205 (7) | 0.0147 (7) | 0.0005 (6) | 0.0011 (6) | 0.0015 (6) |
| C20 | 0.0196 (7) | 0.0192 (7) | 0.0178 (7) | 0.0018 (6) | −0.0006 (6) | 0.0040 (6) |
| C21 | 0.0226 (8) | 0.0164 (7) | 0.0183 (7) | 0.0002 (6) | 0.0002 (6) | −0.0005 (6) |
| C22 | 0.0245 (8) | 0.0202 (7) | 0.0159 (7) | 0.0027 (7) | −0.0007 (6) | 0.0042 (6) |
| C23 | 0.0226 (8) | 0.0219 (8) | 0.0158 (7) | 0.0015 (7) | −0.0013 (6) | 0.0023 (6) |
| C24 | 0.0237 (8) | 0.0215 (7) | 0.0151 (7) | −0.0016 (6) | −0.0008 (6) | 0.0014 (6) |
| C25 | 0.0215 (8) | 0.0227 (7) | 0.0140 (7) | 0.0003 (6) | 0.0002 (6) | −0.0006 (6) |
| C26 | 0.0177 (7) | 0.0217 (7) | 0.0170 (7) | −0.0006 (6) | −0.0011 (6) | 0.0024 (6) |
| C27 | 0.0174 (7) | 0.0263 (8) | 0.0130 (7) | −0.0016 (6) | −0.0006 (6) | 0.0010 (6) |
| C28 | 0.0225 (8) | 0.0245 (8) | 0.0168 (7) | 0.0003 (7) | 0.0003 (6) | −0.0029 (6) |
| C29 | 0.0277 (8) | 0.0229 (8) | 0.0181 (8) | 0.0047 (7) | −0.0028 (6) | 0.0007 (6) |
| C30 | 0.0222 (8) | 0.0269 (8) | 0.0192 (8) | 0.0048 (7) | −0.0010 (6) | −0.0031 (7) |
| C31 | 0.0230 (8) | 0.0264 (8) | 0.0165 (7) | 0.0005 (7) | 0.0029 (6) | 0.0007 (6) |
| C32 | 0.0359 (10) | 0.0292 (9) | 0.0231 (8) | 0.0125 (8) | −0.0005 (8) | −0.0024 (7) |
| C33 | 0.0301 (8) | 0.0225 (8) | 0.0130 (7) | −0.0001 (7) | −0.0046 (6) | −0.0031 (6) |
| O1M | 0.0289 (13) | 0.0360 (14) | 0.0465 (16) | −0.0027 (12) | 0.0055 (12) | 0.0064 (13) |
| C1M | 0.0213 (17) | 0.044 (2) | 0.0359 (19) | −0.0039 (15) | 0.0058 (16) | −0.0010 (18) |
Geometric parameters (Å, º)
| O1—C5 | 1.4426 (18) | C14—H14B | 0.9800 |
| O1—C4 | 1.454 (2) | C14—H14C | 0.9800 |
| O2—C12 | 1.3500 (19) | C15—H15A | 0.9800 |
| O2—C6 | 1.4587 (18) | C15—H15B | 0.9800 |
| O3—C12 | 1.208 (2) | C15—H15C | 0.9800 |
| O4—C26 | 1.3602 (19) | C16—C17 | 1.398 (2) |
| O4—C30 | 1.4321 (19) | C16—C21 | 1.400 (2) |
| O5—C27 | 1.3713 (18) | C17—C18 | 1.382 (2) |
| O5—C31 | 1.4381 (19) | C17—H17 | 0.9500 |
| O6—C28 | 1.3619 (19) | C18—C19 | 1.402 (2) |
| O6—C32 | 1.425 (2) | C18—H18 | 0.9500 |
| N1—C33 | 1.151 (2) | C19—C20 | 1.400 (2) |
| C1—C10 | 1.334 (2) | C19—C22 | 1.462 (2) |
| C1—C2 | 1.508 (2) | C20—C21 | 1.383 (2) |
| C1—H1 | 0.9500 | C20—H20 | 0.9500 |
| C2—C3 | 1.547 (2) | C21—H21 | 0.9500 |
| C2—H2A | 0.9900 | C22—C23 | 1.349 (2) |
| C2—H2B | 0.9900 | C22—H22 | 0.9500 |
| C3—C4 | 1.507 (2) | C23—C33 | 1.440 (2) |
| C3—H3A | 0.9900 | C23—C24 | 1.491 (2) |
| C3—H3B | 0.9900 | C24—C29 | 1.395 (2) |
| C4—C5 | 1.470 (2) | C24—C25 | 1.395 (2) |
| C4—C15 | 1.505 (2) | C25—C26 | 1.391 (2) |
| C5—C6 | 1.506 (2) | C25—H25 | 0.9500 |
| C5—H5 | 1.0000 | C26—C27 | 1.401 (2) |
| C6—C7 | 1.558 (2) | C27—C28 | 1.398 (2) |
| C6—H6 | 1.0000 | C28—C29 | 1.396 (2) |
| C7—C11 | 1.511 (2) | C29—H29 | 0.9500 |
| C7—C8 | 1.544 (2) | C30—H30A | 0.9800 |
| C7—H7 | 1.0000 | C30—H30B | 0.9800 |
| C8—C9 | 1.540 (2) | C30—H30C | 0.9800 |
| C8—H8A | 0.9900 | C31—H31A | 0.9800 |
| C8—H8B | 0.9900 | C31—H31B | 0.9800 |
| C9—C10 | 1.508 (2) | C31—H31C | 0.9800 |
| C9—H9A | 0.9900 | C32—H32A | 0.9800 |
| C9—H9B | 0.9900 | C32—H32B | 0.9800 |
| C10—C14 | 1.505 (2) | C32—H32C | 0.9800 |
| C11—C13 | 1.338 (2) | O1M—C1M | 1.408 (5) |
| C11—C12 | 1.484 (2) | O1M—H1M | 0.8400 |
| C13—C16 | 1.466 (2) | C1M—H1M1 | 0.9800 |
| C13—H13 | 0.9500 | C1M—H1M2 | 0.9800 |
| C14—H14A | 0.9800 | C1M—H1M3 | 0.9800 |
| C5—O1—C4 | 60.96 (10) | C4—C15—H15A | 109.5 |
| C12—O2—C6 | 111.14 (11) | C4—C15—H15B | 109.5 |
| C26—O4—C30 | 117.40 (12) | H15A—C15—H15B | 109.5 |
| C27—O5—C31 | 114.60 (12) | C4—C15—H15C | 109.5 |
| C28—O6—C32 | 117.42 (12) | H15A—C15—H15C | 109.5 |
| C10—C1—C2 | 127.16 (15) | H15B—C15—H15C | 109.5 |
| C10—C1—H1 | 116.4 | C17—C16—C21 | 117.62 (14) |
| C2—C1—H1 | 116.4 | C17—C16—C13 | 123.93 (14) |
| C1—C2—C3 | 111.01 (13) | C21—C16—C13 | 118.42 (14) |
| C1—C2—H2A | 109.4 | C18—C17—C16 | 121.39 (15) |
| C3—C2—H2A | 109.4 | C18—C17—H17 | 119.3 |
| C1—C2—H2B | 109.4 | C16—C17—H17 | 119.3 |
| C3—C2—H2B | 109.4 | C17—C18—C19 | 120.96 (14) |
| H2A—C2—H2B | 108.0 | C17—C18—H18 | 119.5 |
| C4—C3—C2 | 110.34 (14) | C19—C18—H18 | 119.5 |
| C4—C3—H3A | 109.6 | C20—C19—C18 | 117.49 (14) |
| C2—C3—H3A | 109.6 | C20—C19—C22 | 116.35 (14) |
| C4—C3—H3B | 109.6 | C18—C19—C22 | 126.16 (15) |
| C2—C3—H3B | 109.6 | C21—C20—C19 | 121.39 (14) |
| H3A—C3—H3B | 108.1 | C21—C20—H20 | 119.3 |
| O1—C4—C5 | 59.12 (10) | C19—C20—H20 | 119.3 |
| O1—C4—C15 | 112.24 (13) | C20—C21—C16 | 120.93 (14) |
| C5—C4—C15 | 122.56 (15) | C20—C21—H21 | 119.5 |
| O1—C4—C3 | 117.45 (14) | C16—C21—H21 | 119.5 |
| C5—C4—C3 | 116.05 (14) | C23—C22—C19 | 131.73 (15) |
| C15—C4—C3 | 116.40 (14) | C23—C22—H22 | 114.1 |
| O1—C5—C4 | 59.92 (10) | C19—C22—H22 | 114.1 |
| O1—C5—C6 | 121.09 (13) | C22—C23—C33 | 122.00 (14) |
| C4—C5—C6 | 124.78 (14) | C22—C23—C24 | 123.24 (15) |
| O1—C5—H5 | 113.6 | C33—C23—C24 | 114.74 (14) |
| C4—C5—H5 | 113.6 | C29—C24—C25 | 120.25 (14) |
| C6—C5—H5 | 113.6 | C29—C24—C23 | 119.87 (14) |
| O2—C6—C5 | 108.87 (12) | C25—C24—C23 | 119.86 (14) |
| O2—C6—C7 | 106.71 (12) | C26—C25—C24 | 119.76 (15) |
| C5—C6—C7 | 112.02 (12) | C26—C25—H25 | 120.1 |
| O2—C6—H6 | 109.7 | C24—C25—H25 | 120.1 |
| C5—C6—H6 | 109.7 | O4—C26—C25 | 124.05 (14) |
| C7—C6—H6 | 109.7 | O4—C26—C27 | 115.54 (13) |
| C11—C7—C8 | 114.26 (13) | C25—C26—C27 | 120.41 (15) |
| C11—C7—C6 | 102.28 (12) | O5—C27—C28 | 121.77 (14) |
| C8—C7—C6 | 114.66 (13) | O5—C27—C26 | 118.71 (14) |
| C11—C7—H7 | 108.4 | C28—C27—C26 | 119.43 (14) |
| C8—C7—H7 | 108.4 | O6—C28—C29 | 124.43 (15) |
| C6—C7—H7 | 108.4 | O6—C28—C27 | 115.43 (13) |
| C9—C8—C7 | 113.36 (13) | C29—C28—C27 | 120.14 (14) |
| C9—C8—H8A | 108.9 | C24—C29—C28 | 119.87 (15) |
| C7—C8—H8A | 108.9 | C24—C29—H29 | 120.1 |
| C9—C8—H8B | 108.9 | C28—C29—H29 | 120.1 |
| C7—C8—H8B | 108.9 | O4—C30—H30A | 109.5 |
| H8A—C8—H8B | 107.7 | O4—C30—H30B | 109.5 |
| C10—C9—C8 | 113.42 (12) | H30A—C30—H30B | 109.5 |
| C10—C9—H9A | 108.9 | O4—C30—H30C | 109.5 |
| C8—C9—H9A | 108.9 | H30A—C30—H30C | 109.5 |
| C10—C9—H9B | 108.9 | H30B—C30—H30C | 109.5 |
| C8—C9—H9B | 108.9 | O5—C31—H31A | 109.5 |
| H9A—C9—H9B | 107.7 | O5—C31—H31B | 109.5 |
| C1—C10—C14 | 125.32 (15) | H31A—C31—H31B | 109.5 |
| C1—C10—C9 | 120.92 (15) | O5—C31—H31C | 109.5 |
| C14—C10—C9 | 113.76 (14) | H31A—C31—H31C | 109.5 |
| C13—C11—C12 | 118.88 (14) | H31B—C31—H31C | 109.5 |
| C13—C11—C7 | 132.60 (14) | O6—C32—H32A | 109.5 |
| C12—C11—C7 | 108.40 (13) | O6—C32—H32B | 109.5 |
| O3—C12—O2 | 121.34 (14) | H32A—C32—H32B | 109.5 |
| O3—C12—C11 | 128.93 (15) | O6—C32—H32C | 109.5 |
| O2—C12—C11 | 109.63 (13) | H32A—C32—H32C | 109.5 |
| C11—C13—C16 | 130.38 (15) | H32B—C32—H32C | 109.5 |
| C11—C13—H13 | 114.8 | N1—C33—C23 | 177.04 (16) |
| C16—C13—H13 | 114.8 | C1M—O1M—H1M | 109.5 |
| C10—C14—H14A | 109.5 | O1M—C1M—H1M1 | 109.5 |
| C10—C14—H14B | 109.5 | O1M—C1M—H1M2 | 109.5 |
| H14A—C14—H14B | 109.5 | H1M1—C1M—H1M2 | 109.5 |
| C10—C14—H14C | 109.5 | O1M—C1M—H1M3 | 109.5 |
| H14A—C14—H14C | 109.5 | H1M1—C1M—H1M3 | 109.5 |
| H14B—C14—H14C | 109.5 | H1M2—C1M—H1M3 | 109.5 |
| C10—C1—C2—C3 | −107.33 (19) | C11—C13—C16—C17 | −18.2 (3) |
| C1—C2—C3—C4 | 51.42 (18) | C11—C13—C16—C21 | 163.60 (17) |
| C5—O1—C4—C15 | −115.66 (16) | C21—C16—C17—C18 | −3.6 (2) |
| C5—O1—C4—C3 | 105.42 (16) | C13—C16—C17—C18 | 178.17 (16) |
| C2—C3—C4—O1 | −154.06 (13) | C16—C17—C18—C19 | −0.3 (3) |
| C2—C3—C4—C5 | −87.00 (17) | C17—C18—C19—C20 | 3.8 (2) |
| C2—C3—C4—C15 | 68.71 (18) | C17—C18—C19—C22 | −176.49 (17) |
| C4—O1—C5—C6 | 114.86 (17) | C18—C19—C20—C21 | −3.4 (2) |
| C15—C4—C5—O1 | 98.13 (16) | C22—C19—C20—C21 | 176.89 (15) |
| C3—C4—C5—O1 | −107.79 (15) | C19—C20—C21—C16 | −0.6 (2) |
| O1—C4—C5—C6 | −108.93 (16) | C17—C16—C21—C20 | 4.1 (2) |
| C15—C4—C5—C6 | −10.8 (2) | C13—C16—C21—C20 | −177.64 (15) |
| C3—C4—C5—C6 | 143.28 (15) | C20—C19—C22—C23 | −176.47 (17) |
| C12—O2—C6—C5 | 135.46 (13) | C18—C19—C22—C23 | 3.8 (3) |
| C12—O2—C6—C7 | 14.37 (17) | C19—C22—C23—C33 | −4.2 (3) |
| O1—C5—C6—O2 | 44.08 (18) | C19—C22—C23—C24 | 177.52 (16) |
| C4—C5—C6—O2 | 116.99 (15) | C22—C23—C24—C29 | 159.30 (17) |
| O1—C5—C6—C7 | 161.85 (13) | C33—C23—C24—C29 | −19.1 (2) |
| C4—C5—C6—C7 | −125.24 (16) | C22—C23—C24—C25 | −19.2 (3) |
| O2—C6—C7—C11 | −11.56 (16) | C33—C23—C24—C25 | 162.36 (15) |
| C5—C6—C7—C11 | −130.62 (13) | C29—C24—C25—C26 | −2.5 (2) |
| O2—C6—C7—C8 | −135.79 (13) | C23—C24—C25—C26 | 175.96 (15) |
| C5—C6—C7—C8 | 105.15 (15) | C30—O4—C26—C25 | −16.8 (2) |
| C11—C7—C8—C9 | 147.81 (13) | C30—O4—C26—C27 | 164.02 (14) |
| C6—C7—C8—C9 | −94.58 (15) | C24—C25—C26—O4 | −179.51 (15) |
| C7—C8—C9—C10 | 70.67 (17) | C24—C25—C26—C27 | −0.4 (2) |
| C2—C1—C10—C14 | −8.4 (3) | C31—O5—C27—C28 | −63.9 (2) |
| C2—C1—C10—C9 | 171.04 (14) | C31—O5—C27—C26 | 119.53 (16) |
| C8—C9—C10—C1 | −104.35 (17) | O4—C26—C27—O5 | −0.7 (2) |
| C8—C9—C10—C14 | 75.17 (18) | C25—C26—C27—O5 | −179.93 (14) |
| C8—C7—C11—C13 | −54.0 (2) | O4—C26—C27—C28 | −177.44 (14) |
| C6—C7—C11—C13 | −178.52 (17) | C25—C26—C27—C28 | 3.4 (2) |
| C8—C7—C11—C12 | 130.10 (14) | C32—O6—C28—C29 | −2.5 (2) |
| C6—C7—C11—C12 | 5.60 (16) | C32—O6—C28—C27 | 177.21 (15) |
| C6—O2—C12—O3 | 172.42 (15) | O5—C27—C28—O6 | 0.3 (2) |
| C6—O2—C12—C11 | −10.86 (17) | C26—C27—C28—O6 | 176.89 (14) |
| C13—C11—C12—O3 | 2.6 (3) | O5—C27—C28—C29 | 179.97 (15) |
| C7—C11—C12—O3 | 179.18 (17) | C26—C27—C28—C29 | −3.4 (2) |
| C13—C11—C12—O2 | −173.76 (14) | C25—C24—C29—C28 | 2.5 (2) |
| C7—C11—C12—O2 | 2.78 (18) | C23—C24—C29—C28 | −176.03 (15) |
| C12—C11—C13—C16 | 170.76 (16) | O6—C28—C29—C24 | −179.82 (16) |
| C7—C11—C13—C16 | −4.8 (3) | C27—C28—C29—C24 | 0.5 (2) |
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: SJ5404).
References
- Bruker (2006). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Hall, I. H., Lee, K. H., Starnes, C. O., Sumida, Y., Wu, R. Y., Waddell, T. G., Cochran, J. W. & Gerhart, K. G. (1979). J. Pharm. Sci. 68, 537–542. [DOI] [PubMed]
- Han, C., Barrios, F. J., Riofski, M. V. & Colby, D. A. (2009). J. Org. Chem. 74, 7176–7179. [DOI] [PubMed]
- Hanson, R. L., Lardy, H. A. & Kupchan, S. M. (1970). Science, 168, 378–380. [DOI] [PubMed]
- Hehner, S. P., Heinrich, M., Bork, P. M., Vogt, M., Ratter, F., Lehmann, V., Osthoff, K. S., Dröge, W. & Schmitz, M. L. (1998). J. Biol. Chem. 273, 1288–1297. [DOI] [PubMed]
- Hope, H. (1994). Prog. Inorg. Chem. 41, 1–19.
- Kupchan, S. M., Eakin, M. A. & Thomas, A. M. (1971). J. Med. Chem. 14, 1147–1152. [DOI] [PubMed]
- Neelakantan, S., Nasim, S., Guzman, M. L., Jordan, C. T. & Crooks, P. A. (2009). Bioorg. Med. Chem. Lett. 19, 4346–4349. [DOI] [PubMed]
- Oka, D., Nishimura, K., Shiba, M., Nakai, Y., Arai, Y., Nakayama, M., Takayama, H., Inoue, H., Okuyama, A. & Nonomura, N. (2007). Int. J. Cancer, 120, 2576–2581. [DOI] [PubMed]
- Parkin, S. (2000). Acta Cryst. A56, 157–162. [DOI] [PubMed]
- Parkin, S. (2013). CIFFIX, http://xray.uky.edu/people/parkin/programs/ciffix.
- Parkin, S. & Hope, H. (1998). J. Appl. Cryst. 31, 945–953.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Penthala, N. R., Janganati, V., Parkin, S., Varughese, K. I. & Crooks, P. A. (2013a). Acta Cryst. E69, o1709–o1710. [DOI] [PMC free article] [PubMed]
- Penthala, N. R., Sonar, V. N., Horn, J., Leggas, M., Yadlapalli, J. S. & Crooks, P. A. (2013b). Medchemcomm, 4, 1073–1078. [DOI] [PMC free article] [PubMed]
- Ralstin, M. C., Gage, E. A., Yip-Schneider, M. T., Klein, P. J., Wiebke, E. A. & Schmidt, C. M. (2006). Mol. Cancer Res. 4, 387–399. [DOI] [PubMed]
- Sheldrick, G. M. (2008a). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2008b). SADABS University of Göttingen, Germany.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sun, H.-X., Zheng, Q.-F. & Tu, J. (2006). Bioorg. Med. Chem. 14, 1189–1198. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814019333/sj5404sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814019333/sj5404Isup2.hkl
. DOI: 10.1107/S1600536814019333/sj5404fig1.tif
A view of the molecule with displacement ellipsoids drawn at the 50% probability level.
CCDC reference: 1021449
Additional supporting information: crystallographic information; 3D view; checkCIF report


