Abstract
In the title compound, C44H37BN4O2, the dihedral angle between the plane of the porphyrin macrocycle ring system [r.m.s. deviation = 0.159 (1) Å] and those of three phenyl rings are 66.11 (4), 74.75 (4) and 57.00 (4)°. The conformational distortion is characterized by a mixture of ruffled, saddle and in-plane distortion modes. In the crystal, the porphyrin molecules are linked by C—H⋯π interactions into supramolecular chains running along the a-axis direction. A pair of bifurcated N—H⋯(N,N) hydrogen bonds occur across the central region of the macrocycle.
Keywords: crystal structure, porphyrinoid, tetrapyrroles, porphyrins
Related literature
For the structure and conformation of porphyrins, see: Scheidt & Lee (1987 ▶); Jentzen et al. (1997 ▶); Senge (2000 ▶, 2006 ▶). For the synthesis, see: Finnigan et al. (2011 ▶). For the handling of crystals, see Hope (1994 ▶). For related boronyl porphyrin structures, see: Hyslop et al. (1998 ▶); Schwalbe et al. (2012 ▶). For other recent free base porphyrin structures, see: Miranda et al. (2012 ▶); Leonarska et al. (2012 ▶); Senge (2013 ▶).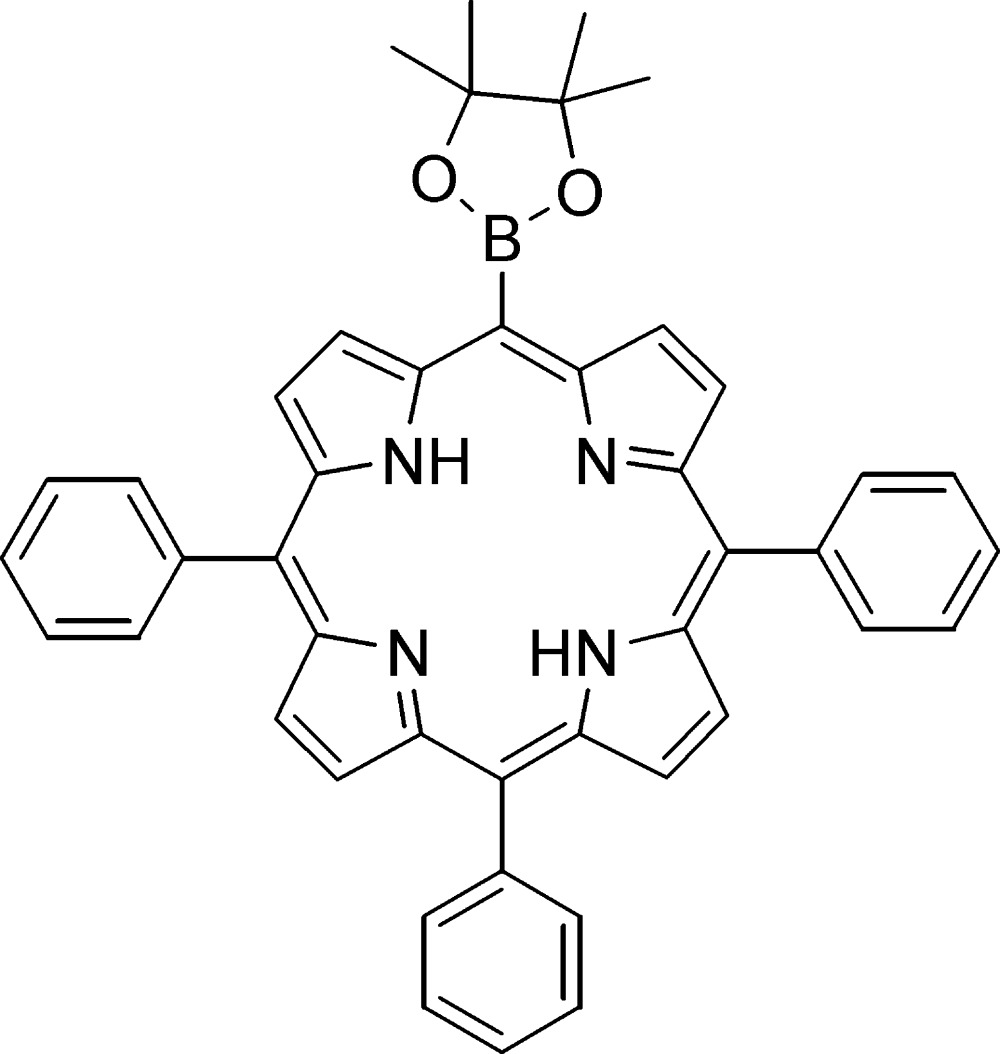
Experimental
Crystal data
C44H37BN4O2
M r = 664.59
Monoclinic,

a = 13.0758 (5) Å
b = 10.5245 (4) Å
c = 27.1358 (10) Å
β = 110.353 (2)°
V = 3501.2 (2) Å3
Z = 4
Cu Kα radiation
μ = 0.61 mm−1
T = 100 K
0.50 × 0.50 × 0.35 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.751, T max = 0.815
25108 measured reflections
5993 independent reflections
5788 reflections with I > 2σ(I)
R int = 0.029
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.103
S = 1.03
5993 reflections
464 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.27 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶) and DIAMOND (Brandenburg, 1998 ▶); software used to prepare material for publication: SHELXTL-Plus (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536814019680/lx2293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814019680/lx2293Isup2.hkl
. DOI: 10.1107/S1600536814019680/lx2293fig1.tif
Molecular structure of the title compound. Thermal ellipsoids are drawn at 50% probability level.
x y z x y z . DOI: 10.1107/S1600536814019680/lx2293fig2.tif
A view of the N—H⋯N and C—H⋯π interactions (dotted lines) in the crystal structure of the title compound. H atoms non-participating in hydrogen-bonding were omitted for clarity. [Symmetry code: (i) x − 1, y, z; (ii) x + 1, y, z.]
CCDC reference: 1022040
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C100–C105 phenyl ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N21—H21⋯N24 | 0.88 | 2.35 | 2.9084 (16) | 122 |
| N21—H21⋯N22 | 0.88 | 2.38 | 2.9266 (16) | 121 |
| N23—H23⋯N24 | 0.88 | 2.34 | 2.8978 (16) | 121 |
| N23—H23⋯N22 | 0.88 | 2.35 | 2.8986 (16) | 120 |
| C153—H153⋯Cg1i | 0.95 | 2.58 | 3.488 (2) | 160 |
Symmetry code: (i)  .
.
Acknowledgments
This work was supported by a grant from Science Foundation Ireland (SFI P·I. 09/IN.1/B2650).
supplementary crystallographic information
S1. Comment
The compound was prepared via borylation of 5-bromo-10,15,20-triphenylporphyrin with 4,4,5,5-tetramethyl-1,3,2-dioxaborolane under dichlorobis(triphenylphosphine)palladium(II) catalysis (Finnigan et al., 2011). Crystallization from CH2Cl2/CH3OH yielded monoclinic crystals without solvent inclusion.
The compound was investigated with regard to its macrocycle conformation. The porphyrin macrocycle ring system, with a mean deviation of 0.159 (1) Å from the least-squares plane defined by the 24 constituent atoms. Three phenyl rings are essentially planar, with a mean deviation of 0.001 (1) (C100–C105), 0.003 (1) (C150–C155) and 0.007 (1) (C200–C205) Å from the least-squares plane defined by the six constituent atoms. The dihedral angle formed by the porphyrin macrocycle ring system and three phenyl rings are 66.11 (4)(C100–C105), 74.75 (4) (C150–C155) and 57.00 (4) (C200–C205) °, respectively. A conformational analysis was performed using the NSD (normal structural decomposition) method developed by Shelnutt and coworkers (Jentzen et al., 1997). The compound exhibits a moderate degree of conformational distortion. The main contributing out-of-plane distortion modes are ruf, sad and wav. Likewise moderate contributions from macrocycle breathing and N-str are observed for the in-plane distortions. Related Zn(II) complexes, e.g. {5,15-diphenyl-10-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)porphyrinato}zinc(II) (Hyslop et al., 1998; Schwalbe et al., 2012) exhibit more planar conformations. Other recent porphyrin free base structures have been reported by Miranda et al. (2012), Leonarska et al. (2012), and Senge (2013).
In the crystal packing (Fig. 2), the porphyrin molecules are linked by C—H···π interactions (Table 1, Cg1 is the centroid of the C100–C105 phenyl ring) into supramolecular chains running along the a axis. Also intramolecular N—H···N hydrogen bonds occur (Table 1).
S2. Experimental
The title compound was prepared as described by Finnigan et al. (2011).
Figures
Fig. 1.

Molecular structure of the title compound. Thermal ellipsoids are drawn at 50% probability level.
Fig. 2.

A view of the N—H···N and C—H···π interactions (dotted lines) in the crystal structure of the title compound. H atoms non-participating in hydrogen-bonding were omitted for clarity. [Symmetry code: (i) x -1, y, z; (ii) x + 1, y, z.]
Crystal data
| C44H37BN4O2 | F(000) = 1400 |
| Mr = 664.59 | Dx = 1.261 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: -P 2ybc | Cell parameters from 9566 reflections |
| a = 13.0758 (5) Å | θ = 4.1–66.8° |
| b = 10.5245 (4) Å | µ = 0.61 mm−1 |
| c = 27.1358 (10) Å | T = 100 K |
| β = 110.353 (2)° | Triangle, purple |
| V = 3501.2 (2) Å3 | 0.50 × 0.50 × 0.35 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 5993 independent reflections |
| Radiation source: sealed tube | 5788 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.029 |
| Detector resolution: 8.33 pixels mm-1 | θmax = 66.8°, θmin = 4.0° |
| φ and ω scans | h = −15→15 |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | k = −8→12 |
| Tmin = 0.751, Tmax = 0.815 | l = −31→32 |
| 25108 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.103 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0402P)2 + 2.0737P] where P = (Fo2 + 2Fc2)/3 |
| 5993 reflections | (Δ/σ)max < 0.001 |
| 464 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. The carbon atoms of one phenyl unit show some degree of thermal librational movement. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.55291 (11) | 0.54316 (12) | 0.72969 (5) | 0.0213 (3) | |
| C2 | 0.63876 (11) | 0.60493 (13) | 0.77064 (5) | 0.0229 (3) | |
| H2 | 0.6384 | 0.6249 | 0.8047 | 0.027* | |
| C3 | 0.72077 (11) | 0.62999 (13) | 0.75218 (5) | 0.0233 (3) | |
| H3 | 0.7872 | 0.6721 | 0.7709 | 0.028* | |
| C4 | 0.68981 (11) | 0.58188 (12) | 0.69939 (5) | 0.0209 (3) | |
| C5 | 0.75153 (11) | 0.58579 (13) | 0.66621 (5) | 0.0221 (3) | |
| C6 | 0.72497 (11) | 0.51900 (13) | 0.61822 (5) | 0.0225 (3) | |
| C7 | 0.80206 (12) | 0.50156 (14) | 0.59095 (6) | 0.0281 (3) | |
| H7 | 0.8718 | 0.5401 | 0.5997 | 0.034* | |
| C8 | 0.75622 (12) | 0.42099 (14) | 0.55110 (6) | 0.0280 (3) | |
| H8 | 0.7883 | 0.3894 | 0.5270 | 0.034* | |
| C9 | 0.64820 (11) | 0.39125 (13) | 0.55173 (5) | 0.0223 (3) | |
| C10 | 0.57290 (11) | 0.31342 (13) | 0.51484 (5) | 0.0219 (3) | |
| C11 | 0.46626 (11) | 0.28910 (13) | 0.51343 (5) | 0.0221 (3) | |
| C12 | 0.38183 (12) | 0.22656 (15) | 0.47235 (6) | 0.0278 (3) | |
| H12 | 0.3891 | 0.1897 | 0.4418 | 0.033* | |
| C13 | 0.28955 (12) | 0.22871 (14) | 0.48442 (6) | 0.0280 (3) | |
| H13 | 0.2207 | 0.1947 | 0.4637 | 0.034* | |
| C14 | 0.31440 (11) | 0.29137 (13) | 0.53390 (5) | 0.0222 (3) | |
| C15 | 0.24417 (11) | 0.31216 (13) | 0.56185 (5) | 0.0226 (3) | |
| C16 | 0.27360 (11) | 0.36879 (13) | 0.61174 (5) | 0.0222 (3) | |
| C17 | 0.19703 (11) | 0.39354 (15) | 0.63825 (6) | 0.0269 (3) | |
| H17 | 0.1209 | 0.3767 | 0.6252 | 0.032* | |
| C18 | 0.25478 (11) | 0.44510 (15) | 0.68516 (6) | 0.0265 (3) | |
| H18 | 0.2272 | 0.4709 | 0.7117 | 0.032* | |
| C19 | 0.36661 (11) | 0.45374 (13) | 0.68739 (5) | 0.0213 (3) | |
| C20 | 0.45034 (11) | 0.50854 (13) | 0.73035 (5) | 0.0212 (3) | |
| N21 | 0.58829 (9) | 0.52924 (11) | 0.68782 (4) | 0.0211 (2) | |
| H21 | 0.5511 | 0.4919 | 0.6579 | 0.025* | |
| N22 | 0.63209 (9) | 0.45164 (11) | 0.59351 (4) | 0.0214 (2) | |
| N23 | 0.42208 (9) | 0.32698 (11) | 0.54992 (4) | 0.0213 (2) | |
| H23 | 0.4573 | 0.3679 | 0.5792 | 0.026* | |
| N24 | 0.37618 (9) | 0.40825 (11) | 0.64213 (4) | 0.0209 (2) | |
| B1 | 0.85661 (13) | 0.67155 (15) | 0.68378 (6) | 0.0242 (3) | |
| C50 | 0.99982 (12) | 0.78798 (15) | 0.73694 (6) | 0.0314 (3) | |
| C51 | 0.99019 (14) | 0.80462 (17) | 0.67871 (7) | 0.0377 (4) | |
| C52 | 0.94995 (14) | 0.89499 (17) | 0.75792 (7) | 0.0405 (4) | |
| H52A | 0.8746 | 0.9086 | 0.7345 | 0.061* | |
| H52B | 0.9922 | 0.9729 | 0.7598 | 0.061* | |
| H52C | 0.9507 | 0.8731 | 0.7931 | 0.061* | |
| C53 | 1.11315 (13) | 0.75979 (18) | 0.77465 (8) | 0.0426 (4) | |
| H53A | 1.1116 | 0.7523 | 0.8104 | 0.064* | |
| H53B | 1.1624 | 0.8289 | 0.7734 | 0.064* | |
| H53C | 1.1390 | 0.6799 | 0.7646 | 0.064* | |
| C54 | 0.9911 (2) | 0.9413 (2) | 0.66153 (9) | 0.0655 (7) | |
| H54A | 0.9874 | 0.9436 | 0.6248 | 0.098* | |
| H54B | 1.0585 | 0.9826 | 0.6838 | 0.098* | |
| H54C | 0.9281 | 0.9862 | 0.6648 | 0.098* | |
| C55 | 1.07129 (17) | 0.7236 (3) | 0.66391 (9) | 0.0624 (6) | |
| H55A | 1.0661 | 0.6352 | 0.6742 | 0.094* | |
| H55B | 1.1453 | 0.7553 | 0.6821 | 0.094* | |
| H55C | 1.0547 | 0.7280 | 0.6258 | 0.094* | |
| C100 | 0.60355 (11) | 0.25149 (13) | 0.47213 (5) | 0.0227 (3) | |
| C101 | 0.60723 (11) | 0.11908 (14) | 0.46904 (6) | 0.0260 (3) | |
| H101 | 0.5935 | 0.0686 | 0.4951 | 0.031* | |
| C102 | 0.63066 (12) | 0.06045 (14) | 0.42833 (6) | 0.0291 (3) | |
| H102 | 0.6330 | −0.0296 | 0.4267 | 0.035* | |
| C103 | 0.65068 (12) | 0.13354 (15) | 0.39013 (6) | 0.0300 (3) | |
| H103 | 0.6664 | 0.0936 | 0.3622 | 0.036* | |
| C104 | 0.64771 (12) | 0.26504 (15) | 0.39283 (6) | 0.0293 (3) | |
| H104 | 0.6617 | 0.3152 | 0.3668 | 0.035* | |
| C105 | 0.62430 (12) | 0.32356 (14) | 0.43358 (6) | 0.0269 (3) | |
| H105 | 0.6224 | 0.4137 | 0.4352 | 0.032* | |
| C150 | 0.12827 (11) | 0.27037 (14) | 0.53697 (5) | 0.0254 (3) | |
| C151 | 0.09046 (14) | 0.16336 (18) | 0.55491 (8) | 0.0473 (5) | |
| H151 | 0.1392 | 0.1135 | 0.5822 | 0.057* | |
| C152 | −0.01833 (16) | 0.1288 (2) | 0.53314 (9) | 0.0565 (6) | |
| H152 | −0.0434 | 0.0548 | 0.5455 | 0.068* | |
| C153 | −0.09030 (13) | 0.20004 (18) | 0.49394 (7) | 0.0399 (4) | |
| H153 | −0.1648 | 0.1759 | 0.4794 | 0.048* | |
| C154 | −0.05394 (12) | 0.30615 (18) | 0.47587 (6) | 0.0374 (4) | |
| H154 | −0.1034 | 0.3560 | 0.4488 | 0.045* | |
| C155 | 0.05526 (12) | 0.34111 (17) | 0.49711 (6) | 0.0326 (4) | |
| H155 | 0.0800 | 0.4143 | 0.4841 | 0.039* | |
| C200 | 0.42271 (11) | 0.53497 (14) | 0.77821 (5) | 0.0227 (3) | |
| C201 | 0.42174 (12) | 0.65766 (15) | 0.79739 (6) | 0.0284 (3) | |
| H201 | 0.4450 | 0.7270 | 0.7815 | 0.034* | |
| C202 | 0.38703 (12) | 0.67918 (17) | 0.83956 (6) | 0.0350 (4) | |
| H202 | 0.3871 | 0.7631 | 0.8525 | 0.042* | |
| C203 | 0.35234 (12) | 0.57919 (18) | 0.86278 (6) | 0.0371 (4) | |
| H203 | 0.3275 | 0.5945 | 0.8912 | 0.044* | |
| C204 | 0.35386 (13) | 0.45689 (17) | 0.84452 (6) | 0.0350 (4) | |
| H204 | 0.3302 | 0.3880 | 0.8605 | 0.042* | |
| C205 | 0.38975 (12) | 0.43456 (15) | 0.80300 (5) | 0.0278 (3) | |
| H205 | 0.3920 | 0.3500 | 0.7912 | 0.033* | |
| O1 | 0.92954 (8) | 0.67730 (10) | 0.73411 (4) | 0.0297 (2) | |
| O2 | 0.88183 (9) | 0.75138 (11) | 0.65026 (4) | 0.0355 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0237 (7) | 0.0168 (6) | 0.0236 (7) | 0.0005 (5) | 0.0084 (5) | 0.0006 (5) |
| C2 | 0.0250 (7) | 0.0209 (7) | 0.0227 (7) | −0.0002 (6) | 0.0082 (5) | −0.0009 (5) |
| C3 | 0.0218 (7) | 0.0200 (7) | 0.0265 (7) | −0.0019 (5) | 0.0064 (5) | −0.0013 (6) |
| C4 | 0.0203 (6) | 0.0155 (6) | 0.0255 (7) | 0.0011 (5) | 0.0063 (5) | 0.0010 (5) |
| C5 | 0.0220 (7) | 0.0174 (7) | 0.0267 (7) | 0.0012 (5) | 0.0083 (5) | 0.0015 (5) |
| C6 | 0.0249 (7) | 0.0176 (7) | 0.0263 (7) | −0.0008 (5) | 0.0106 (6) | 0.0014 (5) |
| C7 | 0.0260 (7) | 0.0281 (8) | 0.0341 (8) | −0.0076 (6) | 0.0155 (6) | −0.0038 (6) |
| C8 | 0.0297 (8) | 0.0282 (8) | 0.0322 (8) | −0.0045 (6) | 0.0186 (6) | −0.0043 (6) |
| C9 | 0.0262 (7) | 0.0178 (7) | 0.0256 (7) | −0.0004 (5) | 0.0122 (6) | 0.0021 (5) |
| C10 | 0.0263 (7) | 0.0177 (7) | 0.0230 (7) | 0.0018 (5) | 0.0101 (6) | 0.0020 (5) |
| C11 | 0.0252 (7) | 0.0186 (7) | 0.0229 (7) | 0.0027 (5) | 0.0087 (5) | 0.0002 (5) |
| C12 | 0.0268 (7) | 0.0308 (8) | 0.0252 (7) | 0.0019 (6) | 0.0080 (6) | −0.0069 (6) |
| C13 | 0.0232 (7) | 0.0301 (8) | 0.0273 (7) | −0.0004 (6) | 0.0047 (6) | −0.0075 (6) |
| C14 | 0.0208 (7) | 0.0205 (7) | 0.0233 (7) | 0.0013 (5) | 0.0050 (5) | −0.0006 (5) |
| C15 | 0.0213 (7) | 0.0206 (7) | 0.0243 (7) | 0.0006 (5) | 0.0058 (5) | 0.0011 (5) |
| C16 | 0.0207 (6) | 0.0225 (7) | 0.0235 (7) | 0.0000 (5) | 0.0077 (5) | 0.0015 (5) |
| C17 | 0.0198 (7) | 0.0363 (8) | 0.0252 (7) | −0.0029 (6) | 0.0084 (6) | 0.0001 (6) |
| C18 | 0.0238 (7) | 0.0339 (8) | 0.0240 (7) | −0.0008 (6) | 0.0111 (6) | −0.0008 (6) |
| C19 | 0.0231 (7) | 0.0194 (7) | 0.0224 (7) | −0.0007 (5) | 0.0091 (5) | 0.0013 (5) |
| C20 | 0.0235 (7) | 0.0168 (6) | 0.0233 (7) | −0.0002 (5) | 0.0083 (5) | 0.0007 (5) |
| N21 | 0.0205 (6) | 0.0200 (6) | 0.0228 (6) | −0.0023 (5) | 0.0074 (4) | −0.0024 (5) |
| N22 | 0.0237 (6) | 0.0178 (6) | 0.0234 (6) | −0.0011 (5) | 0.0091 (5) | −0.0005 (5) |
| N23 | 0.0216 (6) | 0.0212 (6) | 0.0208 (5) | −0.0002 (5) | 0.0070 (4) | −0.0022 (4) |
| N24 | 0.0216 (6) | 0.0201 (6) | 0.0208 (5) | −0.0004 (5) | 0.0072 (4) | −0.0007 (4) |
| B1 | 0.0240 (8) | 0.0198 (8) | 0.0309 (8) | 0.0013 (6) | 0.0121 (7) | −0.0018 (6) |
| C50 | 0.0267 (7) | 0.0292 (8) | 0.0394 (9) | −0.0090 (6) | 0.0130 (7) | −0.0062 (7) |
| C51 | 0.0372 (9) | 0.0385 (9) | 0.0403 (9) | −0.0189 (7) | 0.0172 (7) | −0.0085 (7) |
| C52 | 0.0371 (9) | 0.0374 (9) | 0.0480 (10) | −0.0056 (7) | 0.0161 (8) | −0.0108 (8) |
| C53 | 0.0287 (8) | 0.0416 (10) | 0.0528 (10) | −0.0074 (7) | 0.0082 (8) | −0.0063 (8) |
| C54 | 0.0834 (16) | 0.0572 (13) | 0.0492 (11) | −0.0405 (12) | 0.0147 (11) | 0.0048 (10) |
| C55 | 0.0476 (11) | 0.0894 (17) | 0.0638 (13) | −0.0248 (11) | 0.0364 (10) | −0.0324 (12) |
| C100 | 0.0206 (6) | 0.0224 (7) | 0.0245 (7) | 0.0006 (5) | 0.0071 (5) | −0.0021 (6) |
| C101 | 0.0238 (7) | 0.0224 (7) | 0.0331 (8) | −0.0011 (6) | 0.0115 (6) | −0.0013 (6) |
| C102 | 0.0248 (7) | 0.0221 (7) | 0.0407 (8) | −0.0009 (6) | 0.0116 (6) | −0.0074 (6) |
| C103 | 0.0252 (7) | 0.0353 (9) | 0.0295 (7) | 0.0034 (6) | 0.0096 (6) | −0.0091 (6) |
| C104 | 0.0313 (8) | 0.0327 (8) | 0.0253 (7) | 0.0048 (6) | 0.0116 (6) | 0.0020 (6) |
| C105 | 0.0320 (8) | 0.0226 (7) | 0.0274 (7) | 0.0047 (6) | 0.0118 (6) | 0.0003 (6) |
| C150 | 0.0218 (7) | 0.0286 (8) | 0.0251 (7) | −0.0006 (6) | 0.0073 (6) | −0.0054 (6) |
| C151 | 0.0320 (9) | 0.0420 (10) | 0.0543 (11) | −0.0088 (8) | −0.0021 (8) | 0.0141 (9) |
| C152 | 0.0385 (10) | 0.0521 (12) | 0.0673 (13) | −0.0192 (9) | 0.0037 (9) | 0.0144 (10) |
| C153 | 0.0233 (8) | 0.0526 (11) | 0.0400 (9) | −0.0075 (7) | 0.0063 (7) | −0.0086 (8) |
| C154 | 0.0235 (8) | 0.0570 (11) | 0.0298 (8) | 0.0043 (7) | 0.0068 (6) | 0.0027 (7) |
| C155 | 0.0244 (7) | 0.0438 (9) | 0.0302 (8) | 0.0010 (7) | 0.0102 (6) | 0.0054 (7) |
| C200 | 0.0191 (6) | 0.0251 (7) | 0.0229 (7) | −0.0027 (5) | 0.0059 (5) | −0.0027 (6) |
| C201 | 0.0253 (7) | 0.0268 (8) | 0.0345 (8) | −0.0044 (6) | 0.0120 (6) | −0.0058 (6) |
| C202 | 0.0266 (8) | 0.0413 (9) | 0.0362 (8) | −0.0023 (7) | 0.0099 (6) | −0.0164 (7) |
| C203 | 0.0267 (8) | 0.0623 (11) | 0.0223 (7) | −0.0055 (7) | 0.0087 (6) | −0.0093 (7) |
| C204 | 0.0322 (8) | 0.0495 (10) | 0.0218 (7) | −0.0082 (7) | 0.0075 (6) | 0.0026 (7) |
| C205 | 0.0291 (7) | 0.0294 (8) | 0.0221 (7) | −0.0046 (6) | 0.0055 (6) | 0.0006 (6) |
| O1 | 0.0250 (5) | 0.0272 (5) | 0.0351 (6) | −0.0047 (4) | 0.0080 (4) | 0.0000 (4) |
| O2 | 0.0385 (6) | 0.0351 (6) | 0.0325 (6) | −0.0149 (5) | 0.0119 (5) | −0.0018 (5) |
Geometric parameters (Å, º)
| C1—N21 | 1.3755 (18) | C51—O2 | 1.4685 (19) |
| C1—C20 | 1.3958 (19) | C51—C54 | 1.514 (3) |
| C1—C2 | 1.4317 (19) | C51—C55 | 1.520 (3) |
| C2—C3 | 1.358 (2) | C52—H52A | 0.9800 |
| C2—H2 | 0.9500 | C52—H52B | 0.9800 |
| C3—C4 | 1.4382 (19) | C52—H52C | 0.9800 |
| C3—H3 | 0.9500 | C53—H53A | 0.9800 |
| C4—N21 | 1.3704 (17) | C53—H53B | 0.9800 |
| C4—C5 | 1.4033 (19) | C53—H53C | 0.9800 |
| C5—C6 | 1.413 (2) | C54—H54A | 0.9800 |
| C5—B1 | 1.573 (2) | C54—H54B | 0.9800 |
| C6—N22 | 1.3641 (18) | C54—H54C | 0.9800 |
| C6—C7 | 1.4549 (19) | C55—H55A | 0.9800 |
| C7—C8 | 1.340 (2) | C55—H55B | 0.9800 |
| C7—H7 | 0.9500 | C55—H55C | 0.9800 |
| C8—C9 | 1.4527 (19) | C100—C105 | 1.393 (2) |
| C8—H8 | 0.9500 | C100—C101 | 1.398 (2) |
| C9—N22 | 1.3781 (18) | C101—C102 | 1.389 (2) |
| C9—C10 | 1.399 (2) | C101—H101 | 0.9500 |
| C10—C11 | 1.4050 (19) | C102—C103 | 1.387 (2) |
| C10—C100 | 1.5007 (19) | C102—H102 | 0.9500 |
| C11—N23 | 1.3678 (17) | C103—C104 | 1.387 (2) |
| C11—C12 | 1.428 (2) | C103—H103 | 0.9500 |
| C12—C13 | 1.356 (2) | C104—C105 | 1.390 (2) |
| C12—H12 | 0.9500 | C104—H104 | 0.9500 |
| C13—C14 | 1.428 (2) | C105—H105 | 0.9500 |
| C13—H13 | 0.9500 | C150—C151 | 1.385 (2) |
| C14—N23 | 1.3734 (18) | C150—C155 | 1.385 (2) |
| C14—C15 | 1.3967 (19) | C151—C152 | 1.386 (2) |
| C15—C16 | 1.405 (2) | C151—H151 | 0.9500 |
| C15—C150 | 1.4947 (19) | C152—C153 | 1.372 (3) |
| C16—N24 | 1.3719 (18) | C152—H152 | 0.9500 |
| C16—C17 | 1.4454 (19) | C153—C154 | 1.370 (3) |
| C17—C18 | 1.348 (2) | C153—H153 | 0.9500 |
| C17—H17 | 0.9500 | C154—C155 | 1.391 (2) |
| C18—C19 | 1.4453 (19) | C154—H154 | 0.9500 |
| C18—H18 | 0.9500 | C155—H155 | 0.9500 |
| C19—N24 | 1.3639 (18) | C200—C201 | 1.394 (2) |
| C19—C20 | 1.4151 (19) | C200—C205 | 1.399 (2) |
| C20—C200 | 1.4903 (19) | C201—C202 | 1.389 (2) |
| N21—H21 | 0.8800 | C201—H201 | 0.9500 |
| N23—H23 | 0.8800 | C202—C203 | 1.382 (3) |
| B1—O2 | 1.361 (2) | C202—H202 | 0.9500 |
| B1—O1 | 1.368 (2) | C203—C204 | 1.382 (3) |
| C50—O1 | 1.4690 (18) | C203—H203 | 0.9500 |
| C50—C52 | 1.509 (2) | C204—C205 | 1.383 (2) |
| C50—C53 | 1.509 (2) | C204—H204 | 0.9500 |
| C50—C51 | 1.551 (2) | C205—H205 | 0.9500 |
| N21—C1—C20 | 125.69 (12) | C55—C51—C50 | 112.90 (17) |
| N21—C1—C2 | 106.93 (11) | C50—C52—H52A | 109.5 |
| C20—C1—C2 | 127.31 (13) | C50—C52—H52B | 109.5 |
| C3—C2—C1 | 107.97 (12) | H52A—C52—H52B | 109.5 |
| C3—C2—H2 | 126.0 | C50—C52—H52C | 109.5 |
| C1—C2—H2 | 126.0 | H52A—C52—H52C | 109.5 |
| C2—C3—C4 | 108.31 (12) | H52B—C52—H52C | 109.5 |
| C2—C3—H3 | 125.8 | C50—C53—H53A | 109.5 |
| C4—C3—H3 | 125.8 | C50—C53—H53B | 109.5 |
| N21—C4—C5 | 126.32 (12) | H53A—C53—H53B | 109.5 |
| N21—C4—C3 | 106.57 (11) | C50—C53—H53C | 109.5 |
| C5—C4—C3 | 127.10 (12) | H53A—C53—H53C | 109.5 |
| C4—C5—C6 | 124.40 (13) | H53B—C53—H53C | 109.5 |
| C4—C5—B1 | 117.32 (12) | C51—C54—H54A | 109.5 |
| C6—C5—B1 | 118.28 (12) | C51—C54—H54B | 109.5 |
| N22—C6—C5 | 127.11 (12) | H54A—C54—H54B | 109.5 |
| N22—C6—C7 | 109.88 (12) | C51—C54—H54C | 109.5 |
| C5—C6—C7 | 122.68 (13) | H54A—C54—H54C | 109.5 |
| C8—C7—C6 | 107.20 (12) | H54B—C54—H54C | 109.5 |
| C8—C7—H7 | 126.4 | C51—C55—H55A | 109.5 |
| C6—C7—H7 | 126.4 | C51—C55—H55B | 109.5 |
| C7—C8—C9 | 106.89 (12) | H55A—C55—H55B | 109.5 |
| C7—C8—H8 | 126.6 | C51—C55—H55C | 109.5 |
| C9—C8—H8 | 126.6 | H55A—C55—H55C | 109.5 |
| N22—C9—C10 | 126.00 (12) | H55B—C55—H55C | 109.5 |
| N22—C9—C8 | 109.74 (12) | C105—C100—C101 | 118.56 (13) |
| C10—C9—C8 | 124.26 (12) | C105—C100—C10 | 121.17 (13) |
| C9—C10—C11 | 124.82 (12) | C101—C100—C10 | 120.22 (13) |
| C9—C10—C100 | 119.77 (12) | C102—C101—C100 | 120.81 (14) |
| C11—C10—C100 | 115.40 (12) | C102—C101—H101 | 119.6 |
| N23—C11—C10 | 126.31 (13) | C100—C101—H101 | 119.6 |
| N23—C11—C12 | 107.04 (12) | C103—C102—C101 | 119.94 (14) |
| C10—C11—C12 | 126.57 (13) | C103—C102—H102 | 120.0 |
| C13—C12—C11 | 108.37 (13) | C101—C102—H102 | 120.0 |
| C13—C12—H12 | 125.8 | C102—C103—C104 | 119.86 (14) |
| C11—C12—H12 | 125.8 | C102—C103—H103 | 120.1 |
| C12—C13—C14 | 107.71 (13) | C104—C103—H103 | 120.1 |
| C12—C13—H13 | 126.1 | C103—C104—C105 | 120.14 (14) |
| C14—C13—H13 | 126.1 | C103—C104—H104 | 119.9 |
| N23—C14—C15 | 125.50 (12) | C105—C104—H104 | 119.9 |
| N23—C14—C13 | 107.25 (12) | C104—C105—C100 | 120.69 (14) |
| C15—C14—C13 | 127.24 (13) | C104—C105—H105 | 119.7 |
| C14—C15—C16 | 125.32 (13) | C100—C105—H105 | 119.7 |
| C14—C15—C150 | 117.61 (12) | C151—C150—C155 | 118.59 (14) |
| C16—C15—C150 | 117.06 (12) | C151—C150—C15 | 120.70 (13) |
| N24—C16—C15 | 126.20 (12) | C155—C150—C15 | 120.65 (13) |
| N24—C16—C17 | 110.36 (12) | C150—C151—C152 | 120.19 (17) |
| C15—C16—C17 | 123.44 (12) | C150—C151—H151 | 119.9 |
| C18—C17—C16 | 106.57 (12) | C152—C151—H151 | 119.9 |
| C18—C17—H17 | 126.7 | C153—C152—C151 | 120.83 (18) |
| C16—C17—H17 | 126.7 | C153—C152—H152 | 119.6 |
| C17—C18—C19 | 106.92 (12) | C151—C152—H152 | 119.6 |
| C17—C18—H18 | 126.5 | C154—C153—C152 | 119.55 (15) |
| C19—C18—H18 | 126.5 | C154—C153—H153 | 120.2 |
| N24—C19—C20 | 126.72 (12) | C152—C153—H153 | 120.2 |
| N24—C19—C18 | 110.41 (12) | C153—C154—C155 | 120.11 (15) |
| C20—C19—C18 | 122.80 (12) | C153—C154—H154 | 119.9 |
| C1—C20—C19 | 124.57 (12) | C155—C154—H154 | 119.9 |
| C1—C20—C200 | 119.18 (12) | C150—C155—C154 | 120.73 (15) |
| C19—C20—C200 | 116.19 (12) | C150—C155—H155 | 119.6 |
| C4—N21—C1 | 110.18 (11) | C154—C155—H155 | 119.6 |
| C4—N21—H21 | 124.9 | C201—C200—C205 | 118.42 (13) |
| C1—N21—H21 | 124.9 | C201—C200—C20 | 122.32 (13) |
| C6—N22—C9 | 106.23 (11) | C205—C200—C20 | 119.13 (13) |
| C11—N23—C14 | 109.64 (11) | C202—C201—C200 | 120.45 (14) |
| C11—N23—H23 | 125.2 | C202—C201—H201 | 119.8 |
| C14—N23—H23 | 125.2 | C200—C201—H201 | 119.8 |
| C19—N24—C16 | 105.71 (11) | C203—C202—C201 | 120.33 (15) |
| O2—B1—O1 | 113.15 (13) | C203—C202—H202 | 119.8 |
| O2—B1—C5 | 122.75 (13) | C201—C202—H202 | 119.8 |
| O1—B1—C5 | 124.06 (13) | C202—C203—C204 | 119.86 (14) |
| O1—C50—C52 | 105.50 (12) | C202—C203—H203 | 120.1 |
| O1—C50—C53 | 109.19 (13) | C204—C203—H203 | 120.1 |
| C52—C50—C53 | 110.02 (14) | C203—C204—C205 | 120.11 (15) |
| O1—C50—C51 | 102.03 (11) | C203—C204—H204 | 119.9 |
| C52—C50—C51 | 114.05 (15) | C205—C204—H204 | 119.9 |
| C53—C50—C51 | 115.20 (14) | C204—C205—C200 | 120.80 (15) |
| O2—C51—C54 | 108.07 (16) | C204—C205—H205 | 119.6 |
| O2—C51—C55 | 106.34 (14) | C200—C205—H205 | 119.6 |
| C54—C51—C55 | 111.76 (18) | B1—O1—C50 | 107.07 (12) |
| O2—C51—C50 | 102.37 (11) | B1—O2—C51 | 107.33 (12) |
| C54—C51—C50 | 114.50 (15) | ||
| N21—C1—C2—C3 | 1.93 (15) | C20—C19—N24—C16 | −178.05 (13) |
| C20—C1—C2—C3 | −175.19 (13) | C18—C19—N24—C16 | −1.13 (15) |
| C1—C2—C3—C4 | −1.34 (16) | C15—C16—N24—C19 | −178.43 (13) |
| C2—C3—C4—N21 | 0.26 (15) | C17—C16—N24—C19 | 1.58 (15) |
| C2—C3—C4—C5 | −178.83 (13) | C4—C5—B1—O2 | −134.75 (15) |
| N21—C4—C5—C6 | −9.8 (2) | C6—C5—B1—O2 | 44.7 (2) |
| C3—C4—C5—C6 | 169.07 (13) | C4—C5—B1—O1 | 42.9 (2) |
| N21—C4—C5—B1 | 169.59 (13) | C6—C5—B1—O1 | −137.66 (14) |
| C3—C4—C5—B1 | −11.5 (2) | O1—C50—C51—O2 | 27.64 (15) |
| C4—C5—C6—N22 | 7.2 (2) | C52—C50—C51—O2 | −85.58 (15) |
| B1—C5—C6—N22 | −172.24 (13) | C53—C50—C51—O2 | 145.78 (14) |
| C4—C5—C6—C7 | −165.55 (13) | O1—C50—C51—C54 | 144.32 (16) |
| B1—C5—C6—C7 | 15.0 (2) | C52—C50—C51—C54 | 31.1 (2) |
| N22—C6—C7—C8 | −1.82 (17) | C53—C50—C51—C54 | −97.5 (2) |
| C5—C6—C7—C8 | 172.03 (13) | O1—C50—C51—C55 | −86.27 (16) |
| C6—C7—C8—C9 | 2.27 (17) | C52—C50—C51—C55 | 160.50 (15) |
| C7—C8—C9—N22 | −2.08 (17) | C53—C50—C51—C55 | 31.9 (2) |
| C7—C8—C9—C10 | 177.52 (14) | C9—C10—C100—C105 | −65.26 (18) |
| N22—C9—C10—C11 | 2.9 (2) | C11—C10—C100—C105 | 113.43 (15) |
| C8—C9—C10—C11 | −176.60 (13) | C9—C10—C100—C101 | 117.51 (15) |
| N22—C9—C10—C100 | −178.50 (12) | C11—C10—C100—C101 | −63.80 (17) |
| C8—C9—C10—C100 | 2.0 (2) | C105—C100—C101—C102 | −0.2 (2) |
| C9—C10—C11—N23 | −6.4 (2) | C10—C100—C101—C102 | 177.08 (13) |
| C100—C10—C11—N23 | 175.03 (12) | C100—C101—C102—C103 | 0.0 (2) |
| C9—C10—C11—C12 | 169.88 (14) | C101—C102—C103—C104 | 0.3 (2) |
| C100—C10—C11—C12 | −8.7 (2) | C102—C103—C104—C105 | −0.3 (2) |
| N23—C11—C12—C13 | 0.46 (17) | C103—C104—C105—C100 | 0.0 (2) |
| C10—C11—C12—C13 | −176.36 (14) | C101—C100—C105—C104 | 0.3 (2) |
| C11—C12—C13—C14 | −0.79 (17) | C10—C100—C105—C104 | −177.03 (13) |
| C12—C13—C14—N23 | 0.82 (16) | C14—C15—C150—C151 | 105.95 (18) |
| C12—C13—C14—C15 | −178.41 (14) | C16—C15—C150—C151 | −73.76 (19) |
| N23—C14—C15—C16 | −2.4 (2) | C14—C15—C150—C155 | −77.00 (18) |
| C13—C14—C15—C16 | 176.65 (14) | C16—C15—C150—C155 | 103.29 (16) |
| N23—C14—C15—C150 | 177.87 (13) | C155—C150—C151—C152 | 0.0 (3) |
| C13—C14—C15—C150 | −3.0 (2) | C15—C150—C151—C152 | 177.07 (18) |
| C14—C15—C16—N24 | −2.8 (2) | C150—C151—C152—C153 | −0.5 (3) |
| C150—C15—C16—N24 | 176.88 (13) | C151—C152—C153—C154 | 0.5 (3) |
| C14—C15—C16—C17 | 177.18 (14) | C152—C153—C154—C155 | 0.1 (3) |
| C150—C15—C16—C17 | −3.1 (2) | C151—C150—C155—C154 | 0.6 (2) |
| N24—C16—C17—C18 | −1.47 (17) | C15—C150—C155—C154 | −176.50 (14) |
| C15—C16—C17—C18 | 178.55 (14) | C153—C154—C155—C150 | −0.7 (3) |
| C16—C17—C18—C19 | 0.71 (17) | C1—C20—C200—C201 | 60.41 (19) |
| C17—C18—C19—N24 | 0.24 (17) | C19—C20—C200—C201 | −117.00 (15) |
| C17—C18—C19—C20 | 177.30 (13) | C1—C20—C200—C205 | −123.79 (14) |
| N21—C1—C20—C19 | −0.2 (2) | C19—C20—C200—C205 | 58.80 (17) |
| C2—C1—C20—C19 | 176.38 (13) | C205—C200—C201—C202 | −1.1 (2) |
| N21—C1—C20—C200 | −177.40 (12) | C20—C200—C201—C202 | 174.77 (13) |
| C2—C1—C20—C200 | −0.8 (2) | C200—C201—C202—C203 | −0.4 (2) |
| N24—C19—C20—C1 | 10.7 (2) | C201—C202—C203—C204 | 1.1 (2) |
| C18—C19—C20—C1 | −165.86 (13) | C202—C203—C204—C205 | −0.2 (2) |
| N24—C19—C20—C200 | −172.05 (13) | C203—C204—C205—C200 | −1.4 (2) |
| C18—C19—C20—C200 | 11.4 (2) | C201—C200—C205—C204 | 2.0 (2) |
| C5—C4—N21—C1 | −179.92 (13) | C20—C200—C205—C204 | −174.01 (13) |
| C3—C4—N21—C1 | 0.98 (15) | O2—B1—O1—C50 | 11.02 (16) |
| C20—C1—N21—C4 | 175.39 (13) | C5—B1—O1—C50 | −166.80 (13) |
| C2—C1—N21—C4 | −1.79 (15) | C52—C50—O1—B1 | 95.52 (14) |
| C5—C6—N22—C9 | −173.02 (13) | C53—C50—O1—B1 | −146.28 (13) |
| C7—C6—N22—C9 | 0.49 (15) | C51—C50—O1—B1 | −23.93 (15) |
| C10—C9—N22—C6 | −178.67 (13) | O1—B1—O2—C51 | 8.13 (17) |
| C8—C9—N22—C6 | 0.92 (15) | C5—B1—O2—C51 | −174.01 (13) |
| C10—C11—N23—C14 | 176.90 (13) | C54—C51—O2—B1 | −143.58 (15) |
| C12—C11—N23—C14 | 0.06 (15) | C55—C51—O2—B1 | 96.28 (17) |
| C15—C14—N23—C11 | 178.71 (13) | C50—C51—O2—B1 | −22.37 (16) |
| C13—C14—N23—C11 | −0.54 (15) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C100–C105 phenyl ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N21—H21···N24 | 0.88 | 2.35 | 2.9084 (16) | 122 |
| N21—H21···N22 | 0.88 | 2.38 | 2.9266 (16) | 121 |
| N23—H23···N24 | 0.88 | 2.34 | 2.8978 (16) | 121 |
| N23—H23···N22 | 0.88 | 2.35 | 2.8986 (16) | 120 |
| C153—H153···Cg1i | 0.95 | 2.58 | 3.488 (2) | 160 |
Symmetry code: (i) x−1, y, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: LX2293).
References
- Brandenburg, K. (1998). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2005). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Finnigan, E. M., Rein, R., Solladie, N., Dahms, K., Götz, D. H. G., Bringmann, G. & Senge, M. O. (2011). Tetrahedron, 67, 1126–1134.
- Hope, H. (1994). Prog. Inorg. Chem. 41, 1–19.
- Hyslop, A. G., Kellett, M. A., Iovine, P. M. & Therien, M. J. (1998). J. Am. Chem. Soc. 120, 12676–12677.
- Jentzen, W., Song, X.-Z. & Shelnutt, J. A. (1997). J. Phys. Chem. B, 101, 1684–1699.
- Leonarska, A., Zubko, M., Kuś, P., Kusz, J. & Ratuszna, A. (2012). Acta Cryst. E68, o2797–o2798. [DOI] [PMC free article] [PubMed]
- Miranda, M. D., Ramos Silva, M., Maria, T. M. R., Balakrishna, A. & Sobral, A. J. F. N. (2012). Acta Cryst. E68, o3462–o3463. [DOI] [PMC free article] [PubMed]
- Scheidt, W. R. & Lee, Y. J. (1987). Struct. Bonding (Berlin), 64, 1–70.
- Schwalbe, M., Metzinger, R., Teets, T. S. & Novera, D. G. (2012). Chem. Eur. J. 18, 15449–15458. [DOI] [PubMed]
- Senge, M. O. (2000). The Porphyrin Handbook, edited by K. M. Kadish, K. M. Smith, R. Guilard, Vol. 10, pp. 1–218. San Diego: Academic Press.
- Senge, M. O. (2006). Chem. Commun. pp. 243–256. [DOI] [PubMed]
- Senge, M. O. (2013). Acta Cryst. E69, o1048. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536814019680/lx2293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814019680/lx2293Isup2.hkl
. DOI: 10.1107/S1600536814019680/lx2293fig1.tif
Molecular structure of the title compound. Thermal ellipsoids are drawn at 50% probability level.
x y z x y z . DOI: 10.1107/S1600536814019680/lx2293fig2.tif
A view of the N—H⋯N and C—H⋯π interactions (dotted lines) in the crystal structure of the title compound. H atoms non-participating in hydrogen-bonding were omitted for clarity. [Symmetry code: (i) x − 1, y, z; (ii) x + 1, y, z.]
CCDC reference: 1022040
Additional supporting information: crystallographic information; 3D view; checkCIF report


