Abstract
In the title molecule, C17H12N4OS, the thiazole ring forms a dihedral angle of 10.8 (2)° with the phenyl ring and an angle of 3.1 (3)° with the indole ring system [which has a maximum deviation of 0.035 (2) Å]. The dihedral angle between the planes of the phenyl ring and the indole ring system is 11.5 (1)°. An intramolecular N—H⋯O hydrogen bond is observed. In the crystal, pairs of N—H⋯O hydrogen bonds form inversion dimers with an R 2 2(8) graph-set motif.
Keywords: crystal structure; indolinone; hydrazine; 1,3-thiazole; hydrogen bonding; biological activity
Related literature
For the biological activities of substituted thiazoles, see: Ali et al. (2011 ▶); Bharti et al. (2010 ▶); Kondratieva et al. (2007 ▶). For a related structure, see: Sadık et al. (2004 ▶).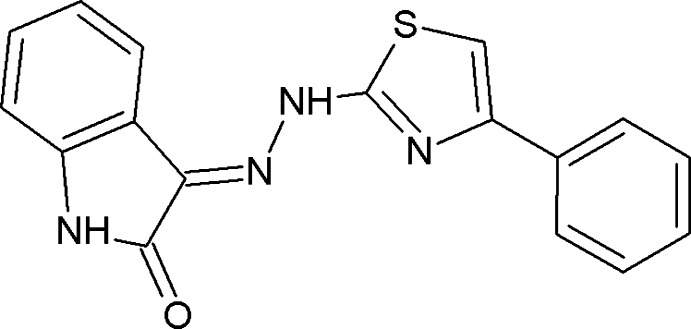
Experimental
Crystal data
C17H12N4OS
M r = 320.37
Monoclinic,

a = 17.7108 (8) Å
b = 5.1411 (2) Å
c = 15.9065 (6) Å
β = 94.706 (3)°
V = 1443.45 (10) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 296 K
0.35 × 0.31 × 0.25 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.887, T max = 0.934
11530 measured reflections
3142 independent reflections
2124 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.109
S = 1.09
3142 reflections
208 parameters
H-atom parameters constrained
Δρmax = 0.20 e Å−3
Δρmin = −0.25 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 1998 ▶); data reduction: SAINT; program(s) used to solve structure: SIR92 (Altomare et al., 1994 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and CAMERON (Watkin et al., 1993 ▶); software used to prepare material for publication: PARST (Nardelli, 1995 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814022715/lh5732sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814022715/lh5732Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814022715/lh5732Isup3.cml
. DOI: 10.1107/S1600536814022715/lh5732fig1.tif
The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level. The dashed line indicates an intramolecular N—H⋯N bond
. DOI: 10.1107/S1600536814022715/lh5732fig2.tif
Part of the crystal structure with hydrogen bonds indicated as dotted lines
CCDC reference: 1029498
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| N2H2O1 | 0.86 | 2.12 | 2.771(2) | 133 |
| N4H4O1i | 0.86 | 2.11 | 2.922(2) | 158 |
Symmetry code: (i)  .
.
Acknowledgments
USIC, Karnatak University, Dharwad, is greatly acknowledged for the single-crystal XRD data collection. BMH is grateful to the UGC for financial support in the form of an RFSMS scholarship.
supplementary crystallographic information
S1. Experimental
S1.1. Synthesis and crystallization
An ethanolic solution of 1.81g (0.01 M) of 2-hydrazino-4-phenylthiazole was added drop wise to an etanolic solution of 1.47g (0.01 M) of isatin with constant stirring. After the complete addition, the reaction mixture was stirred further for 8-9 hrs until the solid separated out from the reaction mixture. The separated solid was filtered and washed with cold alcohol, dried and recrystallized from DMF (Yield: 95 %. MP: 443-446K). Block-shaped colourless crystals were obtained by slow evaporation of a solution of the title compound at room temperature in DMF:water in the ratio 2:1.
S1.2. Refinement
H atoms were placed in idealized positions and refined using a riding-model approximation with N—H = 0.86 Å, C—H = 0.93 Å and with Uiso(H) = 1.2Ueq(N,C).
S2. Comment
Isatin derivatives and compounds containing a thiazole ring are class of organic compounds which have fascinated many synthetic researchers due to their wide range of biological activity ( Ali et al., 2011; Bharti et al., 2010; Kondratieva et al., 2007).
The molecular structure of the title compound is shown in Fig. 1. An intramolecular N—H···O hydrogen bond is observed. The thiazole ring is essentially planar with a maximum deviation of 0.005 (2) Å for atom N1. The thiazole ring (S1/C9/N1/C7/C8) forms dihedral angles of 10.8 (2)° with the phenyl ring (C1–C6) and 3.1 (3)° with the indole ring system (C10—C16/N4/C17, with a maximum deviation of 0.035 (2)Å for atom C17). The dihedral angle between the phenyl ring and the indole ring system is 11.5 (1)Å. In the crystal, pairs of N—H···O hydrogen bonds form inversion dimers (Fig. 2). A closely related structure appears in the literature (Sadik, et al., 2004).
Figures
Fig. 1.

The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level. The dashed line indicates an intramolecular N—H···N bond
Fig. 2.
Part of the crystal structure with hydrogen bonds indicated as dotted lines
Crystal data
| C17H12N4OS | Z = 4 |
| Mr = 320.37 | F(000) = 664 |
| Monoclinic, P21/c | Dx = 1.474 Mg m−3 |
| Hall symbol: -P 2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 17.7108 (8) Å | µ = 0.23 mm−1 |
| b = 5.1411 (2) Å | T = 296 K |
| c = 15.9065 (6) Å | Block, colourless |
| β = 94.706 (3)° | 0.35 × 0.31 × 0.25 mm |
| V = 1443.45 (10) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2124 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.039 |
| φ and ω scans | θmax = 27.1°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −22→22 |
| Tmin = 0.887, Tmax = 0.934 | k = −6→6 |
| 11530 measured reflections | l = −20→20 |
| 3142 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.109 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0451P)2 + 0.0098P] where P = (Fo2 + 2Fc2)/3 |
| 3142 reflections | (Δ/σ)max < 0.001 |
| 208 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.33816 (3) | 0.66660 (12) | 0.39879 (3) | 0.04097 (19) | |
| O1 | 0.08908 (8) | 0.2212 (3) | 0.51706 (8) | 0.0481 (4) | |
| N1 | 0.28727 (8) | 0.7985 (3) | 0.54037 (9) | 0.0325 (4) | |
| N2 | 0.21944 (9) | 0.4599 (3) | 0.46996 (9) | 0.0369 (4) | |
| H2 | 0.1873 | 0.4489 | 0.5075 | 0.044* | |
| N3 | 0.21516 (9) | 0.3013 (3) | 0.40283 (9) | 0.0341 (4) | |
| N4 | 0.05982 (9) | −0.1170 (3) | 0.42364 (9) | 0.0387 (5) | |
| H4 | 0.0225 | −0.1842 | 0.4474 | 0.046* | |
| C1 | 0.43038 (11) | 1.3281 (4) | 0.58140 (12) | 0.0359 (5) | |
| H1 | 0.4504 | 1.335 | 0.5292 | 0.043* | |
| C2 | 0.45583 (11) | 1.5006 (4) | 0.64372 (12) | 0.0419 (5) | |
| H2A | 0.4931 | 1.6214 | 0.6335 | 0.05* | |
| C3 | 0.42631 (12) | 1.4945 (4) | 0.72089 (12) | 0.0410 (5) | |
| H3 | 0.4433 | 1.6113 | 0.7629 | 0.049* | |
| C4 | 0.37150 (12) | 1.3150 (4) | 0.73568 (12) | 0.0415 (5) | |
| H4A | 0.3512 | 1.3113 | 0.7877 | 0.05* | |
| C5 | 0.34649 (11) | 1.1406 (4) | 0.67384 (12) | 0.0376 (5) | |
| H5 | 0.3098 | 1.0188 | 0.6849 | 0.045* | |
| C6 | 0.37525 (10) | 1.1438 (4) | 0.59528 (11) | 0.0304 (5) | |
| C7 | 0.34960 (10) | 0.9552 (4) | 0.52935 (11) | 0.0309 (5) | |
| C8 | 0.38282 (11) | 0.9102 (4) | 0.45680 (11) | 0.0367 (5) | |
| H8 | 0.4247 | 1.0007 | 0.4406 | 0.044* | |
| C9 | 0.27677 (11) | 0.6406 (4) | 0.47676 (11) | 0.0314 (5) | |
| C10 | 0.16106 (11) | 0.1315 (4) | 0.39588 (11) | 0.0319 (5) | |
| C11 | 0.14763 (10) | −0.0560 (4) | 0.32827 (11) | 0.0317 (5) | |
| C12 | 0.18154 (11) | −0.1039 (4) | 0.25430 (12) | 0.0409 (5) | |
| H12 | 0.222 | −0.003 | 0.2397 | 0.049* | |
| C13 | 0.15406 (12) | −0.3045 (4) | 0.20274 (12) | 0.0451 (6) | |
| H13 | 0.1762 | −0.3386 | 0.1528 | 0.054* | |
| C14 | 0.09393 (12) | −0.4556 (4) | 0.22454 (12) | 0.0422 (6) | |
| H14 | 0.0768 | −0.5912 | 0.1893 | 0.051* | |
| C15 | 0.05887 (11) | −0.4091 (4) | 0.29754 (12) | 0.0392 (5) | |
| H15 | 0.0183 | −0.51 | 0.312 | 0.047* | |
| C16 | 0.08638 (10) | −0.2078 (4) | 0.34775 (11) | 0.0319 (5) | |
| C17 | 0.10069 (11) | 0.0896 (4) | 0.45404 (12) | 0.0362 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0424 (3) | 0.0466 (4) | 0.0351 (3) | −0.0080 (3) | 0.0099 (2) | −0.0052 (2) |
| O1 | 0.0479 (9) | 0.0568 (11) | 0.0417 (8) | −0.0142 (8) | 0.0156 (7) | −0.0154 (8) |
| N1 | 0.0296 (9) | 0.0333 (11) | 0.0350 (8) | −0.0022 (8) | 0.0050 (7) | −0.0018 (8) |
| N2 | 0.0329 (10) | 0.0424 (12) | 0.0363 (9) | −0.0091 (9) | 0.0080 (7) | −0.0058 (8) |
| N3 | 0.0328 (9) | 0.0359 (11) | 0.0335 (8) | −0.0026 (9) | 0.0026 (7) | −0.0024 (8) |
| N4 | 0.0354 (10) | 0.0422 (12) | 0.0400 (9) | −0.0129 (9) | 0.0112 (7) | −0.0035 (8) |
| C1 | 0.0343 (11) | 0.0375 (14) | 0.0362 (10) | −0.0027 (10) | 0.0056 (8) | 0.0039 (10) |
| C2 | 0.0377 (12) | 0.0398 (15) | 0.0478 (12) | −0.0074 (11) | 0.0010 (9) | −0.0001 (11) |
| C3 | 0.0436 (13) | 0.0342 (14) | 0.0442 (12) | −0.0026 (11) | −0.0030 (9) | −0.0071 (10) |
| C4 | 0.0454 (13) | 0.0432 (15) | 0.0366 (11) | −0.0002 (12) | 0.0078 (9) | −0.0046 (10) |
| C5 | 0.0359 (12) | 0.0366 (14) | 0.0411 (11) | −0.0069 (11) | 0.0085 (9) | −0.0020 (10) |
| C6 | 0.0291 (11) | 0.0275 (12) | 0.0343 (10) | 0.0043 (10) | 0.0010 (8) | 0.0020 (9) |
| C7 | 0.0289 (10) | 0.0285 (12) | 0.0354 (10) | −0.0006 (10) | 0.0026 (8) | 0.0023 (9) |
| C8 | 0.0345 (11) | 0.0377 (14) | 0.0385 (11) | −0.0086 (10) | 0.0071 (9) | −0.0001 (10) |
| C9 | 0.0290 (11) | 0.0309 (13) | 0.0344 (10) | −0.0004 (10) | 0.0026 (8) | 0.0012 (9) |
| C10 | 0.0282 (11) | 0.0341 (13) | 0.0334 (10) | −0.0010 (10) | 0.0025 (8) | 0.0010 (9) |
| C11 | 0.0275 (10) | 0.0331 (13) | 0.0342 (10) | 0.0012 (10) | 0.0016 (8) | −0.0001 (9) |
| C12 | 0.0326 (12) | 0.0505 (15) | 0.0404 (11) | −0.0049 (11) | 0.0080 (9) | −0.0022 (11) |
| C13 | 0.0370 (12) | 0.0588 (17) | 0.0398 (11) | 0.0042 (12) | 0.0047 (9) | −0.0104 (11) |
| C14 | 0.0381 (12) | 0.0428 (15) | 0.0445 (12) | 0.0027 (11) | −0.0042 (9) | −0.0094 (11) |
| C15 | 0.0343 (12) | 0.0387 (14) | 0.0442 (11) | −0.0016 (11) | 0.0013 (9) | −0.0029 (10) |
| C16 | 0.0279 (11) | 0.0358 (13) | 0.0321 (10) | 0.0026 (10) | 0.0026 (8) | −0.0003 (9) |
| C17 | 0.0322 (11) | 0.0398 (14) | 0.0368 (11) | −0.0016 (11) | 0.0040 (9) | −0.0008 (10) |
Geometric parameters (Å, º)
| S1—C8 | 1.711 (2) | C4—C5 | 1.377 (3) |
| S1—C9 | 1.7203 (19) | C4—H4A | 0.93 |
| O1—C17 | 1.240 (2) | C5—C6 | 1.388 (2) |
| N1—C9 | 1.299 (2) | C5—H5 | 0.93 |
| N1—C7 | 1.389 (2) | C6—C7 | 1.472 (3) |
| N2—N3 | 1.341 (2) | C7—C8 | 1.358 (2) |
| N2—C9 | 1.374 (2) | C8—H8 | 0.93 |
| N2—H2 | 0.86 | C10—C11 | 1.449 (3) |
| N3—C10 | 1.294 (2) | C10—C17 | 1.486 (3) |
| N4—C17 | 1.352 (2) | C11—C12 | 1.386 (2) |
| N4—C16 | 1.411 (2) | C11—C16 | 1.392 (3) |
| N4—H4 | 0.86 | C12—C13 | 1.381 (3) |
| C1—C2 | 1.378 (3) | C12—H12 | 0.93 |
| C1—C6 | 1.391 (3) | C13—C14 | 1.385 (3) |
| C1—H1 | 0.93 | C13—H13 | 0.93 |
| C2—C3 | 1.373 (3) | C14—C15 | 1.382 (3) |
| C2—H2A | 0.93 | C14—H14 | 0.93 |
| C3—C4 | 1.373 (3) | C15—C16 | 1.372 (3) |
| C3—H3 | 0.93 | C15—H15 | 0.93 |
| C8—S1—C9 | 87.69 (9) | C7—C8—S1 | 111.71 (15) |
| C9—N1—C7 | 109.19 (15) | C7—C8—H8 | 124.1 |
| N3—N2—C9 | 117.83 (15) | S1—C8—H8 | 124.1 |
| N3—N2—H2 | 121.1 | N1—C9—N2 | 122.80 (17) |
| C9—N2—H2 | 121.1 | N1—C9—S1 | 117.03 (15) |
| C10—N3—N2 | 118.11 (16) | N2—C9—S1 | 120.17 (14) |
| C17—N4—C16 | 111.08 (16) | N3—C10—C11 | 125.96 (17) |
| C17—N4—H4 | 124.5 | N3—C10—C17 | 127.60 (18) |
| C16—N4—H4 | 124.5 | C11—C10—C17 | 106.44 (17) |
| C2—C1—C6 | 121.10 (18) | C12—C11—C16 | 119.27 (18) |
| C2—C1—H1 | 119.5 | C12—C11—C10 | 133.79 (19) |
| C6—C1—H1 | 119.5 | C16—C11—C10 | 106.93 (16) |
| C3—C2—C1 | 120.2 (2) | C13—C12—C11 | 118.73 (19) |
| C3—C2—H2A | 119.9 | C13—C12—H12 | 120.6 |
| C1—C2—H2A | 119.9 | C11—C12—H12 | 120.6 |
| C2—C3—C4 | 119.63 (19) | C12—C13—C14 | 120.71 (19) |
| C2—C3—H3 | 120.2 | C12—C13—H13 | 119.6 |
| C4—C3—H3 | 120.2 | C14—C13—H13 | 119.6 |
| C3—C4—C5 | 120.37 (19) | C15—C14—C13 | 121.4 (2) |
| C3—C4—H4A | 119.8 | C15—C14—H14 | 119.3 |
| C5—C4—H4A | 119.8 | C13—C14—H14 | 119.3 |
| C4—C5—C6 | 121.01 (19) | C16—C15—C14 | 117.20 (19) |
| C4—C5—H5 | 119.5 | C16—C15—H15 | 121.4 |
| C6—C5—H5 | 119.5 | C14—C15—H15 | 121.4 |
| C5—C6—C1 | 117.73 (18) | C15—C16—C11 | 122.65 (18) |
| C5—C6—C7 | 121.26 (18) | C15—C16—N4 | 128.34 (18) |
| C1—C6—C7 | 121.00 (17) | C11—C16—N4 | 109.01 (17) |
| C8—C7—N1 | 114.38 (17) | O1—C17—N4 | 126.73 (19) |
| C8—C7—C6 | 126.00 (18) | O1—C17—C10 | 126.84 (19) |
| N1—C7—C6 | 119.59 (16) | N4—C17—C10 | 106.42 (17) |
| C9—N2—N3—C10 | −179.69 (17) | N2—N3—C10—C17 | 0.7 (3) |
| C6—C1—C2—C3 | −0.6 (3) | N3—C10—C11—C12 | −3.6 (4) |
| C1—C2—C3—C4 | 0.2 (3) | C17—C10—C11—C12 | 176.1 (2) |
| C2—C3—C4—C5 | 0.4 (3) | N3—C10—C11—C16 | 177.11 (18) |
| C3—C4—C5—C6 | −0.7 (3) | C17—C10—C11—C16 | −3.2 (2) |
| C4—C5—C6—C1 | 0.3 (3) | C16—C11—C12—C13 | −1.0 (3) |
| C4—C5—C6—C7 | 179.04 (18) | C10—C11—C12—C13 | 179.7 (2) |
| C2—C1—C6—C5 | 0.3 (3) | C11—C12—C13—C14 | −0.2 (3) |
| C2—C1—C6—C7 | −178.37 (17) | C12—C13—C14—C15 | 0.9 (3) |
| C9—N1—C7—C8 | 0.9 (2) | C13—C14—C15—C16 | −0.3 (3) |
| C9—N1—C7—C6 | −177.30 (17) | C14—C15—C16—C11 | −0.9 (3) |
| C5—C6—C7—C8 | −167.50 (19) | C14—C15—C16—N4 | 178.32 (18) |
| C1—C6—C7—C8 | 11.2 (3) | C12—C11—C16—C15 | 1.6 (3) |
| C5—C6—C7—N1 | 10.4 (3) | C10—C11—C16—C15 | −178.92 (17) |
| C1—C6—C7—N1 | −170.89 (17) | C12—C11—C16—N4 | −177.75 (17) |
| N1—C7—C8—S1 | −0.5 (2) | C10—C11—C16—N4 | 1.7 (2) |
| C6—C7—C8—S1 | 177.49 (15) | C17—N4—C16—C15 | −178.68 (18) |
| C9—S1—C8—C7 | 0.06 (16) | C17—N4—C16—C11 | 0.7 (2) |
| C7—N1—C9—N2 | 179.57 (17) | C16—N4—C17—O1 | 176.05 (19) |
| C7—N1—C9—S1 | −0.8 (2) | C16—N4—C17—C10 | −2.7 (2) |
| N3—N2—C9—N1 | −178.05 (17) | N3—C10—C17—O1 | 4.6 (3) |
| N3—N2—C9—S1 | 2.4 (2) | C11—C10—C17—O1 | −175.10 (19) |
| C8—S1—C9—N1 | 0.48 (16) | N3—C10—C17—N4 | −176.72 (18) |
| C8—S1—C9—N2 | −179.92 (16) | C11—C10—C17—N4 | 3.6 (2) |
| N2—N3—C10—C11 | −179.72 (16) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1 | 0.86 | 2.12 | 2.771 (2) | 133 |
| N4—H4···O1i | 0.86 | 2.11 | 2.922 (2) | 158 |
Symmetry code: (i) −x, −y, −z+1.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: LH5732).
References
- Ali, M. A., Mirza, A. H., Bakar, H. J. H. A. & Bernhardt, P. V. (2011). Polyhedron, 30, 556–564.
- Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994). J. Appl. Cryst. 27, 435.
- Bharti, S. K., Nath, G., Tilak, R. & Singh, S. K. (2010). Eur. J. Med. Chem. 45, 651–660. [DOI] [PubMed]
- Bruker (1998). SMART and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kondratieva, M. L., Pepeleva, A. V., Belskaia, N. P., Koksharov, A. V., Groundwater, P. V., Robeyns, K., Van Meervelt, L., Dehaen, W., Fan, Z. J. & Bakulev, V. A. (2007). Tetrahedron, 63, 3042–3048.
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Sadık, G., Necmi, D., Ibrahim, Y., Alaaddin, Ç. & Dinçer, M. (2004). Acta Cryst. E60, o889–o891.
- Sheldrick, G. M. (1996). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Watkin, D. M., Pearce, L. & Prout, C. K. (1993). CAMERON. Chemical Crystallography Laboratory, University of Oxford, England.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814022715/lh5732sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814022715/lh5732Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814022715/lh5732Isup3.cml
. DOI: 10.1107/S1600536814022715/lh5732fig1.tif
The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level. The dashed line indicates an intramolecular N—H⋯N bond
. DOI: 10.1107/S1600536814022715/lh5732fig2.tif
Part of the crystal structure with hydrogen bonds indicated as dotted lines
CCDC reference: 1029498
Additional supporting information: crystallographic information; 3D view; checkCIF report



