Two isotypic title compounds comprise a 2-iminopyridine ring fused with a cyclooctane ring. In one compound, the cyclooctane ring adopts a twisted chair–chair conformation, while in the second, this ring adopts a twisted boat–chair conformation.
Keywords: crystal structure, cyclooctapyridine, hydrogen bonding
Abstract
The molecules of the two isotypic title compounds, C25H24BrN3, (I), and C25H24FN3, (II), comprise a 2-iminopyridine ring fused with a cyclooctane ring. In (I), the cyclooctane ring adopts a twisted chair–chair conformation, while in (II), this ring adopts a twisted boat–chair conformation. The dihedral angles between the planes of the pyridine ring and the bromobenzene and phenyl rings are 80.14 (12) and 71.72 (13)°, respectively, in (I). The equivalent angles in (II) are 75.25 (8) and 68.34 (9)°, respectively. In both crystals, inversion dimers linked by pairs of C—H⋯N interactions generate R 2 2(14) loops, which are further connected by weak C—H⋯π interactions, generating (110) sheets.
Chemical context
The pyridine skeleton is of great importance to chemists as well as to biologists as it is found in a large variety of naturally occurring compounds and also in clinically useful molecules having diverse biological activities. Its derivatives are known to possess antimicrobial (Jo et al., 2004 ▶) and antiviral (Mavel et al., 2002 ▶) activities. The heterocyclic 1,4-dihydropyridine ring is a common feature in compounds with various pharmacological activities such as antimicrobial (Hooper et al., 1982 ▶) and antithrombotic (Sunkel et al., 1990 ▶) activities. The chemistry of imines in particular is of special interest in the literature due to their numerous practical applications (Echevarria et al., 1999 ▶). Imines have attracted much attention because of their wide variety of applications in the electronics and photonics fields (Wang et al., 2001 ▶). Imines and their complexes have a variety of applications in the biological, clinical and analytical fields (Singh et al., 1975 ▶; Patel et al., 1999 ▶). Our interest in the preparation of pharmacologically active 2-imino pyridines led us to synthesise the title compounds and we have undertaken the X-ray crystal structure determination of these compounds in order to establish their conformations.
Structural commentary
The structures of compounds (I) and (II) are shown in Figs. 1 ▶ and 2, respectively ▶. The cyclooctane ring adopts a twisted chair–chair conformation in compound (I) and twisted boat–chair conformation (Wiberg, 2003 ▶) in compound (II).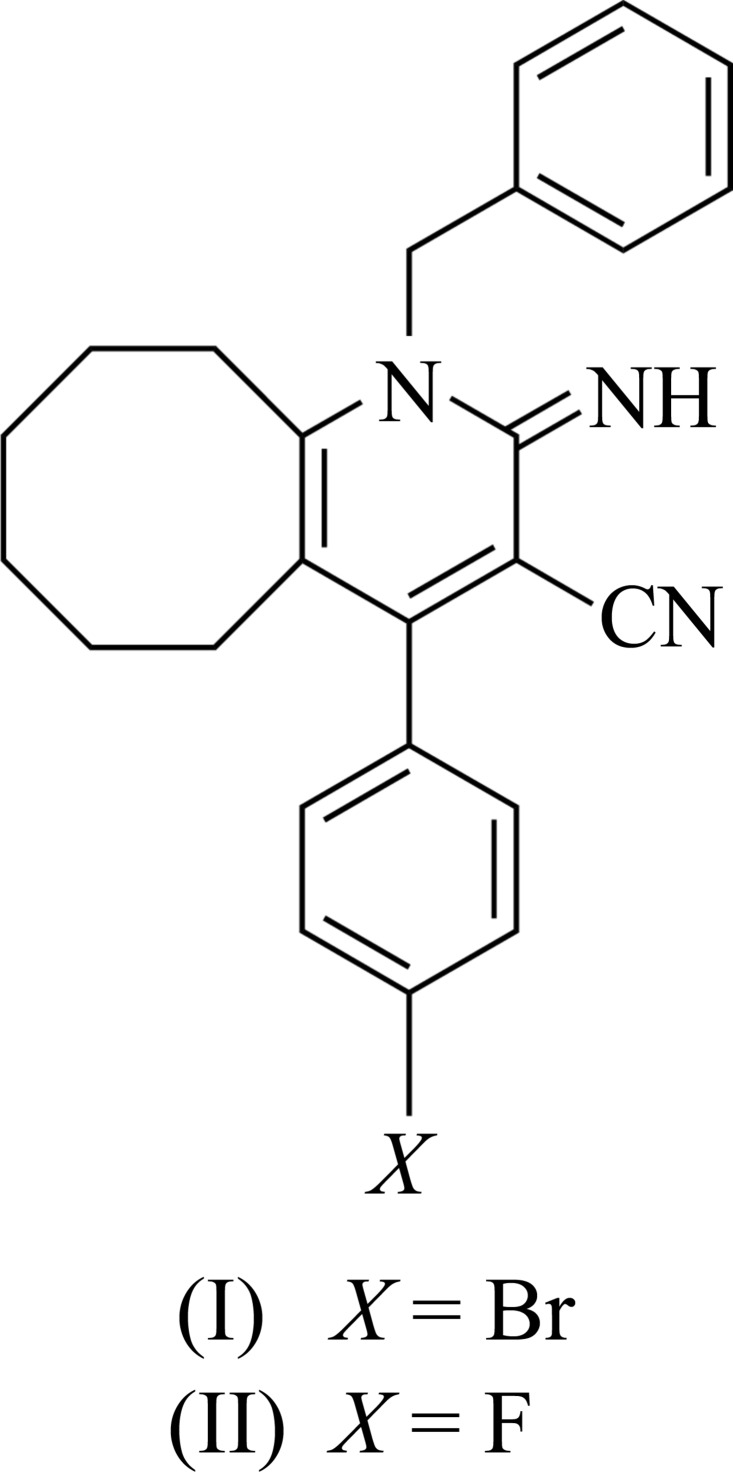
Figure 1.
The molecular structure of (I), showing 50% probability displacement ellipsoids.
Figure 2.
The molecular structure of (II), showing 50% probability displacement ellipsoids.
In both compounds, the imino group is nearly coplanar with the pyridine ring, as indicated by the N1=C1—N3—C5 torsion angle [178.8 (2) for compound (I) and 179.05 (13)° for compound (II)]. Steric hindrances rotate the phenyl (C13–C18) and aromatic (C31–C36) rings out of the plane of the central pyridine ring by 71.72 (13) and 80.14 (12)°, respectively, in compound (I), and by 68.34 (9) and 75.25 (8)°, respectively, in compound (II). Opening up of the N3—C5—C4 angle [121.54 (19)° for compound (I) and 121.29 (13)° for compound (II)] and considerable shortening of the C5—N3 [1.376 (3) Å for compound (I) and 1.3777 (18) Å for compound (II)] bond distance may directly be attributed to the bulky substituents at the ortho position C5. The endocyclic angles of the pyridine ring cover the range 114.29 (18)–123.02 (2)° and 118.86 (13)–123.11 (12)° for compounds (I) and (II) respectively. The C1—N3—C5 angle [122.93 (2) for compound (I) and 123.11 (12)° for compound (II)] is expanded as in pyridine itself [123.9 (3)°; Jin et al., 2005 ▶].
Supramolecular features
In the crystals, pairs of C—H⋯N interactions form  (14) ring motifs (Bernstein et al., 1995 ▶), and the resulting dimers are further connected through weak C—H⋯π interactions involving the phenyl ring as acceptor (Tables 1 ▶ and 2 ▶, Figs. 3 ▶, 4 ▶). In each case, the resulting supramolecular structure is a layer propagating parallel to the (110) plane.
(14) ring motifs (Bernstein et al., 1995 ▶), and the resulting dimers are further connected through weak C—H⋯π interactions involving the phenyl ring as acceptor (Tables 1 ▶ and 2 ▶, Figs. 3 ▶, 4 ▶). In each case, the resulting supramolecular structure is a layer propagating parallel to the (110) plane.
Table 1. Hydrogen-bond geometry (, ) for (I) .
Cg1 is the centroid of the C13C18 phenyl ring.
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C32H32N1i | 0.93 | 2.56 | 3.421(3) | 154 |
| C11H11A Cg1ii | 0.97 | 2.97 | 3.648(3) | 128 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Table 2. Hydrogen-bond geometry (, ) for (II) .
Cg1 is the centroid of the C13C18 phenyl ring.
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C32H32N1i | 0.93 | 2.53 | 3.421(2) | 160 |
| C11H11A Cg1ii | 0.97 | 2.93 | 3.484(2) | 118 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 3.

Partial packing diagram of the title compound (I). Dashed lines represent intermolecular hydrogen bonds and C—H⋯π contacts. For clarity, H atoms not involved in hydrogen bonding have been omitted.
Figure 4.

Partial packing diagram of the title compound (II). Dashed lines represent intermolecular hydrogen bonds and C—H⋯π contacts. For clarity, H atoms not involved in hydrogen bonding have been omitted.
Database survey
Similar structures reported in the literature are 2-methoxy-4-(2-methoxyphenyl)-5,6,7,8,9,10-hexahydrocycloocta[b]pyridine-3-carbonitrile (Vishnupriya et al., 2014a ▶) and 4-(2-fluorophenyl)-2-methoxy-5,6,7,8,9,10-hexahydrocycloocta[b]-pyridine-3-carbonitrile (Vishnupriya et al., 2014b ▶). The twisted conformation of the cyclooctane ring of compound (I) is similar to those found in the related structures. However, the C=NH functional group present in the title compound allows the formation of C—H⋯N hydrogen bonds, which are not present in the above-cited compounds. In the title compounds, the bond lengths in the central pyridine ring span the range 1.369–1.446 Å, which compare well with the range observed in the similar structures (1.314–1.400 Å), but these bonds are systematically longer in the title compounds, due to the substitution of the pyridine N atom by a benzyl group. The bond length of the nitrile group attached to pyridine ring [N2 C38 = 1.137 (3) Å in compound (I) and 1.1426 (19) Å in compound (II)] and the linearity of the cyano moiety [N2 C38—C2 = 176.3 (3) for compound (I) and 175.68 (17)° for compound (II)] have characteristic features that are observed in 3-cyano-2-pyridine derivatives (Hursthouse et al., 1992 ▶; Patel et al., 2002 ▶).
Synthesis and crystallization
The two compounds were prepared in a similar manner using 4-fluoro aldehyde (1 mmol) for compound (I) and 4-bromo aldehyde (1 mmol) for compound (II). A mixture of cyclooctanone (1mmol), respective aldehyde (1 mmol) and malononitrile (1 mmol) were taken in ethanol (10 mL) to which p-toluenesulfonic acid (pTSA) (0.5 mmol) was added. The reaction mixture was heated under reflux for 2–3 h. After completion of the reaction (TLC), the reaction mixture was poured into crushed ice and extracted with ethyl acetate. The excess solvent was removed under vacuum and the residue was subjected to column chromatography using petroleum ether/ethyl acetate mixture (97:3 v/v) as eluent to afford pure product. The product was recrystallized from ethyl acetate, affording colourless crystals of compounds (I) and (II) [m.p. 493 K; yield 91% for (I) and m.p. 473 K; yield 65% for (II)].
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▶. C-bound H atoms were placed in calculated positions and allowed to ride on their carrier atoms, with C—H = 0.93 (aromatic CH) or 0.97 Å (methylene CH2). Imine atom H1 was found in a difference map and refined with a distance restraint in both compounds of N—H = 0.86 (10) Å. Isotropic displacement parameters for H atoms were calculated as U iso = 1.5U eq(C) for CH3 groups and U iso = 1.2U eq(carrier atom) for all other H atoms. The DELU restraint was applied in compound (II).
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C25H24BrN3 | C25H24FN3 |
| M r | 446.38 | 385.47 |
| Crystal system, space group | Triclinic, P

|
Triclinic, P

|
| Temperature (K) | 293 | 293 |
| a, b, c () | 10.2103(3), 10.7643(4), 11.6942(4) | 10.1370(4), 10.2078(3), 11.8238(4) |
| , , () | 101.074(1), 106.726(1), 115.058(1) | 109.688(2), 100.309(2), 111.420(2) |
| V (3) | 1039.46(6) | 1006.73(6) |
| Z | 2 | 2 |
| Radiation type | Mo K | Mo K |
| (mm1) | 1.99 | 0.08 |
| Crystal size (mm) | 0.21 0.19 0.18 | 0.21 0.19 0.18 |
| Data collection | ||
| Diffractometer | Bruker Kappa APEXII | Bruker Kappa APEXII |
| Absorption correction | Multi-scan (SADABS; Bruker, 2004 ▶) | Multi-scan (SADABS; Bruker, 2004 ▶) |
| T min, T max | 0.967, 0.974 | 0.967, 0.974 |
| No. of measured, independent and observed [I > 2(I)] reflections | 25106, 4532, 3830 | 23254, 3752, 2876 |
| R int | 0.027 | 0.022 |
| (sin /)max (1) | 0.639 | 0.606 |
| Refinement | ||
| R[F 2 > 2(F 2)], wR(F 2), S | 0.039, 0.107, 1.03 | 0.039, 0.109, 1.05 |
| No. of reflections | 4532 | 3752 |
| No. of parameters | 266 | 267 |
| No. of restraints | 2 | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| max, min (e 3) | 0.93, 0.87 | 0.17, 0.14 |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S1600536814022016/hb7284sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814022016/hb7284Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S1600536814022016/hb7284IIsup3.hkl
Supporting information file. DOI: 10.1107/S1600536814022016/hb7284Isup4.cml
Supporting information file. DOI: 10.1107/S1600536814022016/hb7284IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
JS and RAN thank the management of The Madura College (Autonomous), Madurai, for their encouragement and support. RRK thanks the University Grants Commission, New Delhi, for funds through Major Research Project F. No. 42–242/2013 (SR).
supplementary crystallographic information
Crystal data
| C25H24FN3 | Z = 2 |
| Mr = 385.47 | F(000) = 408 |
| Triclinic, P1 | Dx = 1.272 Mg m−3 |
| a = 10.1370 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.2078 (3) Å | Cell parameters from 2000 reflections |
| c = 11.8238 (4) Å | θ = 2–31° |
| α = 109.688 (2)° | µ = 0.08 mm−1 |
| β = 100.309 (2)° | T = 293 K |
| γ = 111.420 (2)° | Block, colourless |
| V = 1006.73 (6) Å3 | 0.21 × 0.19 × 0.18 mm |
Data collection
| Bruker Kappa APEXII diffractometer | 2876 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.022 |
| φ and ω scans | θmax = 25.5°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 2004) | h = −12→12 |
| Tmin = 0.967, Tmax = 0.974 | k = −12→12 |
| 23254 measured reflections | l = −14→14 |
| 3752 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.039 | w = 1/[σ2(Fo2) + (0.0436P)2 + 0.3339P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.109 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.17 e Å−3 |
| 3752 reflections | Δρmin = −0.14 e Å−3 |
| 267 parameters | Extinction correction: SHELXL2014/6 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 2 restraints | Extinction coefficient: 0.027 (3) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.38640 (16) | 0.41969 (17) | 0.59999 (13) | 0.0347 (3) | |
| C2 | 0.28977 (16) | 0.44165 (16) | 0.50992 (13) | 0.0343 (3) | |
| C3 | 0.16525 (16) | 0.32042 (16) | 0.40896 (13) | 0.0341 (3) | |
| C4 | 0.12688 (16) | 0.16454 (17) | 0.39051 (13) | 0.0366 (3) | |
| C5 | 0.21646 (16) | 0.13987 (16) | 0.47452 (13) | 0.0345 (3) | |
| C6 | 0.18252 (19) | −0.02211 (18) | 0.46013 (15) | 0.0442 (4) | |
| H6A | 0.0738 | −0.0860 | 0.4268 | 0.053* | |
| H6B | 0.2196 | −0.0159 | 0.5442 | 0.053* | |
| C7 | 0.2506 (2) | −0.1046 (2) | 0.37256 (17) | 0.0562 (5) | |
| H7A | 0.3563 | −0.0323 | 0.3975 | 0.067* | |
| H7B | 0.2469 | −0.1927 | 0.3887 | 0.067* | |
| C8 | 0.1783 (2) | −0.1646 (2) | 0.22930 (17) | 0.0583 (5) | |
| H8A | 0.2219 | −0.2277 | 0.1857 | 0.070* | |
| H8B | 0.0716 | −0.2328 | 0.2046 | 0.070* | |
| C9 | 0.1946 (2) | −0.0406 (2) | 0.18178 (17) | 0.0570 (5) | |
| H9A | 0.2813 | 0.0570 | 0.2441 | 0.068* | |
| H9B | 0.2155 | −0.0717 | 0.1026 | 0.068* | |
| C10 | 0.0585 (2) | −0.0109 (2) | 0.15827 (15) | 0.0564 (5) | |
| H10A | −0.0238 | −0.1036 | 0.0864 | 0.068* | |
| H10B | 0.0830 | 0.0743 | 0.1337 | 0.068* | |
| C11 | 0.00304 (18) | 0.02948 (18) | 0.27088 (15) | 0.0475 (4) | |
| H11A | −0.0777 | 0.0557 | 0.2488 | 0.057* | |
| H11B | −0.0377 | −0.0616 | 0.2872 | 0.057* | |
| C12 | 0.44774 (17) | 0.23479 (19) | 0.65601 (14) | 0.0411 (4) | |
| H12A | 0.5478 | 0.3219 | 0.6876 | 0.049* | |
| H12B | 0.4517 | 0.1414 | 0.6027 | 0.049* | |
| C13 | 0.41019 (17) | 0.21501 (18) | 0.76831 (14) | 0.0409 (4) | |
| C14 | 0.4226 (2) | 0.0975 (2) | 0.79530 (17) | 0.0578 (5) | |
| H14 | 0.4469 | 0.0274 | 0.7406 | 0.069* | |
| C15 | 0.3991 (2) | 0.0836 (3) | 0.9035 (2) | 0.0756 (7) | |
| H15 | 0.4071 | 0.0040 | 0.9208 | 0.091* | |
| C16 | 0.3641 (3) | 0.1865 (3) | 0.98465 (19) | 0.0782 (7) | |
| H16 | 0.3500 | 0.1780 | 1.0580 | 0.094* | |
| C17 | 0.3499 (2) | 0.3013 (2) | 0.95823 (17) | 0.0694 (6) | |
| H17 | 0.3251 | 0.3706 | 1.0133 | 0.083* | |
| C18 | 0.3719 (2) | 0.31600 (19) | 0.85034 (16) | 0.0521 (4) | |
| H18 | 0.3609 | 0.3944 | 0.8328 | 0.063* | |
| C31 | 0.07281 (16) | 0.35321 (17) | 0.31963 (13) | 0.0371 (3) | |
| C32 | 0.12480 (18) | 0.39428 (18) | 0.22997 (15) | 0.0435 (4) | |
| H32 | 0.2186 | 0.4033 | 0.2270 | 0.052* | |
| C33 | 0.0391 (2) | 0.4221 (2) | 0.14484 (16) | 0.0491 (4) | |
| H33 | 0.0734 | 0.4484 | 0.0839 | 0.059* | |
| C34 | −0.09671 (19) | 0.4101 (2) | 0.15238 (16) | 0.0494 (4) | |
| C35 | −0.1521 (2) | 0.3707 (2) | 0.23943 (18) | 0.0586 (5) | |
| H35 | −0.2454 | 0.3637 | 0.2424 | 0.070* | |
| C36 | −0.06628 (19) | 0.3413 (2) | 0.32320 (17) | 0.0526 (4) | |
| H36 | −0.1027 | 0.3133 | 0.3827 | 0.063* | |
| C38 | 0.33469 (17) | 0.60054 (18) | 0.53208 (14) | 0.0395 (3) | |
| N1 | 0.50412 (15) | 0.52710 (17) | 0.69710 (13) | 0.0485 (4) | |
| N2 | 0.37721 (18) | 0.73047 (17) | 0.55698 (14) | 0.0572 (4) | |
| N3 | 0.34211 (13) | 0.26298 (14) | 0.57514 (11) | 0.0346 (3) | |
| F1 | −0.18123 (13) | 0.43791 (15) | 0.06937 (11) | 0.0758 (4) | |
| H1 | 0.520 (2) | 0.6177 (14) | 0.7008 (19) | 0.065 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0340 (7) | 0.0397 (8) | 0.0314 (7) | 0.0162 (6) | 0.0147 (6) | 0.0157 (6) |
| C2 | 0.0359 (7) | 0.0374 (7) | 0.0328 (7) | 0.0173 (6) | 0.0154 (6) | 0.0167 (6) |
| C3 | 0.0353 (7) | 0.0403 (8) | 0.0326 (7) | 0.0194 (6) | 0.0157 (6) | 0.0181 (6) |
| C4 | 0.0347 (8) | 0.0383 (8) | 0.0350 (8) | 0.0157 (6) | 0.0112 (6) | 0.0158 (6) |
| C5 | 0.0360 (7) | 0.0376 (8) | 0.0322 (7) | 0.0162 (6) | 0.0153 (6) | 0.0167 (6) |
| C6 | 0.0515 (9) | 0.0422 (8) | 0.0411 (8) | 0.0199 (7) | 0.0139 (7) | 0.0238 (7) |
| C7 | 0.0739 (12) | 0.0487 (10) | 0.0532 (10) | 0.0376 (9) | 0.0180 (9) | 0.0221 (8) |
| C8 | 0.0773 (13) | 0.0448 (9) | 0.0506 (10) | 0.0314 (9) | 0.0204 (9) | 0.0156 (8) |
| C9 | 0.0743 (12) | 0.0491 (10) | 0.0431 (9) | 0.0251 (9) | 0.0243 (9) | 0.0169 (8) |
| C10 | 0.0729 (12) | 0.0428 (9) | 0.0354 (9) | 0.0186 (9) | 0.0039 (8) | 0.0135 (7) |
| C11 | 0.0421 (9) | 0.0407 (8) | 0.0467 (9) | 0.0137 (7) | 0.0027 (7) | 0.0172 (7) |
| C12 | 0.0405 (8) | 0.0499 (9) | 0.0373 (8) | 0.0251 (7) | 0.0112 (6) | 0.0204 (7) |
| C13 | 0.0387 (8) | 0.0425 (8) | 0.0324 (7) | 0.0133 (7) | 0.0039 (6) | 0.0164 (7) |
| C14 | 0.0618 (11) | 0.0604 (11) | 0.0516 (10) | 0.0290 (9) | 0.0076 (9) | 0.0299 (9) |
| C15 | 0.0747 (14) | 0.0776 (14) | 0.0661 (13) | 0.0202 (12) | 0.0013 (11) | 0.0504 (12) |
| C16 | 0.0799 (15) | 0.0760 (15) | 0.0375 (10) | −0.0012 (12) | 0.0042 (10) | 0.0284 (10) |
| C17 | 0.0788 (14) | 0.0539 (11) | 0.0429 (10) | 0.0053 (10) | 0.0240 (10) | 0.0117 (9) |
| C18 | 0.0616 (11) | 0.0412 (9) | 0.0427 (9) | 0.0140 (8) | 0.0192 (8) | 0.0164 (7) |
| C31 | 0.0391 (8) | 0.0378 (8) | 0.0346 (8) | 0.0190 (6) | 0.0110 (6) | 0.0156 (6) |
| C32 | 0.0418 (8) | 0.0477 (9) | 0.0451 (9) | 0.0207 (7) | 0.0159 (7) | 0.0246 (7) |
| C33 | 0.0565 (10) | 0.0548 (10) | 0.0469 (9) | 0.0266 (8) | 0.0200 (8) | 0.0319 (8) |
| C34 | 0.0564 (10) | 0.0532 (10) | 0.0450 (9) | 0.0318 (8) | 0.0102 (8) | 0.0250 (8) |
| C35 | 0.0529 (10) | 0.0847 (13) | 0.0617 (11) | 0.0458 (10) | 0.0251 (9) | 0.0396 (10) |
| C36 | 0.0530 (10) | 0.0756 (12) | 0.0524 (10) | 0.0389 (9) | 0.0266 (8) | 0.0389 (9) |
| C38 | 0.0427 (8) | 0.0410 (8) | 0.0368 (8) | 0.0197 (7) | 0.0155 (7) | 0.0178 (7) |
| N1 | 0.0435 (8) | 0.0441 (8) | 0.0426 (8) | 0.0135 (7) | 0.0039 (6) | 0.0149 (6) |
| N2 | 0.0693 (10) | 0.0438 (8) | 0.0571 (9) | 0.0241 (7) | 0.0215 (8) | 0.0230 (7) |
| N3 | 0.0360 (6) | 0.0407 (7) | 0.0299 (6) | 0.0189 (5) | 0.0114 (5) | 0.0168 (5) |
| F1 | 0.0799 (8) | 0.1045 (9) | 0.0715 (7) | 0.0590 (7) | 0.0180 (6) | 0.0556 (7) |
Geometric parameters (Å, º)
| C1—N1 | 1.2847 (19) | C12—N3 | 1.4790 (18) |
| C1—N3 | 1.3994 (18) | C12—C13 | 1.501 (2) |
| C1—C2 | 1.446 (2) | C12—H12A | 0.9700 |
| C2—C3 | 1.369 (2) | C12—H12B | 0.9700 |
| C2—C38 | 1.428 (2) | C13—C18 | 1.381 (2) |
| C3—C4 | 1.419 (2) | C13—C14 | 1.382 (2) |
| C3—C31 | 1.4896 (19) | C14—C15 | 1.385 (3) |
| C4—C5 | 1.3737 (19) | C14—H14 | 0.9300 |
| C4—C11 | 1.505 (2) | C15—C16 | 1.366 (3) |
| C5—N3 | 1.3777 (18) | C15—H15 | 0.9300 |
| C5—C6 | 1.502 (2) | C16—C17 | 1.358 (3) |
| C6—C7 | 1.533 (2) | C16—H16 | 0.9300 |
| C6—H6A | 0.9700 | C17—C18 | 1.381 (2) |
| C6—H6B | 0.9700 | C17—H17 | 0.9300 |
| C7—C8 | 1.520 (2) | C18—H18 | 0.9300 |
| C7—H7A | 0.9700 | C31—C36 | 1.380 (2) |
| C7—H7B | 0.9700 | C31—C32 | 1.384 (2) |
| C8—C9 | 1.518 (2) | C32—C33 | 1.382 (2) |
| C8—H8A | 0.9700 | C32—H32 | 0.9300 |
| C8—H8B | 0.9700 | C33—C34 | 1.358 (2) |
| C9—C10 | 1.517 (3) | C33—H33 | 0.9300 |
| C9—H9A | 0.9700 | C34—F1 | 1.3570 (18) |
| C9—H9B | 0.9700 | C34—C35 | 1.363 (2) |
| C10—C11 | 1.525 (2) | C35—C36 | 1.382 (2) |
| C10—H10A | 0.9700 | C35—H35 | 0.9300 |
| C10—H10B | 0.9700 | C36—H36 | 0.9300 |
| C11—H11A | 0.9700 | C38—N2 | 1.1426 (19) |
| C11—H11B | 0.9700 | N1—H1 | 0.864 (9) |
| N1—C1—N3 | 118.86 (13) | C10—C11—H11B | 109.2 |
| N1—C1—C2 | 126.92 (14) | H11A—C11—H11B | 107.9 |
| N3—C1—C2 | 114.22 (12) | N3—C12—C13 | 115.71 (12) |
| C3—C2—C38 | 121.57 (13) | N3—C12—H12A | 108.4 |
| C3—C2—C1 | 123.31 (13) | C13—C12—H12A | 108.4 |
| C38—C2—C1 | 115.11 (13) | N3—C12—H12B | 108.4 |
| C2—C3—C4 | 119.20 (13) | C13—C12—H12B | 108.4 |
| C2—C3—C31 | 119.87 (13) | H12A—C12—H12B | 107.4 |
| C4—C3—C31 | 120.92 (13) | C18—C13—C14 | 118.55 (15) |
| C5—C4—C3 | 118.86 (13) | C18—C13—C12 | 122.21 (14) |
| C5—C4—C11 | 120.91 (13) | C14—C13—C12 | 119.14 (15) |
| C3—C4—C11 | 119.70 (13) | C13—C14—C15 | 120.31 (19) |
| C4—C5—N3 | 121.29 (13) | C13—C14—H14 | 119.8 |
| C4—C5—C6 | 121.54 (13) | C15—C14—H14 | 119.8 |
| N3—C5—C6 | 117.16 (12) | C16—C15—C14 | 120.2 (2) |
| C5—C6—C7 | 115.02 (13) | C16—C15—H15 | 119.9 |
| C5—C6—H6A | 108.5 | C14—C15—H15 | 119.9 |
| C7—C6—H6A | 108.5 | C17—C16—C15 | 119.92 (19) |
| C5—C6—H6B | 108.5 | C17—C16—H16 | 120.0 |
| C7—C6—H6B | 108.5 | C15—C16—H16 | 120.0 |
| H6A—C6—H6B | 107.5 | C16—C17—C18 | 120.6 (2) |
| C8—C7—C6 | 117.39 (15) | C16—C17—H17 | 119.7 |
| C8—C7—H7A | 108.0 | C18—C17—H17 | 119.7 |
| C6—C7—H7A | 108.0 | C17—C18—C13 | 120.41 (18) |
| C8—C7—H7B | 108.0 | C17—C18—H18 | 119.8 |
| C6—C7—H7B | 108.0 | C13—C18—H18 | 119.8 |
| H7A—C7—H7B | 107.2 | C36—C31—C32 | 118.80 (14) |
| C9—C8—C7 | 116.07 (15) | C36—C31—C3 | 121.03 (13) |
| C9—C8—H8A | 108.3 | C32—C31—C3 | 120.16 (13) |
| C7—C8—H8A | 108.3 | C33—C32—C31 | 120.83 (15) |
| C9—C8—H8B | 108.3 | C33—C32—H32 | 119.6 |
| C7—C8—H8B | 108.3 | C31—C32—H32 | 119.6 |
| H8A—C8—H8B | 107.4 | C34—C33—C32 | 118.37 (15) |
| C10—C9—C8 | 115.10 (16) | C34—C33—H33 | 120.8 |
| C10—C9—H9A | 108.5 | C32—C33—H33 | 120.8 |
| C8—C9—H9A | 108.5 | F1—C34—C33 | 118.67 (15) |
| C10—C9—H9B | 108.5 | F1—C34—C35 | 118.52 (16) |
| C8—C9—H9B | 108.5 | C33—C34—C35 | 122.81 (15) |
| H9A—C9—H9B | 107.5 | C34—C35—C36 | 118.38 (16) |
| C9—C10—C11 | 115.73 (14) | C34—C35—H35 | 120.8 |
| C9—C10—H10A | 108.3 | C36—C35—H35 | 120.8 |
| C11—C10—H10A | 108.3 | C31—C36—C35 | 120.80 (16) |
| C9—C10—H10B | 108.3 | C31—C36—H36 | 119.6 |
| C11—C10—H10B | 108.3 | C35—C36—H36 | 119.6 |
| H10A—C10—H10B | 107.4 | N2—C38—C2 | 175.68 (17) |
| C4—C11—C10 | 112.25 (13) | C1—N1—H1 | 109.5 (13) |
| C4—C11—H11A | 109.2 | C5—N3—C1 | 123.11 (12) |
| C10—C11—H11A | 109.2 | C5—N3—C12 | 120.92 (12) |
| C4—C11—H11B | 109.2 | C1—N3—C12 | 115.68 (12) |
| N1—C1—C2—C3 | −179.54 (14) | C14—C15—C16—C17 | 1.1 (3) |
| N3—C1—C2—C3 | 0.16 (19) | C15—C16—C17—C18 | −0.6 (3) |
| N1—C1—C2—C38 | 1.5 (2) | C16—C17—C18—C13 | −0.6 (3) |
| N3—C1—C2—C38 | −178.81 (12) | C14—C13—C18—C17 | 1.4 (3) |
| C38—C2—C3—C4 | 179.19 (13) | C12—C13—C18—C17 | −174.81 (16) |
| C1—C2—C3—C4 | 0.3 (2) | C2—C3—C31—C36 | −105.63 (18) |
| C38—C2—C3—C31 | −0.3 (2) | C4—C3—C31—C36 | 74.87 (19) |
| C1—C2—C3—C31 | −179.22 (13) | C2—C3—C31—C32 | 75.25 (18) |
| C2—C3—C4—C5 | −0.2 (2) | C4—C3—C31—C32 | −104.25 (17) |
| C31—C3—C4—C5 | 179.26 (13) | C36—C31—C32—C33 | −0.4 (2) |
| C2—C3—C4—C11 | −172.00 (13) | C3—C31—C32—C33 | 178.75 (14) |
| C31—C3—C4—C11 | 7.5 (2) | C31—C32—C33—C34 | 0.8 (2) |
| C3—C4—C5—N3 | −0.3 (2) | C32—C33—C34—F1 | 179.75 (15) |
| C11—C4—C5—N3 | 171.40 (13) | C32—C33—C34—C35 | −0.6 (3) |
| C3—C4—C5—C6 | −179.76 (13) | F1—C34—C35—C36 | 179.53 (16) |
| C11—C4—C5—C6 | −8.1 (2) | C33—C34—C35—C36 | −0.2 (3) |
| C4—C5—C6—C7 | 87.41 (18) | C32—C31—C36—C35 | −0.4 (3) |
| N3—C5—C6—C7 | −92.12 (16) | C3—C31—C36—C35 | −179.49 (16) |
| C5—C6—C7—C8 | −73.7 (2) | C34—C35—C36—C31 | 0.6 (3) |
| C6—C7—C8—C9 | 67.0 (2) | C4—C5—N3—C1 | 0.7 (2) |
| C7—C8—C9—C10 | −99.2 (2) | C6—C5—N3—C1 | −179.73 (12) |
| C8—C9—C10—C11 | 54.5 (2) | C4—C5—N3—C12 | −172.79 (13) |
| C5—C4—C11—C10 | −89.02 (17) | C6—C5—N3—C12 | 6.73 (19) |
| C3—C4—C11—C10 | 82.57 (17) | N1—C1—N3—C5 | 179.05 (13) |
| C9—C10—C11—C4 | 53.53 (19) | C2—C1—N3—C5 | −0.67 (18) |
| N3—C12—C13—C18 | −47.2 (2) | N1—C1—N3—C12 | −7.09 (19) |
| N3—C12—C13—C14 | 136.67 (15) | C2—C1—N3—C12 | 173.18 (11) |
| C18—C13—C14—C15 | −0.9 (3) | C13—C12—N3—C5 | −88.06 (16) |
| C12—C13—C14—C15 | 175.42 (16) | C13—C12—N3—C1 | 97.94 (15) |
| C13—C14—C15—C16 | −0.3 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C13–C18 phenyl ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C32—H32···N1i | 0.93 | 2.53 | 3.421 (2) | 160 |
| C11—H11A···Cg1ii | 0.97 | 2.93 | 3.484 (2) | 118 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z+1.
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2004). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Echevarria, A., Nascimento, M. G., d, G., Gerônimo, V., Miller, J. & Giesbrecht, A. (1999). J. Braz. Chem. Soc. 10, 60–64.
- Hooper, D. C., Wolfson, J. S., McHugh, G. L., Winters, M. B. & Swartz, M. N. (1982). Antimicrob. Agents Chemother. 22, 662–671. [DOI] [PMC free article] [PubMed]
- Hursthouse, M. B., Karaulov, A. I., Ciechanowicz-Rutkowska, M., Kolasa, A. & Zankowska-Jasińska, W. (1992). Acta Cryst. C48, 1257–1260.
- Jin, Z.-M., Shun, N., Lü, Y.-P., Hu, M.-L. & Shen, L. (2005). Acta Cryst. C61, m43–m45. [DOI] [PubMed]
- Jo, Y. W., Im, W. B., Rhee, J. K., Shim, M. J., Kim, W. B. & Choi, E. C. (2004). Bioorg. Med. Chem. 12, 5909–5915. [DOI] [PubMed]
- Mavel, S., Renou, J., Galtier, C., Allouchi, H., Snoeck, R., Andrei, G., De Clercq, E., Balzarini, J. & Gueiffier, A. (2002). Bioorg. Med. Chem. 10, 941–946. [DOI] [PubMed]
- Patel, U. H., Dave, C. G., Jotani, M. M. & Shah, H. C. (2002). Acta Cryst. C58, o191–o192. [DOI] [PubMed]
- Patel, P. R., Thaker, B. T. & Zele, S. (1999). Indian J. Chem. Sect. A, 38, 563–566.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Singh, P., Goel, R. L. & Singh, B. P. J. (1975). Indian Chem, 52, 958–959Yoeong.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sunkel, C. E., de Casa-Juana, M. F., Santos, L., Gómez, M. M., Villarroya, M., González-Morales, M. A., Priego, J. G. & Ortega, M. P. (1990). J. Med. Chem. 33, 3205–3210. [DOI] [PubMed]
- Vishnupriya, R., Suresh, J., Maharani, S., Kumar, R. R. & Lakshman, P. L. N. (2014a). Acta Cryst. E70, o656. [DOI] [PMC free article] [PubMed]
- Vishnupriya, R., Suresh, J., Maharani, S., Kumar, R. R. & Lakshman, P. L. N. (2014b). Acta Cryst. E70, o872. [DOI] [PMC free article] [PubMed]
- Wang, X., Shen, Y., Pan, Y. & Liang, Y. (2001). Langmuir, 17, 3162–3167.
- Wiberg, K. B. (2003). J. Org. Chem. 68, 9322–9329. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S1600536814022016/hb7284sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814022016/hb7284Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S1600536814022016/hb7284IIsup3.hkl
Supporting information file. DOI: 10.1107/S1600536814022016/hb7284Isup4.cml
Supporting information file. DOI: 10.1107/S1600536814022016/hb7284IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report




