Abstract
In the title compound, C16H12O5, synthesized from isopthaloyl chloride and 2′-hydroxyacetophenone, the dihedral angle between the planes of the aromatic rings is 71.37 (9)°. In the crystal, carboxylic acid inversion dimers generate R 2 2(8) loops. The dimers are linked by C—H⋯O interactions, generating (101) sheets.
Keywords: crystal structure, 3-[(2-acetylphenoxy)carbonyl]benzoic acid, hydrogen bonding, 2′-hydroxyacetophenone, isopthaloyl chloride
Related literature
For related structures, see: Derissen (1974 ▶); Tanimoto et al. (1973 ▶).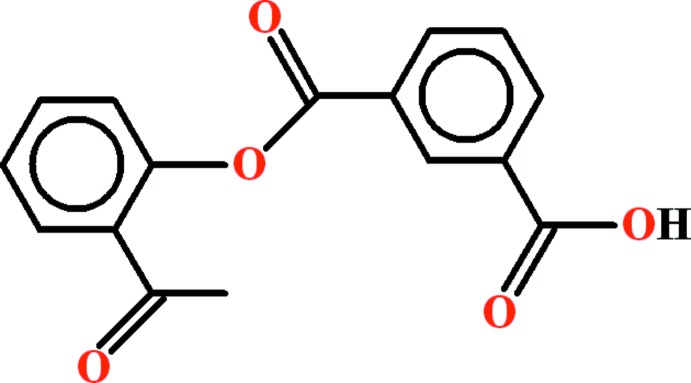
Experimental
Crystal data
C16H12O5
M r = 284.26
Monoclinic,
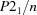
a = 13.5081 (10) Å
b = 7.4743 (6) Å
c = 13.9421 (11) Å
β = 106.671 (3)°
V = 1348.48 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 296 K
0.38 × 0.28 × 0.25 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.963, T max = 0.977
10280 measured reflections
2655 independent reflections
1971 reflections with I > 2σ(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.115
S = 1.03
2655 reflections
192 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.20 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814021904/hb7293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814021904/hb7293Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814021904/hb7293Isup3.cml
. DOI: 10.1107/S1600536814021904/hb7293fig1.tif
View of the title compound with displacement ellipsoids drawn at the 50% probability level.
PLATON . DOI: 10.1107/S1600536814021904/hb7293fig2.tif
The partial packing (PLATON; Spek, 2009), which shows that molecules form dimers which are interlinked.
CCDC reference: 1027627
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| O1H1O2i | 0.82 | 1.84 | 2.6623(18) | 175 |
| C4H4O5ii | 0.93 | 2.58 | 3.257(3) | 130 |
Symmetry codes: (i) 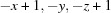 ; (ii)
; (ii) 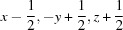 .
.
Acknowledgments
The authors are grateful to the University of Malakand, Khyber Pakhtunkhwa, Pakistan, for provision of laboratory facilities for carrying out this research work.
supplementary crystallographic information
S1. Comment
The title compound (I), (Fig. 1) has been synthesized for forming different metal complexes. The crystal structures of isophthalic acid and acetophenone have been published by (Derissen, 1974) and (Tanimoto, et al., 1973) which are related to the title compound (I).
In (I) the group A (C1—C8/O1—O4) being like a part of isophthalic acid and benzene ring attached to it B (C9—C13) are almost planar with r. m. s. deviation of 0.0308 and 0.0034 Å, respectively. The dihedral angle between A/B is 71.98 (5)°. The acetaldehyde group C (O5/C15/C16) attached to ring B is of course planar. The dihedral angle between B/C is 9.56 (23)°. The molecules are dimerized due to coventional H-bondings of O—H···O type (Table 1, Fig. 2) forming R22(8) loop. The dimers are further interlinked due to C—H···O bondings where C—H is of benzene containing carboxylate and O is of acetaldehyde group.
S2. Experimental
Isopthaloyl chloride (25 mmol) and 2'-hydroxyacetophenone (35 mmol) were refluxed in the aquauos solution of pyridine for 30 min. The mixture was cooled to room temperature and added to a beaker containing 2 N HCl. The crushed ice was added and stirred vigorously. The precipitate formed were obtained though filteration. The column chromatography was done ethyl acetate:n-hexane (4:6) to obtain the pure product. Light yellow prisms were obtained after two days.
S3. Refinement
All H atoms were geometrically placed [(O–H = 0.82 Å (hydroxyl), C–H = 0.93 Å (aromatic) and C–H = 0.96 Å (methyl) and refined as riding with with Uiso(H) = xUeq(C, O), where x = 1.5 for hydroxy & methyl and x = 1.2 for aromatic H-atoms.
Figures
Fig. 1.
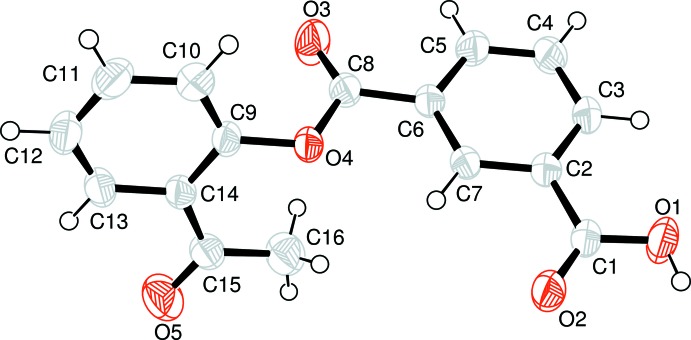
View of the title compound with displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
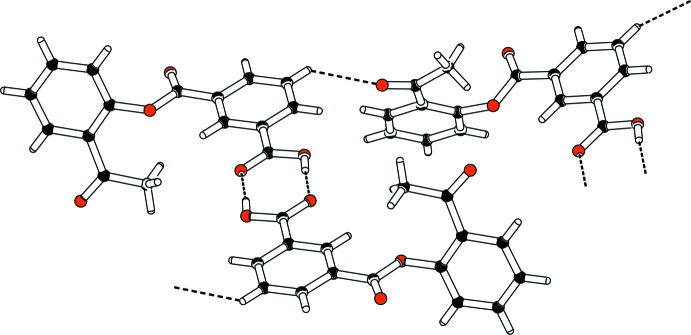
The partial packing (PLATON; Spek, 2009), which shows that molecules form dimers which are interlinked.
Crystal data
| C16H12O5 | F(000) = 592 |
| Mr = 284.26 | Dx = 1.400 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.5081 (10) Å | Cell parameters from 1971 reflections |
| b = 7.4743 (6) Å | θ = 1.9–26.0° |
| c = 13.9421 (11) Å | µ = 0.11 mm−1 |
| β = 106.671 (3)° | T = 296 K |
| V = 1348.48 (18) Å3 | Prism, light yellow |
| Z = 4 | 0.38 × 0.28 × 0.25 mm |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 2655 independent reflections |
| Radiation source: fine-focus sealed tube | 1971 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.022 |
| Detector resolution: 7.50 pixels mm-1 | θmax = 26.0°, θmin = 1.9° |
| ω scans | h = −15→16 |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | k = −6→9 |
| Tmin = 0.963, Tmax = 0.977 | l = −17→17 |
| 10280 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.115 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0505P)2 + 0.4011P] where P = (Fo2 + 2Fc2)/3 |
| 2655 reflections | (Δ/σ)max < 0.001 |
| 192 parameters | Δρmax = 0.18 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.36130 (10) | 0.0311 (2) | 0.47366 (9) | 0.0704 (5) | |
| H1 | 0.4166 | 0.0276 | 0.5177 | 0.106* | |
| O2 | 0.46474 (9) | −0.0058 (2) | 0.37694 (9) | 0.0627 (4) | |
| O3 | 0.14695 (9) | −0.0066 (2) | −0.05814 (9) | 0.0669 (4) | |
| O4 | 0.31564 (8) | −0.05274 (16) | 0.01634 (7) | 0.0411 (3) | |
| O5 | 0.48190 (13) | 0.2965 (2) | −0.11562 (12) | 0.0767 (5) | |
| C1 | 0.37753 (13) | 0.0154 (2) | 0.38694 (11) | 0.0435 (4) | |
| C2 | 0.28422 (12) | 0.0221 (2) | 0.29912 (11) | 0.0370 (4) | |
| C3 | 0.18668 (13) | 0.0478 (2) | 0.31086 (12) | 0.0435 (4) | |
| H3 | 0.1791 | 0.0628 | 0.3746 | 0.052* | |
| C4 | 0.10086 (13) | 0.0512 (3) | 0.22840 (13) | 0.0505 (5) | |
| H4 | 0.0355 | 0.0673 | 0.2366 | 0.061* | |
| C5 | 0.11211 (13) | 0.0306 (3) | 0.13359 (13) | 0.0471 (4) | |
| H5 | 0.0542 | 0.0336 | 0.0780 | 0.056* | |
| C6 | 0.20966 (12) | 0.0052 (2) | 0.12066 (11) | 0.0368 (4) | |
| C7 | 0.29593 (12) | 0.0005 (2) | 0.20355 (11) | 0.0370 (4) | |
| H7 | 0.3613 | −0.0169 | 0.1955 | 0.044* | |
| C8 | 0.21703 (12) | −0.0183 (2) | 0.01684 (12) | 0.0404 (4) | |
| C9 | 0.33430 (12) | −0.0900 (2) | −0.07617 (11) | 0.0381 (4) | |
| C10 | 0.30562 (14) | −0.2564 (3) | −0.11818 (13) | 0.0485 (4) | |
| H10 | 0.2692 | −0.3350 | −0.0892 | 0.058* | |
| C11 | 0.33147 (15) | −0.3054 (3) | −0.20375 (13) | 0.0560 (5) | |
| H11 | 0.3132 | −0.4178 | −0.2319 | 0.067* | |
| C12 | 0.38433 (14) | −0.1876 (3) | −0.24715 (13) | 0.0552 (5) | |
| H12 | 0.4012 | −0.2199 | −0.3049 | 0.066* | |
| C13 | 0.41194 (13) | −0.0225 (3) | −0.20493 (12) | 0.0476 (5) | |
| H13 | 0.4474 | 0.0559 | −0.2351 | 0.057* | |
| C14 | 0.38845 (12) | 0.0318 (2) | −0.11770 (11) | 0.0375 (4) | |
| C15 | 0.42394 (13) | 0.2153 (2) | −0.07883 (12) | 0.0456 (4) | |
| C16 | 0.38994 (17) | 0.3003 (3) | 0.00334 (16) | 0.0614 (5) | |
| H16A | 0.4174 | 0.4195 | 0.0148 | 0.092* | |
| H16B | 0.4148 | 0.2311 | 0.0635 | 0.092* | |
| H16C | 0.3158 | 0.3051 | −0.0154 | 0.092* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0455 (8) | 0.1379 (14) | 0.0272 (6) | 0.0069 (9) | 0.0097 (5) | −0.0055 (7) |
| O2 | 0.0363 (7) | 0.1189 (13) | 0.0323 (6) | 0.0023 (7) | 0.0089 (5) | −0.0024 (7) |
| O3 | 0.0391 (7) | 0.1245 (13) | 0.0328 (7) | 0.0139 (7) | 0.0037 (5) | 0.0015 (7) |
| O4 | 0.0325 (6) | 0.0653 (8) | 0.0264 (5) | −0.0005 (5) | 0.0097 (4) | −0.0007 (5) |
| O5 | 0.0916 (11) | 0.0748 (10) | 0.0791 (10) | −0.0290 (9) | 0.0495 (9) | −0.0073 (8) |
| C1 | 0.0401 (10) | 0.0629 (12) | 0.0286 (8) | −0.0013 (8) | 0.0116 (7) | −0.0012 (7) |
| C2 | 0.0356 (9) | 0.0454 (10) | 0.0305 (8) | −0.0018 (7) | 0.0104 (6) | 0.0008 (6) |
| C3 | 0.0430 (10) | 0.0594 (11) | 0.0321 (8) | −0.0023 (8) | 0.0169 (7) | −0.0031 (7) |
| C4 | 0.0338 (9) | 0.0763 (13) | 0.0449 (10) | 0.0017 (9) | 0.0171 (8) | −0.0054 (9) |
| C5 | 0.0329 (9) | 0.0706 (13) | 0.0360 (9) | 0.0009 (8) | 0.0071 (7) | −0.0027 (8) |
| C6 | 0.0324 (8) | 0.0483 (10) | 0.0300 (8) | −0.0011 (7) | 0.0097 (6) | 0.0008 (7) |
| C7 | 0.0313 (8) | 0.0500 (10) | 0.0312 (8) | −0.0018 (7) | 0.0113 (6) | 0.0011 (7) |
| C8 | 0.0317 (8) | 0.0574 (11) | 0.0311 (8) | 0.0010 (7) | 0.0074 (7) | 0.0027 (7) |
| C9 | 0.0317 (8) | 0.0567 (11) | 0.0256 (7) | 0.0028 (7) | 0.0077 (6) | −0.0007 (7) |
| C10 | 0.0474 (10) | 0.0557 (11) | 0.0418 (9) | −0.0069 (9) | 0.0118 (8) | −0.0027 (8) |
| C11 | 0.0544 (11) | 0.0617 (13) | 0.0480 (10) | −0.0025 (9) | 0.0082 (9) | −0.0173 (9) |
| C12 | 0.0499 (11) | 0.0804 (15) | 0.0376 (9) | 0.0033 (10) | 0.0161 (8) | −0.0135 (9) |
| C13 | 0.0411 (9) | 0.0692 (13) | 0.0362 (9) | 0.0021 (8) | 0.0172 (7) | 0.0005 (8) |
| C14 | 0.0296 (8) | 0.0522 (10) | 0.0307 (8) | 0.0036 (7) | 0.0087 (6) | 0.0012 (7) |
| C15 | 0.0404 (9) | 0.0555 (11) | 0.0419 (9) | −0.0006 (8) | 0.0135 (8) | 0.0047 (8) |
| C16 | 0.0714 (14) | 0.0531 (12) | 0.0682 (13) | −0.0077 (10) | 0.0335 (11) | −0.0133 (10) |
Geometric parameters (Å, º)
| O1—C1 | 1.2946 (19) | C6—C8 | 1.489 (2) |
| O1—H1 | 0.8200 | C7—H7 | 0.9300 |
| O2—C1 | 1.236 (2) | C9—C10 | 1.382 (2) |
| O3—C8 | 1.1953 (19) | C9—C14 | 1.394 (2) |
| O4—C8 | 1.3587 (19) | C10—C11 | 1.385 (2) |
| O4—C9 | 1.4111 (17) | C10—H10 | 0.9300 |
| O5—C15 | 1.215 (2) | C11—C12 | 1.379 (3) |
| C1—C2 | 1.485 (2) | C11—H11 | 0.9300 |
| C2—C3 | 1.387 (2) | C12—C13 | 1.372 (3) |
| C2—C7 | 1.395 (2) | C12—H12 | 0.9300 |
| C3—C4 | 1.379 (2) | C13—C14 | 1.402 (2) |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| C4—C5 | 1.382 (2) | C14—C15 | 1.502 (2) |
| C4—H4 | 0.9300 | C15—C16 | 1.493 (2) |
| C5—C6 | 1.393 (2) | C16—H16A | 0.9600 |
| C5—H5 | 0.9300 | C16—H16B | 0.9600 |
| C6—C7 | 1.386 (2) | C16—H16C | 0.9600 |
| C1—O1—H1 | 109.5 | C10—C9—O4 | 117.60 (14) |
| C8—O4—C9 | 118.33 (12) | C14—C9—O4 | 120.23 (15) |
| O2—C1—O1 | 122.65 (15) | C9—C10—C11 | 119.60 (17) |
| O2—C1—C2 | 121.50 (14) | C9—C10—H10 | 120.2 |
| O1—C1—C2 | 115.84 (14) | C11—C10—H10 | 120.2 |
| C3—C2—C7 | 120.03 (15) | C12—C11—C10 | 119.96 (18) |
| C3—C2—C1 | 121.19 (14) | C12—C11—H11 | 120.0 |
| C7—C2—C1 | 118.78 (14) | C10—C11—H11 | 120.0 |
| C4—C3—C2 | 120.27 (15) | C13—C12—C11 | 119.85 (16) |
| C4—C3—H3 | 119.9 | C13—C12—H12 | 120.1 |
| C2—C3—H3 | 119.9 | C11—C12—H12 | 120.1 |
| C3—C4—C5 | 119.91 (15) | C12—C13—C14 | 122.08 (17) |
| C3—C4—H4 | 120.0 | C12—C13—H13 | 119.0 |
| C5—C4—H4 | 120.0 | C14—C13—H13 | 119.0 |
| C4—C5—C6 | 120.44 (15) | C9—C14—C13 | 116.61 (16) |
| C4—C5—H5 | 119.8 | C9—C14—C15 | 126.71 (14) |
| C6—C5—H5 | 119.8 | C13—C14—C15 | 116.68 (15) |
| C7—C6—C5 | 119.71 (14) | O5—C15—C16 | 119.23 (18) |
| C7—C6—C8 | 122.18 (14) | O5—C15—C14 | 118.82 (16) |
| C5—C6—C8 | 118.11 (14) | C16—C15—C14 | 121.95 (15) |
| C6—C7—C2 | 119.65 (15) | C15—C16—H16A | 109.5 |
| C6—C7—H7 | 120.2 | C15—C16—H16B | 109.5 |
| C2—C7—H7 | 120.2 | H16A—C16—H16B | 109.5 |
| O3—C8—O4 | 122.75 (15) | C15—C16—H16C | 109.5 |
| O3—C8—C6 | 125.75 (15) | H16A—C16—H16C | 109.5 |
| O4—C8—C6 | 111.49 (13) | H16B—C16—H16C | 109.5 |
| C10—C9—C14 | 121.90 (14) | ||
| O2—C1—C2—C3 | −179.32 (18) | C5—C6—C8—O4 | −176.17 (15) |
| O1—C1—C2—C3 | 1.4 (3) | C8—O4—C9—C10 | −74.90 (19) |
| O2—C1—C2—C7 | 1.2 (3) | C8—O4—C9—C14 | 110.95 (17) |
| O1—C1—C2—C7 | −178.08 (17) | C14—C9—C10—C11 | 0.3 (3) |
| C7—C2—C3—C4 | 0.4 (3) | O4—C9—C10—C11 | −173.72 (15) |
| C1—C2—C3—C4 | −179.14 (17) | C9—C10—C11—C12 | −0.8 (3) |
| C2—C3—C4—C5 | −0.6 (3) | C10—C11—C12—C13 | 0.5 (3) |
| C3—C4—C5—C6 | 0.4 (3) | C11—C12—C13—C14 | 0.2 (3) |
| C4—C5—C6—C7 | 0.0 (3) | C10—C9—C14—C13 | 0.4 (2) |
| C4—C5—C6—C8 | 179.31 (17) | O4—C9—C14—C13 | 174.31 (13) |
| C5—C6—C7—C2 | −0.3 (3) | C10—C9—C14—C15 | −179.79 (16) |
| C8—C6—C7—C2 | −179.53 (15) | O4—C9—C14—C15 | −5.9 (2) |
| C3—C2—C7—C6 | 0.1 (2) | C12—C13—C14—C9 | −0.7 (2) |
| C1—C2—C7—C6 | 179.60 (15) | C12—C13—C14—C15 | 179.48 (16) |
| C9—O4—C8—O3 | −5.1 (3) | C9—C14—C15—O5 | 170.50 (17) |
| C9—O4—C8—C6 | 175.91 (14) | C13—C14—C15—O5 | −9.7 (2) |
| C7—C6—C8—O3 | −175.87 (18) | C9—C14—C15—C16 | −9.6 (3) |
| C5—C6—C8—O3 | 4.9 (3) | C13—C14—C15—C16 | 170.19 (17) |
| C7—C6—C8—O4 | 3.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.82 | 1.84 | 2.6623 (18) | 175 |
| C4—H4···O5ii | 0.93 | 2.58 | 3.257 (3) | 130 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) x−1/2, −y+1/2, z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: HB7293).
References
- Bruker (2005). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Derissen, J. L. (1974). Acta Cryst. B30, 2764–2765.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tanimoto, Y., Kobayashi, H., Nagakura, S. & Saito, Y. (1973). Acta Cryst. B29, 1822–1826.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814021904/hb7293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814021904/hb7293Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814021904/hb7293Isup3.cml
. DOI: 10.1107/S1600536814021904/hb7293fig1.tif
View of the title compound with displacement ellipsoids drawn at the 50% probability level.
PLATON . DOI: 10.1107/S1600536814021904/hb7293fig2.tif
The partial packing (PLATON; Spek, 2009), which shows that molecules form dimers which are interlinked.
CCDC reference: 1027627
Additional supporting information: crystallographic information; 3D view; checkCIF report


