In the title compound, the mean plane of the cyclohexane ring is almost normal to the benzene ring and to the mean plane of the 1,2,4-thiadiazinane ring. In the crystal, molecules are linked by N—H⋯O hydrogen bonds, forming chains along [10 ], which are in turn linked via C—H⋯π interactions, forming sheets parallel to (010).
], which are in turn linked via C—H⋯π interactions, forming sheets parallel to (010).
Keywords: crystal structure, benzothiadiazine, hydrogen bonding, chirality
Abstract
In the title compound, C14H19BrN2O2S, the 1,2,4-thiadiazinane ring adopts an envelope conformation with the N atom (attached to the sulfonyl group) as the flap, while the cyclohexane ring adopts a chair conformation. The mean plane of the cyclohexane ring is almost normal to the benzene ring and the mean plane of the 1,2,4-thiadiazinane ring, making dihedral angles of 70.4 (2) and 71.43 (19)°, respectively. Furthermore, the dihedral angle between the benzene ring and the mean plane of the 1,2,4-thiadiazinane ring is 4.91 (18)°. The molecular structure is stabilized by an intramolecular C—H⋯O hydrogen bond, which encloses an S(6) ring motif. In the crystal, molecules are linked by N—H⋯O hydrogen bonds into chains along [10-1], forming a C(6) graph-set motif. These chains are interconnected via C—H⋯π interactions, leading to chains along [-101], so finally forming sheets parallel to (010).
Chemical context
The sulfonamide class of drugs have been widely reported for their antibacterial and antifungal activities (Trujillo et al., 2009 ▶). 1,2,4-Benzothiadiazine 1,1-dioxides are used as antihypertensive, diuretic, antidiabetic, glutaminergic neuro modulators (Cordi et al., 1996 ▶) and K-channel inhibitors (Di Bella et al., 1983 ▶). Furthermore, benzothiadiazine-3-one 1,1-dioxide and its derivatives have been reported for their potential hypoglycemic (Scozzafava et al., 2003 ▶), anticancer and anti-HIV activities (Casini et al., 2002 ▶), and they have also been reported to serve as selective antagonists of CXR2 (Hayao et al., 1968 ▶). In addition, 2-substituted-2H-1,2,4-benzothiadiazine-3(4H)one 1,1-dioxides have been found to exhibit varying degrees of sedative and hypotensive activities (Khelili et al., 2012 ▶). A number of benzothiadiazine 1,1-dioxide derivatives have recently been reported to display numerous biological activities (Tullio et al., 2011 ▶). 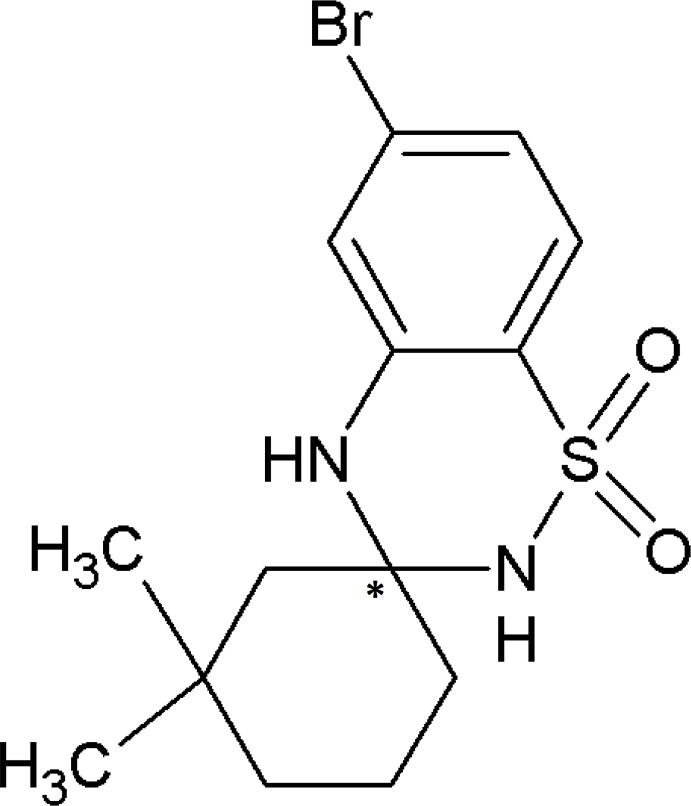
In view of their broad spectrum of biological activities, and in a continuation of our work on this class of compound, we have synthesized the title compound, (1), and report herein on its spectroscopic analysis and crystal structure.
Structural commentary
The molecular structure of the title molecule is shown in Fig. 1 ▶. The relative configuration of the asymmetric center is R for atom C7. The cyclohexane ring (C7–C12) adopts a chair conformation, confirmed by the puckering amplitude of Q = 0.4285 Å. The 1,2,4-thiadiazinane ring (N1/S1/C4/C3/N2/C7) adopts an envelope conformation with the flap atom N1 deviating by 0.565 (3) Å from the mean plane defined by atoms C7/N2/C3/C4/S1 [maximum deviation = 0.033 (1) Å for atom S1]. The mean plane of the cyclohexane ring is almost normal to the benzene ring (C1–C6) and the mean plane of the 1,2,4-thiadiazinane ring, making dihedral angles of 70.4 (2) and 71.43 (19)°, respectively. The dihedral angle between the benzene ring and the mean plane of the 1,2,4-thiadiazinane ring is 4.91 (18)°. The molecular structure is stabilized by an intramolecular C—H⋯O hydrogen bond, which forms an S(6) ring motif (Table 1 ▶ and Fig. 1 ▶).
Figure 1.
A view of the molecular structure of the title molecule, showing the atom labelling. Displacement ellipsoids are drawn at the 50% probability level. The C—H⋯O hydrogen bond is shown as a dashed line (see Table 1 ▶ for details).
Table 1. Hydrogen-bond geometry (, ).
Cg is the centroid of the C1C6 ring.
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C12H12AO1 | 0.97 | 2.40 | 3.066(5) | 126 |
| N2HN2O1i | 0.86 | 2.26 | 3.101(5) | 166 |
| C11H11A Cg ii | 0.97 | 2.58 | 3.506(5) | 160 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Supramolecular features
In the crystal, molecules are linked by N—H⋯O hydrogen bonds (Table 1 ▶ and Fig. 2 ▶), forming chains with a C(6) graph-set motif along [10 ]. The chains are linked via structure-directing C—H⋯π interactions, leading to the formation of C(6) chains along [
]. The chains are linked via structure-directing C—H⋯π interactions, leading to the formation of C(6) chains along [ 01]. These interactions lead to the formation of sheets parallel to (010) (Table 1 ▶ and Fig. 2 ▶).
01]. These interactions lead to the formation of sheets parallel to (010) (Table 1 ▶ and Fig. 2 ▶).
Figure 2.
A view along the a axis of the crystal packing of the title compound. Hydrogen bonds are shown as thin blue lines (see Table 1 ▶ for details; H atoms not involved in hydrogen bonding have been omitted for clarity).
Database survey
In two similar structures, namely 6-bromo-4H-spiro[1,2,4-benzothiadiazine-3,1′-cyclobutane] 1,1-dioxide, (2) (Shinoj Kumar, 2014a ▶, and 6-bromo-1′-ethyl-4H-spiro[1,2,4-benzothiadiazine-3,4′-piperidine] 1,1-dioxide, (3) (Shinoj Kumar, 2014b ▶, the 1,2,4-thiadiazinane rings adopt a twisted chair conformation, in contrast to the envelope conformation observed in (1). In (2), the dihedral angle between the benzene ring and the mean plane of the cyclobutyl ring is 73.76 (1)°, while that between the benzene ring and the mean plane of the 1,2,4-thiadiazinane ring is 4.72 (1)°, and that between the mean plane of the cyclobutyl ring and the mean plane of the 1,2,4-thiadiazinane ring is 78.44 (1)°. In (3), the same dihedral angles are similar, being 73.61 (1), 6.73 (1) and 73.81 (1)°, respectively. These angles are also similar to those observed in the title compound, (1).
Synthesis and crystallization
To a cooled solution of 2-amino-4-bromobenzene sulfonamide (5 g, 19.9 mmol) and anhydrous magnesium sulfate (MgSO4; 3.5 g, 29.88 mmol) in dry toluene (60 ml), 3,3-dimethylcyclohexanone (22 mmol) was added followed by slow addition of polyphosphoric acid anhydride (T3P; 19 ml, 29.88 mmol, 50% solution in ethyl acetate). The reaction mixture was then refluxed in a sealed tube at 393 K for 6 h. It was cooled to 283 K and neutralized with saturated sodium bicarbonate solution (100 ml). The crude product was extracted with ethyl acetate (100 ml) and was finally washed with brine solution (50 ml). The organic phase was dried over anhydrous sodium sulfate and concentrated to give the crude product as a brown solid. It was then dissolved in a minimum amount of ethyl acetate (25 ml) and stirred for 1h in an ice-cooled bath, filtered and washed with cold ethyl acetate (10 ml × 2) to give pure compound (1) (4.5 g, 75% yield) as a white solid. Colourless prisms of the title compound were obtained by slow evaporation of a solution of the compound in ethyl acetate.
Spectroscopic characterization
The IR spectra of the title compound exhibits strong bands at 1374 cm−1 due to asymmetric (S=O) stretching and a band at 1165 cm−1 due to symmetric (S=O) stretching. Further, a single band appearing at 3110 cm−1 is due to the secondary N—H group of the sulfonamide, and a band at 3308 cm−1 confirms the cyclization of sulfonamide through condensation with the ketone. Appearance of bands in the range of 2970–2815 cm−1 is assigned to the C—H stretching is due to the presence of the saturated hydrocarbons. The 1H NMR spectrum shows peaks at δ 7.53 (s, 1H, SO2NH), 6.934–6.930 (d, 1H, Ar-H), 7.37–7.35 (d, 1H, Ar-H), 3.33 (s, 1H, NH), 2.51–1.28 (m, 9H, CH2), 0.9–1.1 (s, 6H, CH3). The 13C NMR spectrum shows peaks at δ 144 (C1), 119 (C2), 126 (C3), 127 (C4), 119 (C5), 118 (C6), 117 (C7), 71 (C8), 47 (C9), 36 (C10), 33 (C11), 31 (C12), 18 (C13 and C14). The LC–MS spectrum shows the appearance of molecular ion peaks at m/z 358 and 357 values, confirming the structure of the compound.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▶. The NH hydrogens were located in a difference Fourier map. N- and C-bound H atoms were included in calculated positions (N—H = 0.86 and C—H = 0.93–0.97 Å) and allowed to ride on their parent atoms, with U iso(H) = 1.5U eq(C) for methyl H atoms and 1.2U eq(N,C) for other H atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C14H19BrN2O2S |
| M r | 359.28 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 293 |
| a, b, c () | 6.4316(7), 24.263(3), 10.0829(13) |
| () | 105.604(9) |
| V (3) | 1515.5(3) |
| Z | 4 |
| Radiation type | Cu K |
| (mm1) | 5.01 |
| Crystal size (mm) | 0.44 0.24 0.19 |
| Data collection | |
| Diffractometer | Bruker APEXII |
| Absorption correction | Multi-scan (SADABS; Bruker, 2009 ▶) |
| T min, T max | 0.271, 0.386 |
| No. of measured, independent and observed [I > 2(I)] reflections | 11574, 2515, 1860 |
| R int | 0.081 |
| (sin /)max (1) | 0.586 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.049, 0.154, 0.94 |
| No. of reflections | 2515 |
| No. of parameters | 183 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 0.61, 0.61 |
Supplementary Material
Crystal structure: contains datablock(s) 1, Global. DOI: 10.1107/S1600536814022417/su2797sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S1600536814022417/su27971sup2.hkl
CCDC reference: 1028895
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge the IOE X-ray diffractometer facility, University of Mysore, Mysore, for the data collection. PPSK, PAS, SS and DBAK are thankful to Tumkur University for providing the laboratory and instrumental facilities to carry out this work.
supplementary crystallographic information
Crystal data
| C14H19BrN2O2S | Dx = 1.575 Mg m−3 |
| Mr = 359.28 | Melting point: 418 K |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 6.4316 (7) Å | Cell parameters from 123 reflections |
| b = 24.263 (3) Å | θ = 7.1–64.6° |
| c = 10.0829 (13) Å | µ = 5.01 mm−1 |
| β = 105.604 (9)° | T = 293 K |
| V = 1515.5 (3) Å3 | Prism, colourless |
| Z = 4 | 0.44 × 0.24 × 0.19 mm |
| F(000) = 736 |
Data collection
| Bruker APEXII diffractometer | 1860 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.081 |
| Graphite monochromator | θmax = 64.6°, θmin = 7.1° |
| phi and φ scans | h = −7→6 |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | k = −28→27 |
| Tmin = 0.271, Tmax = 0.386 | l = −11→11 |
| 11574 measured reflections | 1 standard reflections every 1 reflections |
| 2515 independent reflections | intensity decay: 1% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.154 | H-atom parameters constrained |
| S = 0.94 | w = 1/[σ2(Fo2) + (0.1058P)2 + 0.1836P] where P = (Fo2 + 2Fc2)/3 |
| 2515 reflections | (Δ/σ)max < 0.001 |
| 183 parameters | Δρmax = 0.61 e Å−3 |
| 0 restraints | Δρmin = −0.61 e Å−3 |
Special details
| Experimental. Melting points were determined in open capillaries and are uncorrected. The molecular structures of the synthesized compounds were established using IR, 1H NMR, 13C NMR and LC-MS studies. Solid state FT-IR Spectra were recorded as KBr discs on Jasco FT-IR Spectrometer. 1H NMR and 13C NMR were recorded in DMSO at 399.13 MHz and 75.50 MHz respectively on Bruker model avance II. All the chemical shifts were reported in parts per million (ppm) using tetramethyl silane (TMS) as an internal standard. Mass spectra of the compounds were recordedon Shimadzu LC-2010EV with ESI probe. |
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.0489 (7) | 0.09465 (19) | 0.8681 (5) | 0.0449 (11) | |
| C2 | 0.0873 (7) | 0.14863 (18) | 0.8441 (4) | 0.0398 (10) | |
| H2 | 0.1873 | 0.1575 | 0.7959 | 0.048* | |
| C3 | −0.0244 (6) | 0.19093 (16) | 0.8922 (4) | 0.0336 (9) | |
| C4 | −0.1634 (6) | 0.17472 (17) | 0.9715 (4) | 0.0334 (9) | |
| C5 | −0.2014 (7) | 0.11957 (18) | 0.9908 (4) | 0.0419 (10) | |
| H5 | −0.2992 | 0.1101 | 1.0401 | 0.050* | |
| C6 | −0.0993 (8) | 0.07862 (19) | 0.9394 (5) | 0.0465 (11) | |
| H6 | −0.1275 | 0.0416 | 0.9514 | 0.056* | |
| C7 | −0.0697 (6) | 0.29279 (16) | 0.9203 (4) | 0.0327 (9) | |
| C8 | −0.1071 (7) | 0.33969 (17) | 0.8148 (4) | 0.0392 (10) | |
| H8A | −0.2288 | 0.3297 | 0.7384 | 0.047* | |
| H8B | 0.0186 | 0.3419 | 0.7792 | 0.047* | |
| C9 | −0.1502 (7) | 0.39760 (18) | 0.8637 (5) | 0.0444 (11) | |
| C10 | 0.0144 (8) | 0.41054 (18) | 1.0010 (5) | 0.0491 (11) | |
| H10A | −0.0274 | 0.4444 | 1.0382 | 0.059* | |
| H10B | 0.1547 | 0.4165 | 0.9848 | 0.059* | |
| C11 | 0.0326 (8) | 0.36432 (19) | 1.1071 (4) | 0.0464 (11) | |
| H11A | 0.1388 | 0.3744 | 1.1918 | 0.056* | |
| H11B | −0.1053 | 0.3595 | 1.1277 | 0.056* | |
| C12 | 0.0987 (6) | 0.31045 (18) | 1.0527 (4) | 0.0378 (10) | |
| H12A | 0.1127 | 0.2819 | 1.1220 | 0.045* | |
| H12B | 0.2379 | 0.3150 | 1.0339 | 0.045* | |
| C13 | −0.3793 (8) | 0.4038 (2) | 0.8792 (6) | 0.0579 (13) | |
| H13A | −0.3914 | 0.3838 | 0.9590 | 0.087* | |
| H13B | −0.4809 | 0.3893 | 0.7988 | 0.087* | |
| H13C | −0.4094 | 0.4420 | 0.8895 | 0.087* | |
| C14 | −0.1192 (10) | 0.4391 (2) | 0.7572 (6) | 0.0657 (15) | |
| H14A | −0.1452 | 0.4756 | 0.7856 | 0.098* | |
| H14B | −0.2187 | 0.4311 | 0.6697 | 0.098* | |
| H14C | 0.0259 | 0.4366 | 0.7492 | 0.098* | |
| N1 | −0.2810 (5) | 0.27879 (14) | 0.9457 (3) | 0.0342 (8) | |
| HN1 | −0.3930 | 0.2988 | 0.9117 | 0.041* | |
| N2 | 0.0041 (5) | 0.24419 (14) | 0.8593 (4) | 0.0373 (8) | |
| HN2 | 0.0712 | 0.2499 | 0.7974 | 0.045* | |
| O1 | −0.1804 (5) | 0.23557 (14) | 1.1797 (3) | 0.0442 (8) | |
| O2 | −0.5208 (4) | 0.21181 (13) | 1.0131 (3) | 0.0469 (8) | |
| S1 | −0.29684 (15) | 0.22511 (4) | 1.03879 (10) | 0.0352 (3) | |
| Br1 | 0.20573 (10) | 0.03945 (2) | 0.80348 (7) | 0.0701 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.043 (3) | 0.043 (3) | 0.047 (3) | −0.001 (2) | 0.010 (2) | −0.002 (2) |
| C2 | 0.041 (2) | 0.038 (2) | 0.044 (2) | −0.0026 (19) | 0.0185 (19) | −0.0035 (18) |
| C3 | 0.035 (2) | 0.031 (2) | 0.033 (2) | −0.0009 (17) | 0.0070 (16) | 0.0000 (16) |
| C4 | 0.032 (2) | 0.040 (2) | 0.030 (2) | −0.0002 (17) | 0.0097 (16) | 0.0006 (16) |
| C5 | 0.048 (3) | 0.045 (2) | 0.036 (2) | −0.002 (2) | 0.0182 (19) | 0.0046 (19) |
| C6 | 0.057 (3) | 0.037 (2) | 0.046 (3) | −0.001 (2) | 0.013 (2) | 0.008 (2) |
| C7 | 0.030 (2) | 0.034 (2) | 0.035 (2) | 0.0003 (17) | 0.0117 (16) | −0.0038 (16) |
| C8 | 0.046 (2) | 0.041 (3) | 0.032 (2) | −0.006 (2) | 0.0119 (18) | −0.0009 (18) |
| C9 | 0.052 (3) | 0.037 (2) | 0.044 (3) | −0.003 (2) | 0.013 (2) | 0.0022 (19) |
| C10 | 0.058 (3) | 0.037 (3) | 0.049 (3) | −0.004 (2) | 0.010 (2) | −0.010 (2) |
| C11 | 0.050 (3) | 0.048 (3) | 0.034 (2) | −0.002 (2) | 0.0005 (19) | −0.009 (2) |
| C12 | 0.030 (2) | 0.043 (2) | 0.037 (2) | −0.0027 (18) | 0.0044 (17) | 0.0009 (18) |
| C13 | 0.049 (3) | 0.055 (3) | 0.067 (3) | 0.014 (2) | 0.009 (2) | 0.001 (3) |
| C14 | 0.091 (4) | 0.052 (3) | 0.051 (3) | 0.000 (3) | 0.013 (3) | 0.011 (2) |
| N1 | 0.0267 (16) | 0.040 (2) | 0.0370 (19) | 0.0074 (14) | 0.0098 (14) | 0.0041 (15) |
| N2 | 0.0420 (19) | 0.0343 (19) | 0.044 (2) | −0.0006 (15) | 0.0265 (16) | −0.0026 (15) |
| O1 | 0.0478 (18) | 0.060 (2) | 0.0258 (15) | 0.0024 (15) | 0.0112 (13) | −0.0019 (13) |
| O2 | 0.0286 (15) | 0.058 (2) | 0.0562 (19) | −0.0040 (14) | 0.0153 (13) | 0.0033 (15) |
| S1 | 0.0322 (5) | 0.0435 (6) | 0.0322 (6) | 0.0004 (4) | 0.0126 (4) | 0.0007 (4) |
| Br1 | 0.0868 (5) | 0.0414 (4) | 0.0966 (6) | 0.0091 (3) | 0.0495 (4) | −0.0070 (3) |
Geometric parameters (Å, º)
| C1—C2 | 1.366 (6) | C9—C10 | 1.533 (6) |
| C1—C6 | 1.394 (6) | C10—C11 | 1.533 (6) |
| C1—Br1 | 1.894 (5) | C10—H10A | 0.9700 |
| C2—C3 | 1.412 (6) | C10—H10B | 0.9700 |
| C2—H2 | 0.9300 | C11—C12 | 1.522 (6) |
| C3—N2 | 1.359 (5) | C11—H11A | 0.9700 |
| C3—C4 | 1.407 (5) | C11—H11B | 0.9700 |
| C4—C5 | 1.383 (6) | C12—H12A | 0.9700 |
| C4—S1 | 1.733 (4) | C12—H12B | 0.9700 |
| C5—C6 | 1.368 (6) | C13—H13A | 0.9600 |
| C5—H5 | 0.9300 | C13—H13B | 0.9600 |
| C6—H6 | 0.9300 | C13—H13C | 0.9600 |
| C7—N2 | 1.466 (5) | C14—H14A | 0.9600 |
| C7—N1 | 1.489 (5) | C14—H14B | 0.9600 |
| C7—C8 | 1.532 (6) | C14—H14C | 0.9600 |
| C7—C12 | 1.537 (5) | N1—S1 | 1.624 (3) |
| C8—C9 | 1.539 (6) | N1—HN1 | 0.8600 |
| C8—H8A | 0.9700 | N2—HN2 | 0.8600 |
| C8—H8B | 0.9700 | O1—S1 | 1.439 (3) |
| C9—C13 | 1.530 (6) | O2—S1 | 1.430 (3) |
| C9—C14 | 1.523 (6) | ||
| C2—C1—C6 | 122.7 (4) | C9—C10—H10B | 109.1 |
| C2—C1—Br1 | 118.6 (3) | C11—C10—H10B | 109.1 |
| C6—C1—Br1 | 118.8 (4) | H10A—C10—H10B | 107.8 |
| C1—C2—C3 | 120.2 (4) | C12—C11—C10 | 110.7 (4) |
| C1—C2—H2 | 119.9 | C12—C11—H11A | 109.5 |
| C3—C2—H2 | 119.9 | C10—C11—H11A | 109.5 |
| N2—C3—C2 | 119.5 (4) | C12—C11—H11B | 109.5 |
| N2—C3—C4 | 123.6 (4) | C10—C11—H11B | 109.5 |
| C2—C3—C4 | 116.9 (4) | H11A—C11—H11B | 108.1 |
| C5—C4—C3 | 120.9 (4) | C11—C12—C7 | 110.7 (3) |
| C5—C4—S1 | 120.2 (3) | C11—C12—H12A | 109.5 |
| C3—C4—S1 | 118.8 (3) | C7—C12—H12A | 109.5 |
| C6—C5—C4 | 121.9 (4) | C11—C12—H12B | 109.5 |
| C6—C5—H5 | 119.0 | C7—C12—H12B | 109.5 |
| C4—C5—H5 | 119.0 | H12A—C12—H12B | 108.1 |
| C5—C6—C1 | 117.2 (4) | C9—C13—H13A | 109.5 |
| C5—C6—H6 | 121.4 | C9—C13—H13B | 109.5 |
| C1—C6—H6 | 121.4 | H13A—C13—H13B | 109.5 |
| N2—C7—N1 | 107.6 (3) | C9—C13—H13C | 109.5 |
| N2—C7—C8 | 108.3 (3) | H13A—C13—H13C | 109.5 |
| N1—C7—C8 | 108.0 (3) | H13B—C13—H13C | 109.5 |
| N2—C7—C12 | 110.9 (3) | C9—C14—H14A | 109.5 |
| N1—C7—C12 | 112.1 (3) | C9—C14—H14B | 109.5 |
| C8—C7—C12 | 109.8 (3) | H14A—C14—H14B | 109.5 |
| C7—C8—C9 | 117.6 (3) | C9—C14—H14C | 109.5 |
| C7—C8—H8A | 107.9 | H14A—C14—H14C | 109.5 |
| C9—C8—H8A | 107.9 | H14B—C14—H14C | 109.5 |
| C7—C8—H8B | 107.9 | C7—N1—S1 | 119.0 (3) |
| C9—C8—H8B | 107.9 | C7—N1—HN1 | 120.5 |
| H8A—C8—H8B | 107.2 | S1—N1—HN1 | 120.5 |
| C13—C9—C14 | 108.7 (4) | C3—N2—C7 | 125.6 (3) |
| C13—C9—C10 | 109.8 (4) | C3—N2—HN2 | 117.2 |
| C14—C9—C10 | 108.2 (4) | C7—N2—HN2 | 117.2 |
| C13—C9—C8 | 112.6 (4) | O2—S1—O1 | 116.72 (18) |
| C14—C9—C8 | 107.9 (4) | O2—S1—N1 | 107.04 (18) |
| C10—C9—C8 | 109.6 (4) | O1—S1—N1 | 109.40 (19) |
| C9—C10—C11 | 112.7 (4) | O2—S1—C4 | 110.51 (19) |
| C9—C10—H10A | 109.1 | O1—S1—C4 | 109.26 (18) |
| C11—C10—H10A | 109.1 | N1—S1—C4 | 103.01 (17) |
| C6—C1—C2—C3 | 0.2 (7) | C9—C10—C11—C12 | 58.9 (5) |
| Br1—C1—C2—C3 | −179.2 (3) | C10—C11—C12—C7 | −60.5 (5) |
| C1—C2—C3—N2 | −175.4 (4) | N2—C7—C12—C11 | 173.9 (3) |
| C1—C2—C3—C4 | 3.8 (6) | N1—C7—C12—C11 | −65.8 (4) |
| N2—C3—C4—C5 | 173.8 (4) | C8—C7—C12—C11 | 54.3 (4) |
| C2—C3—C4—C5 | −5.3 (6) | N2—C7—N1—S1 | 55.6 (4) |
| N2—C3—C4—S1 | −3.3 (5) | C8—C7—N1—S1 | 172.3 (3) |
| C2—C3—C4—S1 | 177.6 (3) | C12—C7—N1—S1 | −66.6 (4) |
| C3—C4—C5—C6 | 2.9 (6) | C2—C3—N2—C7 | −168.8 (4) |
| S1—C4—C5—C6 | −180.0 (3) | C4—C3—N2—C7 | 12.1 (6) |
| C4—C5—C6—C1 | 1.1 (7) | N1—C7—N2—C3 | −36.3 (5) |
| C2—C1—C6—C5 | −2.7 (7) | C8—C7—N2—C3 | −152.8 (4) |
| Br1—C1—C6—C5 | 176.7 (3) | C12—C7—N2—C3 | 86.6 (5) |
| N2—C7—C8—C9 | −170.4 (3) | C7—N1—S1—O2 | −163.1 (3) |
| N1—C7—C8—C9 | 73.3 (4) | C7—N1—S1—O1 | 69.6 (3) |
| C12—C7—C8—C9 | −49.2 (5) | C7—N1—S1—C4 | −46.5 (3) |
| C7—C8—C9—C13 | −75.8 (5) | C5—C4—S1—O2 | −44.6 (4) |
| C7—C8—C9—C14 | 164.3 (4) | C3—C4—S1—O2 | 132.5 (3) |
| C7—C8—C9—C10 | 46.7 (5) | C5—C4—S1—O1 | 85.1 (4) |
| C13—C9—C10—C11 | 74.3 (5) | C3—C4—S1—O1 | −97.7 (3) |
| C14—C9—C10—C11 | −167.3 (4) | C5—C4—S1—N1 | −158.6 (3) |
| C8—C9—C10—C11 | −49.9 (5) | C3—C4—S1—N1 | 18.5 (3) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C1–C6 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12A···O1 | 0.97 | 2.40 | 3.066 (5) | 126 |
| N2—HN2···O1i | 0.86 | 2.26 | 3.101 (5) | 166 |
| C11—H11A···Cgii | 0.97 | 2.58 | 3.506 (5) | 160 |
Symmetry codes: (i) x+1/2, −y+1/2, z−1/2; (ii) x+1/2, −y+1/2, z+1/2.
References
- Bruker (2009). APEX2, SADABS, SAINT-Plus and XPREP. Bruker AXS Inc., Madison, Wisconsin, USA.
- Casini, A., Scozzafava, A., Mastrolorenzo, A. & Supuran, C. (2002). Curr. Cancer Drug Targets, 2, 55–75. [DOI] [PubMed]
- Cordi, A., Spedding, M., Serkiz, B., Lepagnol, J., Desos, P. & Morain, P. (1996). Chem. Abstr. 124, 261085.
- Di Bella, M., Monzani, A., Andrisano, M. G., Fabio, U. & Quaglio, G. P. (1983). Farmaco, 38, 466–472.
- Hayao, S., Strycker, W. G., Phillips, B. & Fujimori, H. (1968). J. Med. Chem. 11, 1246–1248. [DOI] [PubMed]
- Khelili, S., Kihal, N., Yekhlef, M., de Tullio, P., Lebrun, P. & Pirotte, B. (2012). Eur. J. Med. Chem. 54, 873–878. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Scozzafava, A., Owa, T., Mastrolorenzo, A. & Supuran, C. T. (2003). Curr. Med. Chem. 10, 925–953. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shinoj Kumar, P. P., Suchetan, P. A., Sreenivasa, S., Naveen, S., Lokanath, N. K. & Aruna Kumar, D. B. (2014a). Private communication (deposition number 1023519). CCDC, Cambridge, England.
- Shinoj Kumar, P. P., Suchetan, P. A., Sreenivasa, S., Naveen, S., Lokanath, N. K. & Aruna Kumar, D. B. (2014b). Private communication (deposition number 1023520). CCDC, Cambridge, England.
- Trujillo, J. I., Kiefer, J. R., Huang, W., Thorarensen, A., Xing, L., Caspers, N. L., Day, J. E., Mathis, K. J., Kretzmer, K. K., Reitz, B. A., Weinberg, R. A., Stegeman, R. A., Wrightstone, A., Christine, L., Compton, R. & Li, X. (2009). Bioorg. Med. Chem. Lett. 19, 908–911. [DOI] [PubMed]
- Tullio, P. de, Servais, A.-C., Fillet, M., Gillotin, F., Somers, F., Chiap, P., Lebrun, P. & Pirotte, B. (2011). J. Med. Chem. 54, 8353–8361. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, Global. DOI: 10.1107/S1600536814022417/su2797sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S1600536814022417/su27971sup2.hkl
CCDC reference: 1028895
Additional supporting information: crystallographic information; 3D view; checkCIF report




