The molecular structure of a non-symmetric structure based on a tetrasubstituted ethylenediamine backbone consists of three hydroxybenzyl groups and one 2-hydroxy-5-methylbenzaldehyde group bonded to the N atoms of the diamine unit. The ethylenediamine skeleton shows a regular extended conformation, while the phenol arms are randomly oriented but governed by hydrogen bonds.
Keywords: crystal structure, non-symmetrical compound, tetrasubstituted ethylenediamine, phenol-arm substituents
Abstract
The non-symmetric title molecule, C32H34N2O5, is based on a tetrasubstituted ethylenediamine backbone. The molecular structure consists of three hydroxybenzyl groups and one 2-hydroxy-5-methylbenzaldehyde group bonded to the N atoms of the diamine unit. The ethylenediamine skeleton shows a regular extended conformation, while the spatial orientation of the phenol arms is governed by hydrogen bonds. In the 2-hydroxy-5-methylbenzaldehyde group, an intramolecular S(6) O—H⋯O hydrogen bond is observed between the alcohol and aldehyde functions, and the neighbouring phenol arm participates in an intramolecular S(6) O—H⋯N hydrogen bond. The third phenol group is involved in a bifurcated intramolecular hydrogen bond with graph-set notation S(6) for O—H⋯N and O—H⋯O intramolecular hydrogen bonds between neighbouring amine and phenol arms, respectively. Finally, the fourth phenol group acts as an acceptor in a bifurcated intramolecular hydrogen bond and also acts as donor in an intermolecular hydrogen bond, which connects inversion-related molecules into dimers with R 4 4(8) ring motifs.
Chemical context
The preparation of non-symmetric compounds has always been of interest in organic synthesis, as well as in coordination chemistry. Compounds containing tetrasubstituted ethylenediamine groups have attracted significant interest because of their coordination versatility towards metal ions, their easy preparation and their biological activity (Musa et al., 2014 ▶). With respect to medical applications, high in vitro cytotoxic activity of free ethylenediamine-type compounds against different types of cancer cells, such as HL-60 leukemic and B16 human melanoma cells lines, has been reported (Dencic et al., 2012 ▶; Lazić et al., 2010 ▶). In addition, metal complexes containing substituted ethylenediamine have also found valuable applications in pharmacological research as potential anticancer agents (Ansari et al., 2009 ▶), radiopharmaceuticals for tumor imaging (Boros et al., 2011 ▶; Price et al., 2012 ▶) and artificial nucleases (Raman et al., 2011 ▶). In this paper, we report the synthesis and crystal structure of the non-symmetric molecule 3-[({2-[bis(2-hydroxybenzyl)amino]ethyl}(2-hydroxybenzyl)amino)methyl]-2-hydroxy-5-methylbenzaldehyde, (I), which is a potential hexadentate ligand with an N2O4-donor set which could stabilize complexes containing high-oxidation-state metal ions, such as TcIII, GaIII and InIII ions, that are widely used in radiopharmaceuticals for diagnostic imaging and related research. 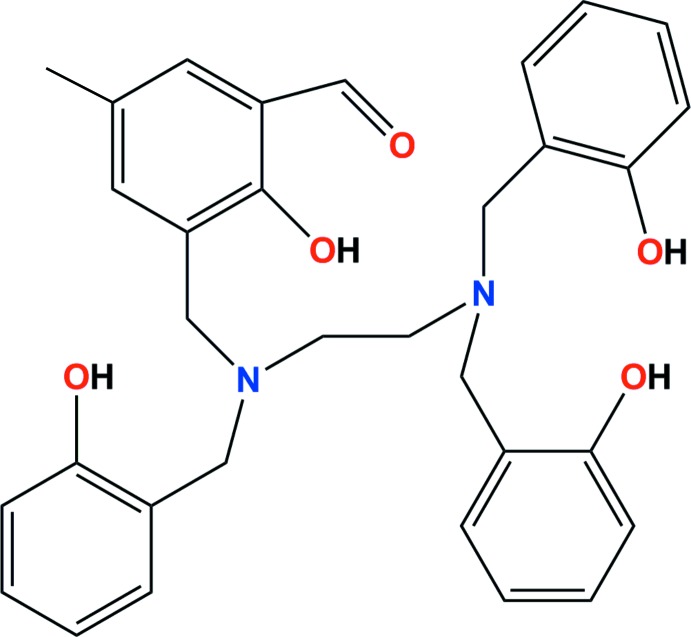
Structural commentary
Compound (I) is a non-symmetric molecule based on a tetrasubstituted ethylenediamine backbone (Fig. 1 ▶). The structure consists of three hydroxybenzyl groups and one 2-hydroxy-5-methylbenzaldehyde group bonded to nitrogen atoms of the diamine unit. The ethylenediamine skeleton shows a regular extended ‘zigzag’ conformation [with an N1—C2—C3—N4 torsion angle of 174.78 (13)°], while the pendant phenol arms are randomly oriented but governed by hydrogen bonds (Table 1 ▶). Three intramolecular hydrogen bonds with an S(6) graph-set motif are observed in the molecular structure of (I) (Fig. 2 ▶). One of these occurs between the neighbouring alcohol and aldehyde groups. In addition, intramolecular O—H⋯N and O—H⋯O interactions, which include bifurcated hydrogen bonds, are observed, involving O—H functions as donors and the amine sites and one phenolic oxygen atom as acceptors. All bond lengths and angles found for (I) are in the expected range for organic compounds (Bruno et al., 2004 ▶).
Figure 1.
The molecular structure of (I), with displacement ellipsoids drawn at the 40% probability level.
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| O10H10O20i | 0.93 | 1.80 | 2.7230(16) | 177 |
| O20H20N1 | 0.94 | 1.75 | 2.5928(17) | 148 |
| O20H20O10 | 0.94 | 2.43 | 3.0362(19) | 122 |
| O30H30N4 | 0.93 | 1.94 | 2.784(2) | 149 |
| O40H40O41 | 0.93 | 1.76 | 2.6146(19) | 151 |
Symmetry code: (i)  .
.
Figure 2.
The intramolecular hydrogen bonds (dashed lines) observed in (I).
Supramolecular features
In the crystal of (I), inversion dimers with  (8) ring motifs are formed by pairs of O—H⋯O hydrogen bonds (Fig. 3 ▶, Table 1 ▶). The approximate planes of the ring motifs of the dimers are arranged as stacks along [010] (Fig. 4 ▶). No π–π stacking interactions are observed.
(8) ring motifs are formed by pairs of O—H⋯O hydrogen bonds (Fig. 3 ▶, Table 1 ▶). The approximate planes of the ring motifs of the dimers are arranged as stacks along [010] (Fig. 4 ▶). No π–π stacking interactions are observed.
Figure 3.
An inversion dimer of (I) formed by intermolecular O—H⋯O hydrogen bonds (dashed lines). [Symmetry code: (′) −x + 1, −y, −z.]
Figure 4.

Partial packing of (I), showing dimers stacked along [010].
Database survey
A search for similar structures in the current version of the Cambridge Structural Database (Version 5.35, November 2013; Groom & Allen, 2014 ▶) resulted in four entries but only three different structures: (i) HUNDIE (CCDC 727272) and HUNDOK (CCDC 727273) (Boyle et al., 2009 ▶); (ii) USODUC (CCDC 809654) (Wang et al., 2011a ▶) and (iii) USODUC01 (CCDC 809654) (Wang et al., 2011b ▶). All of these structures are symmetric molecules and the phenol groups have an additional one or two substituents in the para and ortho positions with respect to the O–H function. As observed in (I), the spatial orientations of the phenol arms are influenced by intra- and intermolecular hydrogen bonding. There are no significant differences in the geometrical parameters; however, the crystal packing shows distinguishable three-dimensional arrangements due to differences in molecular symmetry and intermolecular interactions.
Synthesis and crystallization
The title compound was obtained from a nucleophilic substitution reaction between N,N,N′-tris(2-hydroxybenzyl)-1,2-diaminoethane (Schmitt et al., 2002 ▶) and chloromethyl-4-methyl-6-formylphenol. These precursors were prepared following the methodologies already described in the literature (Schmitt et al., 2002 ▶; Thoer et al., 1988 ▶). A solution of 2-chloromethyl-4-methyl-6-formylphenol (1.19 g, 6.6 mmol) in tetrahydrofuran (40 ml) was added slowly to a cooled solution of N,N,N′-tris(2-hydroxybenzyl)-1,2-diaminoethane (2.50 g, 6.6 mmol) in tetrahydrofuran (40 ml) containing triethylamine (0.96 ml, 6.6 mmol). The reaction was kept cooled during addition time, and the resulting solution stirred for 24 h. Yellow mixture oil/solid was obtained after evaporation of the solvent. A solution of this mixture in CH2Cl2 (50 ml) was washed with a saturated solution of NaHCO3 (3 × 50 ml) and filtered off in the presence of NaSO4. The solvent was removed, and a straw-yellow solid was obtained. This solid was refluxed in n-hexane/CHCl3 (1:1, 100 ml). After cooling the solid was filtered off, washed with n-hexane (80 ml), dried and recrystallized from an ethyl acetate solution to afford 3-[({2-[bis(2-hydroxybenzyl)amino]ethyl}(2-hydroxybenzyl)amino)methyl]-2-hydroxy-5-methylbenzaldehyde, (I).
The formation of (I) was indicated by the presence of the band at 1655 cm−1 in the IR spectrum, which is typical for stretching vibrations ν(C=O) of free aldehyde. In the 1H NMR spectrum, the signal at 9.81 p.p.m. related to one aldehyde proton is further evidence for product formation. Yield 90%, m.p. 444.8–445.4 K. IR (KBr, cm−1): ν(O—H) 3273, ν(C—Har and C—Halif) 3042–2718, ν(C=O)1655, ν(C=C) 1615–1457, δ(O—H) 1365, δ(C—O) 1252, δ(C—Har) 757; 1H NMR (400 MHz, CDCl3) (δ, p.p.m.): 2.29 (s, 3H, CH3), 2.78 (s, 4H, CH2-en), 3.58 (s, 2H, CH2), 3.61–3.77 (m, 6 H, CH2), 6.69–6.87 (m, 6H, CHar), 6.91 (d, 2H, CHar), 6.99 (d, 2 H, CHar), 7.07–7.19 (m, 2H, CHar), 7.24 (d, 2H, CHar), 9.81 (s, 1H, CHald); 13C NMR (400 MHz, DMSO-d 6, δ p.p.m.): 20.0, 48.6, 48.8, 53.5, 54.3, 115.2, 121.7, 122.7, 123.2, 124.5, 127.6, 128.4, 128.7, 129.8, 130.8, 136.7, 156.2, 156.5, 158.7, 191.6. Negative HPLC/ESI–MS (m/z): [M−H] calculated for C32H35N2O5 −, 527.25; found, 527.19. Colourless blocks were grown by slow evaporation of the solvent from a saturated solution of (I) in ethyl acetate.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 2 ▶. H atoms were placed in idealized positions with distances of 0.95 (CHAr), 0.99 (CH2) or 0.98 Å (CH3) with U iso = 1.2U eq(C) or 1.5U eq(Cmethyl). The hydrogen atoms of the alcohol groups were located from a Fourier difference map and treated with a riding-model approximation with U iso(H) = 1.5U eq(O).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C32H34N2O5 |
| M r | 526.61 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 190 |
| a, b, c () | 10.1635(5), 11.0440(6), 13.5439(7) |
| , , () | 113.549(2), 98.381(2), 99.451(3) |
| V (3) | 1336.64(12) |
| Z | 2 |
| Radiation type | Mo K |
| (mm1) | 0.09 |
| Crystal size (mm) | 0.15 0.08 0.04 |
| Data collection | |
| Diffractometer | Bruker APEXII DUO |
| No. of measured, independent and observed [I > 2(I)] reflections | 17177, 8122, 5175 |
| R int | 0.031 |
| (sin /)max (1) | 0.715 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.057, 0.166, 1.02 |
| No. of reflections | 8122 |
| No. of parameters | 353 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 0.42, 0.25 |
Supplementary Material
Crystal structure: contains datablock(s) general, I. DOI: 10.1107/S1600536814024465/lh5739sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814024465/lh5739Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814024465/lh5739Isup3.mol
Supporting information file. DOI: 10.1107/S1600536814024465/lh5739Isup4.cml
CCDC reference: 1033129
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Financiadora de Estudos e Projetos (FINEP) for support.
supplementary crystallographic information
Crystal data
| C32H34N2O5 | Z = 2 |
| Mr = 526.61 | F(000) = 560 |
| Triclinic, P1 | Dx = 1.308 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.1635 (5) Å | Cell parameters from 4159 reflections |
| b = 11.0440 (6) Å | θ = 2.4–30.3° |
| c = 13.5439 (7) Å | µ = 0.09 mm−1 |
| α = 113.549 (2)° | T = 190 K |
| β = 98.381 (2)° | Prismatic, colourless |
| γ = 99.451 (3)° | 0.15 × 0.08 × 0.04 mm |
| V = 1336.64 (12) Å3 |
Data collection
| Bruker APEXII DUO diffractometer | 5175 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.031 |
| Graphite monochromator | θmax = 30.6°, θmin = 1.7° |
| φ and ω scans | h = −13→14 |
| 17177 measured reflections | k = −15→15 |
| 8122 independent reflections | l = −9→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.166 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0806P)2 + 0.2575P] where P = (Fo2 + 2Fc2)/3 |
| 8122 reflections | (Δ/σ)max < 0.001 |
| 353 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.55546 (13) | 0.19576 (13) | 0.30195 (10) | 0.0231 (3) | |
| C2 | 0.46867 (17) | 0.29324 (17) | 0.32893 (12) | 0.0268 (3) | |
| H2A | 0.5193 | 0.3775 | 0.3954 | 0.032* | |
| H2B | 0.3853 | 0.2536 | 0.3460 | 0.032* | |
| C3 | 0.42744 (17) | 0.32788 (16) | 0.23213 (12) | 0.0258 (3) | |
| H3A | 0.5117 | 0.3735 | 0.2200 | 0.031* | |
| H3B | 0.3865 | 0.2416 | 0.1646 | 0.031* | |
| N4 | 0.33030 (13) | 0.41522 (13) | 0.24463 (11) | 0.0258 (3) | |
| C10 | 0.69979 (16) | 0.26500 (17) | 0.31867 (13) | 0.0280 (3) | |
| H10A | 0.7441 | 0.3071 | 0.3987 | 0.034* | |
| H10B | 0.7017 | 0.3390 | 0.2948 | 0.034* | |
| C11 | 0.78117 (16) | 0.17145 (16) | 0.25636 (13) | 0.0263 (3) | |
| C12 | 0.74930 (16) | 0.11211 (18) | 0.14067 (13) | 0.0288 (3) | |
| C13 | 0.82640 (18) | 0.02915 (19) | 0.08148 (15) | 0.0350 (4) | |
| H13 | 0.8023 | −0.0122 | 0.0028 | 0.042* | |
| C14 | 0.93856 (18) | 0.0067 (2) | 0.13728 (16) | 0.0368 (4) | |
| H14 | 0.9915 | −0.0497 | 0.0967 | 0.044* | |
| C15 | 0.97344 (18) | 0.06622 (19) | 0.25181 (16) | 0.0350 (4) | |
| H15 | 1.0507 | 0.0515 | 0.2901 | 0.042* | |
| C16 | 0.89474 (17) | 0.14765 (18) | 0.31045 (14) | 0.0312 (4) | |
| H16 | 0.9188 | 0.1880 | 0.3891 | 0.037* | |
| C20 | 0.54425 (17) | 0.11170 (17) | 0.36403 (13) | 0.0268 (3) | |
| H20A | 0.5551 | 0.1718 | 0.4434 | 0.032* | |
| H20B | 0.6192 | 0.0640 | 0.3576 | 0.032* | |
| C21 | 0.40800 (16) | 0.00815 (16) | 0.32108 (13) | 0.0259 (3) | |
| C22 | 0.34574 (17) | −0.05756 (17) | 0.20764 (14) | 0.0296 (3) | |
| C23 | 0.22507 (19) | −0.15869 (19) | 0.16750 (17) | 0.0394 (4) | |
| H23 | 0.1832 | −0.2018 | 0.0905 | 0.047* | |
| C24 | 0.1654 (2) | −0.1970 (2) | 0.23937 (19) | 0.0457 (5) | |
| H24 | 0.0838 | −0.2678 | 0.2113 | 0.055* | |
| C25 | 0.2237 (2) | −0.1331 (2) | 0.35144 (19) | 0.0439 (5) | |
| H25 | 0.1822 | −0.1592 | 0.4007 | 0.053* | |
| C26 | 0.34370 (18) | −0.03025 (19) | 0.39182 (16) | 0.0338 (4) | |
| H26 | 0.3827 | 0.0149 | 0.4693 | 0.041* | |
| C30 | 0.20423 (18) | 0.36075 (19) | 0.27170 (17) | 0.0364 (4) | |
| H30A | 0.2235 | 0.3827 | 0.3516 | 0.044* | |
| H30B | 0.1767 | 0.2604 | 0.2293 | 0.044* | |
| C31 | 0.08900 (18) | 0.41958 (19) | 0.24471 (18) | 0.0404 (4) | |
| C32 | 0.0497 (2) | 0.4116 (2) | 0.13898 (18) | 0.0453 (5) | |
| C33 | −0.0650 (2) | 0.4543 (2) | 0.1098 (2) | 0.0613 (7) | |
| H33 | −0.0926 | 0.4460 | 0.0368 | 0.074* | |
| C34 | −0.1378 (2) | 0.5087 (2) | 0.1875 (3) | 0.0637 (7) | |
| H34 | −0.2174 | 0.5359 | 0.1672 | 0.076* | |
| C35 | −0.0980 (2) | 0.5243 (2) | 0.2931 (3) | 0.0603 (7) | |
| H35 | −0.1472 | 0.5660 | 0.3467 | 0.072* | |
| C36 | 0.0148 (2) | 0.4792 (2) | 0.3227 (2) | 0.0499 (5) | |
| H36 | 0.0416 | 0.4889 | 0.3962 | 0.060* | |
| C40 | 0.39032 (17) | 0.55858 (16) | 0.32520 (14) | 0.0286 (3) | |
| H40A | 0.4334 | 0.5613 | 0.3967 | 0.034* | |
| H40B | 0.3159 | 0.6066 | 0.3376 | 0.034* | |
| C41 | 0.49612 (16) | 0.63311 (16) | 0.28855 (13) | 0.0247 (3) | |
| C42 | 0.62638 (16) | 0.70468 (16) | 0.35788 (12) | 0.0260 (3) | |
| C43 | 0.71949 (16) | 0.78163 (17) | 0.32493 (13) | 0.0284 (3) | |
| C44 | 0.68364 (17) | 0.78221 (17) | 0.22159 (13) | 0.0293 (3) | |
| H44 | 0.7473 | 0.8341 | 0.1998 | 0.035* | |
| C45 | 0.55741 (18) | 0.70888 (17) | 0.15050 (13) | 0.0290 (3) | |
| C46 | 0.46489 (17) | 0.63763 (17) | 0.18737 (13) | 0.0277 (3) | |
| H46 | 0.3762 | 0.5900 | 0.1406 | 0.033* | |
| C47 | 0.85585 (19) | 0.8554 (2) | 0.39565 (15) | 0.0385 (4) | |
| H47 | 0.9159 | 0.9059 | 0.3704 | 0.046* | |
| C48 | 0.5214 (2) | 0.7027 (2) | 0.03654 (14) | 0.0406 (4) | |
| H48A | 0.5719 | 0.7864 | 0.0368 | 0.061* | |
| H48B | 0.4226 | 0.6935 | 0.0152 | 0.061* | |
| H48C | 0.5462 | 0.6241 | −0.0166 | 0.061* | |
| O10 | 0.64008 (13) | 0.13999 (14) | 0.09024 (10) | 0.0368 (3) | |
| H10 | 0.6275 | 0.1024 | 0.0139 | 0.044* | |
| O20 | 0.40399 (13) | −0.02278 (13) | 0.13465 (9) | 0.0364 (3) | |
| H20 | 0.4684 | 0.0616 | 0.1734 | 0.044* | |
| O30 | 0.12438 (17) | 0.36127 (17) | 0.06209 (13) | 0.0586 (4) | |
| H30 | 0.2124 | 0.3804 | 0.1050 | 0.070* | |
| O40 | 0.66072 (13) | 0.69757 (14) | 0.45562 (10) | 0.0380 (3) | |
| H40 | 0.7520 | 0.7475 | 0.4859 | 0.046* | |
| O41 | 0.89821 (14) | 0.85675 (16) | 0.48535 (11) | 0.0491 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0239 (6) | 0.0241 (6) | 0.0259 (6) | 0.0070 (5) | 0.0077 (5) | 0.0144 (5) |
| C2 | 0.0319 (8) | 0.0270 (8) | 0.0272 (7) | 0.0117 (7) | 0.0117 (6) | 0.0142 (6) |
| C3 | 0.0303 (8) | 0.0247 (8) | 0.0257 (7) | 0.0085 (6) | 0.0090 (6) | 0.0128 (6) |
| N4 | 0.0230 (6) | 0.0211 (6) | 0.0359 (7) | 0.0052 (5) | 0.0086 (5) | 0.0144 (5) |
| C10 | 0.0237 (8) | 0.0273 (8) | 0.0306 (8) | 0.0022 (6) | 0.0043 (6) | 0.0125 (6) |
| C11 | 0.0215 (7) | 0.0259 (8) | 0.0335 (8) | 0.0023 (6) | 0.0066 (6) | 0.0162 (7) |
| C12 | 0.0231 (8) | 0.0338 (9) | 0.0340 (8) | 0.0060 (7) | 0.0087 (6) | 0.0189 (7) |
| C13 | 0.0317 (9) | 0.0410 (10) | 0.0359 (9) | 0.0095 (8) | 0.0128 (7) | 0.0182 (8) |
| C14 | 0.0283 (9) | 0.0389 (10) | 0.0515 (10) | 0.0116 (8) | 0.0183 (8) | 0.0235 (9) |
| C15 | 0.0233 (8) | 0.0389 (10) | 0.0512 (10) | 0.0082 (7) | 0.0082 (7) | 0.0278 (9) |
| C16 | 0.0243 (8) | 0.0341 (9) | 0.0363 (8) | 0.0027 (7) | 0.0034 (6) | 0.0193 (7) |
| C20 | 0.0279 (8) | 0.0285 (8) | 0.0280 (7) | 0.0070 (6) | 0.0057 (6) | 0.0165 (6) |
| C21 | 0.0248 (8) | 0.0242 (8) | 0.0342 (8) | 0.0090 (6) | 0.0088 (6) | 0.0165 (6) |
| C22 | 0.0294 (8) | 0.0240 (8) | 0.0371 (8) | 0.0085 (7) | 0.0097 (7) | 0.0136 (7) |
| C23 | 0.0314 (9) | 0.0274 (9) | 0.0484 (10) | 0.0037 (7) | 0.0034 (8) | 0.0088 (8) |
| C24 | 0.0287 (9) | 0.0303 (10) | 0.0757 (14) | 0.0027 (8) | 0.0142 (9) | 0.0218 (10) |
| C25 | 0.0371 (10) | 0.0409 (11) | 0.0705 (14) | 0.0126 (9) | 0.0256 (10) | 0.0352 (10) |
| C26 | 0.0336 (9) | 0.0351 (9) | 0.0443 (9) | 0.0121 (7) | 0.0149 (7) | 0.0255 (8) |
| C30 | 0.0277 (9) | 0.0319 (9) | 0.0552 (11) | 0.0045 (7) | 0.0136 (8) | 0.0242 (8) |
| C31 | 0.0225 (8) | 0.0279 (9) | 0.0699 (13) | 0.0010 (7) | 0.0098 (8) | 0.0226 (9) |
| C32 | 0.0336 (10) | 0.0280 (10) | 0.0612 (12) | 0.0057 (8) | 0.0007 (9) | 0.0106 (9) |
| C33 | 0.0442 (13) | 0.0331 (11) | 0.0821 (17) | 0.0056 (10) | −0.0162 (11) | 0.0127 (11) |
| C34 | 0.0277 (10) | 0.0339 (12) | 0.113 (2) | 0.0043 (9) | 0.0014 (12) | 0.0225 (13) |
| C35 | 0.0338 (11) | 0.0353 (11) | 0.116 (2) | 0.0094 (9) | 0.0324 (13) | 0.0314 (13) |
| C36 | 0.0362 (11) | 0.0367 (11) | 0.0832 (15) | 0.0069 (9) | 0.0258 (10) | 0.0292 (11) |
| C40 | 0.0279 (8) | 0.0253 (8) | 0.0348 (8) | 0.0062 (6) | 0.0134 (7) | 0.0135 (7) |
| C41 | 0.0255 (7) | 0.0207 (7) | 0.0305 (7) | 0.0079 (6) | 0.0109 (6) | 0.0114 (6) |
| C42 | 0.0273 (8) | 0.0266 (8) | 0.0261 (7) | 0.0077 (6) | 0.0079 (6) | 0.0125 (6) |
| C43 | 0.0243 (8) | 0.0284 (8) | 0.0317 (8) | 0.0038 (6) | 0.0058 (6) | 0.0134 (7) |
| C44 | 0.0302 (8) | 0.0262 (8) | 0.0360 (8) | 0.0048 (7) | 0.0112 (7) | 0.0175 (7) |
| C45 | 0.0336 (9) | 0.0260 (8) | 0.0300 (8) | 0.0092 (7) | 0.0077 (6) | 0.0140 (6) |
| C46 | 0.0253 (8) | 0.0251 (8) | 0.0311 (8) | 0.0057 (6) | 0.0032 (6) | 0.0119 (6) |
| C47 | 0.0283 (9) | 0.0394 (10) | 0.0428 (10) | −0.0015 (8) | 0.0034 (7) | 0.0184 (8) |
| C48 | 0.0518 (12) | 0.0422 (11) | 0.0324 (9) | 0.0116 (9) | 0.0078 (8) | 0.0215 (8) |
| O10 | 0.0338 (7) | 0.0523 (8) | 0.0295 (6) | 0.0177 (6) | 0.0082 (5) | 0.0200 (6) |
| O20 | 0.0418 (7) | 0.0324 (7) | 0.0269 (6) | 0.0004 (6) | 0.0070 (5) | 0.0086 (5) |
| O30 | 0.0575 (10) | 0.0587 (10) | 0.0500 (9) | 0.0222 (8) | −0.0007 (7) | 0.0155 (8) |
| O40 | 0.0342 (7) | 0.0497 (8) | 0.0323 (6) | 0.0037 (6) | 0.0048 (5) | 0.0235 (6) |
| O41 | 0.0367 (7) | 0.0579 (9) | 0.0432 (8) | −0.0025 (7) | −0.0056 (6) | 0.0230 (7) |
Geometric parameters (Å, º)
| N1—C2 | 1.471 (2) | C26—H26 | 0.9500 |
| N1—C10 | 1.478 (2) | C30—C31 | 1.497 (3) |
| N1—C20 | 1.4828 (19) | C30—H30A | 0.9900 |
| C2—C3 | 1.528 (2) | C30—H30B | 0.9900 |
| C2—H2A | 0.9900 | C31—C32 | 1.392 (3) |
| C2—H2B | 0.9900 | C31—C36 | 1.398 (3) |
| C3—N4 | 1.471 (2) | C32—O30 | 1.370 (3) |
| C3—H3A | 0.9900 | C32—C33 | 1.390 (3) |
| C3—H3B | 0.9900 | C33—C34 | 1.372 (4) |
| N4—C40 | 1.476 (2) | C33—H33 | 0.9500 |
| N4—C30 | 1.483 (2) | C34—C35 | 1.361 (4) |
| C10—C11 | 1.500 (2) | C34—H34 | 0.9500 |
| C10—H10A | 0.9900 | C35—C36 | 1.392 (3) |
| C10—H10B | 0.9900 | C35—H35 | 0.9500 |
| C11—C16 | 1.395 (2) | C36—H36 | 0.9500 |
| C11—C12 | 1.397 (2) | C40—C41 | 1.507 (2) |
| C12—O10 | 1.361 (2) | C40—H40A | 0.9900 |
| C12—C13 | 1.389 (2) | C40—H40B | 0.9900 |
| C13—C14 | 1.387 (3) | C41—C46 | 1.384 (2) |
| C13—H13 | 0.9500 | C41—C42 | 1.400 (2) |
| C14—C15 | 1.383 (3) | C42—O40 | 1.3556 (18) |
| C14—H14 | 0.9500 | C42—C43 | 1.407 (2) |
| C15—C16 | 1.389 (2) | C43—C44 | 1.397 (2) |
| C15—H15 | 0.9500 | C43—C47 | 1.456 (2) |
| C16—H16 | 0.9500 | C44—C45 | 1.380 (2) |
| C20—C21 | 1.509 (2) | C44—H44 | 0.9500 |
| C20—H20A | 0.9900 | C45—C46 | 1.400 (2) |
| C20—H20B | 0.9900 | C45—C48 | 1.505 (2) |
| C21—C26 | 1.393 (2) | C46—H46 | 0.9500 |
| C21—C22 | 1.401 (2) | C47—O41 | 1.222 (2) |
| C22—O20 | 1.369 (2) | C47—H47 | 0.9500 |
| C22—C23 | 1.385 (2) | C48—H48A | 0.9800 |
| C23—C24 | 1.382 (3) | C48—H48B | 0.9800 |
| C23—H23 | 0.9500 | C48—H48C | 0.9800 |
| C24—C25 | 1.377 (3) | O10—H10 | 0.9269 |
| C24—H24 | 0.9500 | O20—H20 | 0.9386 |
| C25—C26 | 1.390 (3) | O30—H30 | 0.9332 |
| C25—H25 | 0.9500 | O40—H40 | 0.9349 |
| C2—N1—C10 | 111.44 (12) | C25—C26—H26 | 119.3 |
| C2—N1—C20 | 112.03 (12) | C21—C26—H26 | 119.3 |
| C10—N1—C20 | 111.35 (12) | N4—C30—C31 | 111.34 (14) |
| N1—C2—C3 | 110.69 (12) | N4—C30—H30A | 109.4 |
| N1—C2—H2A | 109.5 | C31—C30—H30A | 109.4 |
| C3—C2—H2A | 109.5 | N4—C30—H30B | 109.4 |
| N1—C2—H2B | 109.5 | C31—C30—H30B | 109.4 |
| C3—C2—H2B | 109.5 | H30A—C30—H30B | 108.0 |
| H2A—C2—H2B | 108.1 | C32—C31—C36 | 118.15 (19) |
| N4—C3—C2 | 116.14 (12) | C32—C31—C30 | 120.33 (18) |
| N4—C3—H3A | 108.3 | C36—C31—C30 | 121.5 (2) |
| C2—C3—H3A | 108.3 | O30—C32—C33 | 119.1 (2) |
| N4—C3—H3B | 108.3 | O30—C32—C31 | 120.03 (18) |
| C2—C3—H3B | 108.3 | C33—C32—C31 | 120.9 (2) |
| H3A—C3—H3B | 107.4 | C34—C33—C32 | 119.3 (3) |
| C3—N4—C40 | 114.03 (13) | C34—C33—H33 | 120.3 |
| C3—N4—C30 | 112.54 (12) | C32—C33—H33 | 120.3 |
| C40—N4—C30 | 109.74 (13) | C35—C34—C33 | 121.2 (2) |
| N1—C10—C11 | 113.43 (13) | C35—C34—H34 | 119.4 |
| N1—C10—H10A | 108.9 | C33—C34—H34 | 119.4 |
| C11—C10—H10A | 108.9 | C34—C35—C36 | 119.9 (2) |
| N1—C10—H10B | 108.9 | C34—C35—H35 | 120.0 |
| C11—C10—H10B | 108.9 | C36—C35—H35 | 120.0 |
| H10A—C10—H10B | 107.7 | C35—C36—C31 | 120.4 (2) |
| C16—C11—C12 | 117.97 (15) | C35—C36—H36 | 119.8 |
| C16—C11—C10 | 121.94 (15) | C31—C36—H36 | 119.8 |
| C12—C11—C10 | 119.96 (14) | N4—C40—C41 | 113.41 (13) |
| O10—C12—C13 | 122.45 (15) | N4—C40—H40A | 108.9 |
| O10—C12—C11 | 116.65 (15) | C41—C40—H40A | 108.9 |
| C13—C12—C11 | 120.90 (15) | N4—C40—H40B | 108.9 |
| C14—C13—C12 | 119.98 (16) | C41—C40—H40B | 108.9 |
| C14—C13—H13 | 120.0 | H40A—C40—H40B | 107.7 |
| C12—C13—H13 | 120.0 | C46—C41—C42 | 118.20 (14) |
| C15—C14—C13 | 120.14 (17) | C46—C41—C40 | 120.76 (14) |
| C15—C14—H14 | 119.9 | C42—C41—C40 | 120.98 (14) |
| C13—C14—H14 | 119.9 | O40—C42—C41 | 119.10 (14) |
| C14—C15—C16 | 119.55 (16) | O40—C42—C43 | 121.17 (14) |
| C14—C15—H15 | 120.2 | C41—C42—C43 | 119.73 (14) |
| C16—C15—H15 | 120.2 | C44—C43—C42 | 119.95 (15) |
| C15—C16—C11 | 121.45 (16) | C44—C43—C47 | 119.64 (15) |
| C15—C16—H16 | 119.3 | C42—C43—C47 | 120.34 (15) |
| C11—C16—H16 | 119.3 | C45—C44—C43 | 121.23 (15) |
| N1—C20—C21 | 111.67 (12) | C45—C44—H44 | 119.4 |
| N1—C20—H20A | 109.3 | C43—C44—H44 | 119.4 |
| C21—C20—H20A | 109.3 | C44—C45—C46 | 117.50 (15) |
| N1—C20—H20B | 109.3 | C44—C45—C48 | 121.42 (16) |
| C21—C20—H20B | 109.3 | C46—C45—C48 | 121.06 (16) |
| H20A—C20—H20B | 107.9 | C41—C46—C45 | 123.31 (15) |
| C26—C21—C22 | 117.90 (16) | C41—C46—H46 | 118.3 |
| C26—C21—C20 | 121.25 (15) | C45—C46—H46 | 118.3 |
| C22—C21—C20 | 120.77 (14) | O41—C47—C43 | 124.37 (17) |
| O20—C22—C23 | 119.00 (16) | O41—C47—H47 | 117.8 |
| O20—C22—C21 | 120.26 (15) | C43—C47—H47 | 117.8 |
| C23—C22—C21 | 120.74 (17) | C45—C48—H48A | 109.5 |
| C24—C23—C22 | 120.06 (18) | C45—C48—H48B | 109.5 |
| C24—C23—H23 | 120.0 | H48A—C48—H48B | 109.5 |
| C22—C23—H23 | 120.0 | C45—C48—H48C | 109.5 |
| C25—C24—C23 | 120.42 (18) | H48A—C48—H48C | 109.5 |
| C25—C24—H24 | 119.8 | H48B—C48—H48C | 109.5 |
| C23—C24—H24 | 119.8 | C12—O10—H10 | 112.6 |
| C24—C25—C26 | 119.46 (18) | C22—O20—H20 | 109.6 |
| C24—C25—H25 | 120.3 | C32—O30—H30 | 103.6 |
| C26—C25—H25 | 120.3 | C42—O40—H40 | 104.9 |
| C25—C26—C21 | 121.40 (18) | ||
| C10—N1—C2—C3 | 81.42 (15) | C40—N4—C30—C31 | 72.17 (19) |
| C20—N1—C2—C3 | −153.07 (13) | N4—C30—C31—C32 | 54.0 (2) |
| N1—C2—C3—N4 | 174.78 (13) | N4—C30—C31—C36 | −128.35 (18) |
| C2—C3—N4—C40 | 72.09 (17) | C36—C31—C32—O30 | 176.19 (18) |
| C2—C3—N4—C30 | −53.74 (18) | C30—C31—C32—O30 | −6.1 (3) |
| C2—N1—C10—C11 | −159.44 (12) | C36—C31—C32—C33 | −3.7 (3) |
| C20—N1—C10—C11 | 74.67 (16) | C30—C31—C32—C33 | 174.00 (18) |
| N1—C10—C11—C16 | −117.78 (16) | O30—C32—C33—C34 | −178.0 (2) |
| N1—C10—C11—C12 | 66.33 (19) | C31—C32—C33—C34 | 1.9 (3) |
| C16—C11—C12—O10 | −178.35 (15) | C32—C33—C34—C35 | 1.5 (3) |
| C10—C11—C12—O10 | −2.3 (2) | C33—C34—C35—C36 | −2.9 (3) |
| C16—C11—C12—C13 | 1.6 (2) | C34—C35—C36—C31 | 1.0 (3) |
| C10—C11—C12—C13 | 177.67 (15) | C32—C31—C36—C35 | 2.3 (3) |
| O10—C12—C13—C14 | 178.56 (17) | C30—C31—C36—C35 | −175.38 (17) |
| C11—C12—C13—C14 | −1.4 (3) | C3—N4—C40—C41 | 67.55 (17) |
| C12—C13—C14—C15 | 0.3 (3) | C30—N4—C40—C41 | −165.16 (14) |
| C13—C14—C15—C16 | 0.5 (3) | N4—C40—C41—C46 | 54.7 (2) |
| C14—C15—C16—C11 | −0.3 (3) | N4—C40—C41—C42 | −128.20 (16) |
| C12—C11—C16—C15 | −0.8 (2) | C46—C41—C42—O40 | −177.66 (14) |
| C10—C11—C16—C15 | −176.73 (15) | C40—C41—C42—O40 | 5.2 (2) |
| C2—N1—C20—C21 | 72.32 (16) | C46—C41—C42—C43 | 1.9 (2) |
| C10—N1—C20—C21 | −162.11 (13) | C40—C41—C42—C43 | −175.28 (15) |
| N1—C20—C21—C26 | −146.22 (15) | O40—C42—C43—C44 | 177.11 (15) |
| N1—C20—C21—C22 | 37.1 (2) | C41—C42—C43—C44 | −2.4 (2) |
| C26—C21—C22—O20 | 179.39 (15) | O40—C42—C43—C47 | 0.2 (3) |
| C20—C21—C22—O20 | −3.8 (2) | C41—C42—C43—C47 | −179.35 (16) |
| C26—C21—C22—C23 | −0.9 (2) | C42—C43—C44—C45 | 0.3 (3) |
| C20—C21—C22—C23 | 175.91 (16) | C47—C43—C44—C45 | 177.31 (17) |
| O20—C22—C23—C24 | 179.02 (16) | C43—C44—C45—C46 | 2.1 (2) |
| C21—C22—C23—C24 | −0.7 (3) | C43—C44—C45—C48 | −176.43 (16) |
| C22—C23—C24—C25 | 1.4 (3) | C42—C41—C46—C45 | 0.7 (2) |
| C23—C24—C25—C26 | −0.4 (3) | C40—C41—C46—C45 | 177.87 (15) |
| C24—C25—C26—C21 | −1.2 (3) | C44—C45—C46—C41 | −2.7 (3) |
| C22—C21—C26—C25 | 1.8 (2) | C48—C45—C46—C41 | 175.86 (16) |
| C20—C21—C26—C25 | −174.92 (16) | C44—C43—C47—O41 | −177.20 (19) |
| C3—N4—C30—C31 | −159.71 (15) | C42—C43—C47—O41 | −0.3 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O10—H10···O20i | 0.93 | 1.80 | 2.7230 (16) | 177 |
| O20—H20···N1 | 0.94 | 1.75 | 2.5928 (17) | 148 |
| O20—H20···O10 | 0.94 | 2.43 | 3.0362 (19) | 122 |
| O30—H30···N4 | 0.93 | 1.94 | 2.784 (2) | 149 |
| O40—H40···O41 | 0.93 | 1.76 | 2.6146 (19) | 151 |
Symmetry code: (i) −x+1, −y, −z.
References
- Ansari, K. I., Kasiri, S., Grant, J. D. & Mandal, S. S. (2009). Dalton Trans. pp. 8525–8531. [DOI] [PubMed]
- Boros, E., Ferreira, C. L., Patrick, B. O., Adam, M. J. & Orvig, C. (2011). Nucl. Med. Biol. 38, 1165–1174. [DOI] [PubMed]
- Boyle, T. J., Pratt, H. D. III, Ottley, L. A. M., Alam, T. M., McIntyre, S. K., Rodriguez, M. A., Farrell, J. & Campana, C. F. (2009). Inorg. Chem. 48, 9191–9204. [DOI] [PubMed]
- Bruker (2009). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruno, I. J., Cole, J. C., Kessler, M., Luo, J., Motherwell, W. D. S., Purkis, L. H., Smith, B. R., Taylor, R., Cooper, R. I., Harris, S. E. & Orpen, A. G. (2004). J. Chem. Inf. Comput. Sci. 44, 2133–2144. [DOI] [PubMed]
- Dencic, S. M., Poljarevic, J., Vilimanovich, U., Bogdanovic, A., Isakovic, A. J., Stevovic, T. K., Dulovic, M., Zogovic, N., Isakovic, A. M., Grguric-Sipka, S., Bumbasirevic, V., Sabo, T., Trajkovic, V. & Markovic, I. (2012). Chem. Res. Toxicol. 25, 931–939. [DOI] [PubMed]
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Lazić, J. M., Vučićević, L., Grgurić-Šipka, S., Janjetović, K., Kaluđerović, G. N., Misirkić, M., Gruden-Pavlović, M., Popadić, D., Paschke, R., Trajković, V. & Sabo, T. J. (2010). ChemMedChem, 5, 881–889. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Musa, M. A., Badisa, V. L. D. & Latinwo, L. M. (2014). Anticancer Res. 34, 1601–1607. [PubMed]
- Price, E. W., Cawthray, J. F., Bailey, G. A., Ferreira, C. L., Boros, E., Adam, M. J. & Orvig, C. (2012). J. Am. Chem. Soc. 134, 8670–8683. [DOI] [PubMed]
- Raman, N., Selvan, A. & Sudharsan, S. (2011). Spectrochim. Acta Part A, 79, 873–883. [DOI] [PubMed]
- Schmitt, H., Lomoth, R., Magnuson, A., Park, J., Fryxelius, J., Kritikos, M., Mårtensson, J., Hammarström, L., Sun, L. & Åkermark, B. (2002). Chem. Eur. J. 8, 3757–3768. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Thoer, A., Denis, G., Delmas, M. & Gaset, A. (1988). Synth. Commun. 18, 2095–2101.
- Wang, N.-S., Wang, Y.-T., Guo, X.-K. & Li, T.-D. (2011a). Acta Cryst. E67, o1438. [DOI] [PMC free article] [PubMed]
- Wang, N. S., Wang, Y. T., Li, J. Y. & Li, T. D. (2011b). Chin. J. Org. Chem. 31, 1703–1706.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) general, I. DOI: 10.1107/S1600536814024465/lh5739sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814024465/lh5739Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814024465/lh5739Isup3.mol
Supporting information file. DOI: 10.1107/S1600536814024465/lh5739Isup4.cml
CCDC reference: 1033129
Additional supporting information: crystallographic information; 3D view; checkCIF report





