Abstract
Neural crest arises at the neural plate border, expresses a core set of regulatory genes, and produces a diverse array of cell types including ectomesenchyme derivatives that elaborate the vertebrate head1,2. The evolution of neural crest has been postulated as a key event leading to the appearance of new cell types that fostered the transition from filter feeding to active predation in ancestral vertebrates3. However, the origin of neural crest remains controversial, since homologous cell types have not been unambiguously identified in non-vertebrate chordates1,4. Here we show that the tunicate Ciona intestinalis possesses a cephalic melanocyte lineage (a9.49) similar to neural crest that can be reprogrammed into migrating ectomesenchyme by the targeted misexpression of Twist. Our results suggest that the neural crest melanocyte regulatory network predated the divergence of tunicates and vertebrates. We propose that the co-option of mesenchyme determinants, such as Twist, into the neural plate ectoderm was crucial for the emergence of the vertebrate “new head”3.
Whole-genome phylogenetic analyses place the tunicates as the true sister clade to vertebrates5, and consequently they are well suited for investigating the evolutionary origins of neural crest. In a previous report on the mangrove tunicate, a migratory cell population originating from the vicinity of the neural tube was likened to neural crest6. However, subsequent studies of eleven additional tunicates provided unequivocal evidence that these cells arise from the mesoderm flanking the neural tube7. It was then suggested that a mesoderm-derived mesenchyme lineage (A7.6) in Ciona possessed some of the properties of neural crest8, although these cells do not arise from the neural plate border and lack expression of key neural crest regulatory genes.
We present evidence that the a9.49 cell lineage of Ciona embryos represents a rudimentary neural crest. It arises at the neural plate border and expresses several neural plate border genes, as well as a number of neural crest specification genes, including Id, Snail, Ets, and FoxD8-13 (Fig. 1a, Supplementary Fig. 1). In vertebrates, Mitf directly activates several target genes required for melanogenesis of neural crest-derived melanocytes, including Tyr and TRP14. In tunicates, Mitf is expressed in the a9.49 lineage15, which can be labeled via electroporation of a Mitf reporter plasmid (Mitf>GFP) (Fig. 1b). The posterior daughters of the lineage (a10.97) intercalate at the dorsal midline and form the gravity-sensing otolith and melanocyte of the light detecting ocellus (Fig. 1c)16. We sought to understand the basis for the differential specification of these pigmented cells.
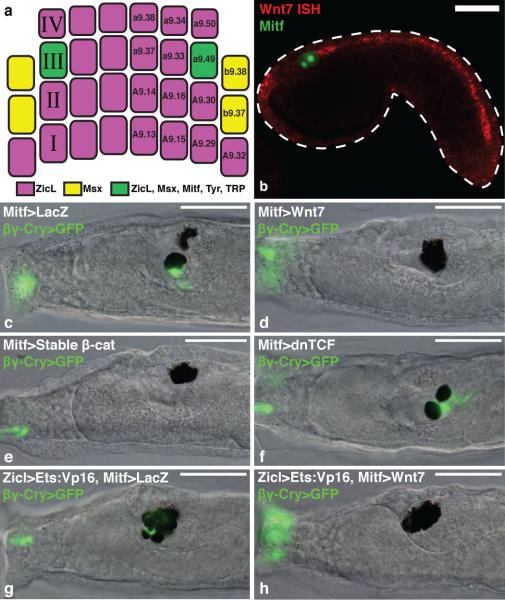
Figure 1. Wnt signaling promotes ocellus formation.
a, Gastrula stage schematic indicates the lineage specific expression of enhancers in this study. b, Tailbud electroporated with Mitf>LacZ detected with an antibody (green), and hybridized with a Wnt7 probe (red). c-f, Larvae electroporated with βγ-crystallin>GFP marks the otolith and anterior palps. c, Co-electroporated with Mitf>LacZ (166/196 had an otolith and ocellus). d, Co-electroporated with Mitf>Wnt7 (172/205 had two ocelli). e, Co-electroporated with Mitf>stable β-catenin (189/205 had two ocelli). f, Coelectroporated with Mitf>dnTCF (100/205 had two otoliths). g,h, Larvae electroporated with Zicl>Ets:VP16 and βγ-crystallin>GFP. g, Co-electroporated with Mitf>LacZ (75/100 had extra otoliths). h, Co-electroporated with Mitf>Wnt7 (only 11/100 had extra otoliths). Scale bars, 50 μm.
Wnt signaling plays a conserved role in neural crest induction, and promotes melanocyte formation from cephalic neural crest in zebrafish17. Both a10.97 cells express Tcf/Lef, the transcriptional effector of Wnt signaling13, thus Wnt might also play a role in Ciona melanogenesis. We found that Wnt7 is expressed along the dorsal midline just posterior to the presumptive ocellus (Fig. 1b), suggesting that it might serve as a positional cue to trigger differentiation of the posterior a10.97 melanocyte.
Wnt signaling was selectively perturbed in the a9.49 lineage using the Mitf enhancer (Fig. 1c-f). A βγ-crystallin reporter was used to distinguish the melanocytes since it is expressed in the otolith but not the ocellus (Fig. 1c). Both pigmented precursors were converted to ocelli upon misexpression of Wnt7 (Fig. 1d). A similar transformation was observed upon targeted expression of a stabilized form of β-catenin, the coactivator of Tcf (Fig. 1e). In contrast, misexpression of a dominant-negative form of Tcf (Mitf>dnTCF) produced the reciprocal transformation: both a10.97 melanocytes differentiated into otoliths and expressed the βγ-crystallin reporter (Fig. 1f).
Supernumerary otoliths were induced by the expression of a constitutively active form of the Ets1/2 transcription factor (Fig. 1g and Supplementary Fig. 2). Coelectroporation of Mitf>Wnt7 transformed these extra otoliths into ocelli (Fig. 1h). These results suggest that Wnt7 signaling specifies the ocellus and suppresses the development of the otolith. To determine the underlying mechanism we sought to identify neural crest specification genes that are selectively activated in the presumptive ocellus in response to Wnt7 signaling.
In vertebrates, Foxd3 has been shown to repress melanogenesis of neural crest cells via downregulation of Mitf14,18. In Ciona, FoxD is directly activated by the accumulation of nuclear β-catenin in the early embryo, indicating a potential link between Wnt signaling and FoxD expression19. We found that FoxD is selectively expressed in the presumptive ocellus, (Fig. 2a, b) adjacent to the expression of Wnt7 in the dorsal midline (Fig. 1b). A FoxD enhancer recapitulates this expression in the presumptive ocellus (Fig. 2c), and is dependent on Wnt signaling, as expression is lost in the presence of dnTCF (Fig. 2d).
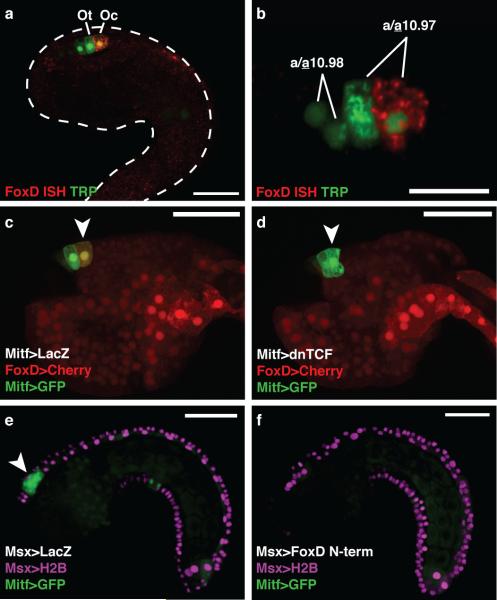
Figure 2. FoxD represses Mitf in the ocellus.
a, Tailbud electroporated with TRP>LacZ detected with an antibody (green) marking the precursors of the otolith and ocellus, and hybridized with a FoxD probe (red). b, FoxD is expressed in the posterior a10.97 cell. c-d, Tailbuds electroporated with FoxD>mCherry and Mitf>GFP. Arrowheads mark the presumptive ocellus. c, Co-electroporated with Mitf>LacZ (126/180 expressed mCherry). d, Co-electroporated with Mitf>dnTCF (only 30/180 expressed mCherry). e-f, Tailbuds electroporated with Msx>H2B mCherry, and Mitf>GFP. Arrowhead shows GFP expression in a9.49 derivatives. e, Co-electroporated with Msx>LacZ. f, Co-electroporated with Msx>FoxD N-term. Scale bars 50 μm (a, c-f); 25 μm (b).
To investigate the role of FoxD in melanogenesis, we expressed variants of FoxD in the midline of the CNS, including the a9.49 lineage, using 5’ regulatory sequences from the Msx gene (Fig. 1a, Supplementary Fig. 3). Targeted expression of either full-length FoxD or the N-terminal third of FoxD (non-DNA binding) abolished expression of the Mitf>GFP reporter gene (Fig. 2e, f, Supplementary Fig. 3). However, misexpression of a constitutive repressor form of FoxD (DNA binding domain fused to a WRPW repressor motif) had little effect on Mitf expression (Supplementary Fig. 3). These results suggest that FoxD represses Mitf independent of its DNA binding domain, which is consistent with its mode of regulation in avian embryos, where Foxd3-mediated repression of Mitf is thought to occur through the sequestration of the transcriptional activator Pax318.
Our results suggest a simple gene regulatory network (Wnt7→FoxD⊣Mitf) for the differential specification of the otolith and ocellus in the Ciona tadpole (Supplementary Fig. 4). Both a10.97 cells express Mitf prior to neurulation and during the convergence of the two cells along the dorsal midline of the anterior neural tube. Subsequently, the posterior a10.97 cell receives a localized Wnt7 signal and activates FoxD, which attenuates Mitf leading to diminished pigmentation in the ocellus. Mitf expression is sustained in the anterior a10.97 cell, which forms the densely pigmented otolith.
Zebrafish employ a remarkably similar mechanism to specify neural crest-derived pigmented melanophores and iridophores, which derive from a common bi-potent Mitf+ progenitor14. The conservation of this network strengthens the argument that the a9.49 lineage of Ciona represents a rudimentary neural crest. However, the a9.49 lineage lacks some of the defining properties of cephalic neural crest, such as long-range migration and the potential to form ectomesenchyme derivatives.
We therefore sought to identify vertebrate neural crest determinants that are not expressed in the Ciona a9.49 lineage. In vertebrates, the craniofacial mesenchyme is derived from primary mesoderm and the ectomesenchyme arising from cephalic neural crest2,20. Both sources of cranial mesenchyme express the conserved mesodermal determinant Twist21 and produce diverse cranial tissues including muscle, cartilage and bone. In tetrapods, it appears that only the cephalic neural crest expresses Twist and produces ectomesenchyme21-23, whereas trunk neural crest lacks Twist expression and generates non-ectomesenchymal derivatives (e.g. neurons, glia and melanocytes)2. Disruption of Twist activity causes severe cephalic neural crest phenotypes, including defects in cell migration and survival, as well as morphological defects of the skull vault and heart21,22,24.
There are three Twist-related genes in Ciona. In this study we focused on the one most similar to Twist1 in vertebrates (Supplementary Fig. 5). In Ciona, Twist is expressed solely in mesoderm-derived mesenchyme (Fig. 3a). It is not expressed in any region of the neural plate, including the a9.49 lineage. Twist-expressing mesoderm undergoes long-range migration (Fig. 3b) and produces a number of diverse tissues in juveniles and adults, including body wall muscles, tunic cells (which populate the protective covering of the adult), and blood cells25. The migration and differentiation of these mesoderm tissues are inhibited when Twist expression is reduced25.
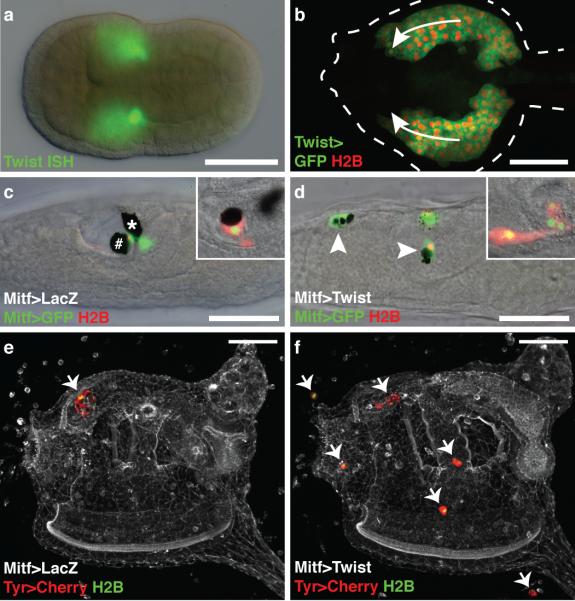
Figure 3. Twist reprograms the a9.49 lineage.
a, Neurula hybridized with a Twist probe. b, Tailbud during mesenchyme migration (arrows) co-electroporated with Twist>GFP and Twist>H2B mCherry. c,d, Larvae electroporated with Mitf>GFP, and Mitf>H2B mCherry. Insets show lineage marked with Tyr>mCherry, and Tyr>H2B YFP. c, Co-electroporated with Mitf>LacZ. # and * mark the otolith and ocellus, respectively. d, Co-electroporated with Mitf>Twist. Arrowheads indicate ectopic position of a9.49 derivatives. e,f, Juveniles electroporated Tyr>mCherry, and Tyr>H2B YFP. Arrows indentify the position of a9.49 derivatives. e, Co-electroporated with Mitf>LacZ. f, Coelectroporated with Mitf>Twist. Scale bars, 50 μm.
To determine whether ectomesenchyme could be formed in Ciona, we misexpressed Twist in the a9.49 lineage using the Mitf enhancer (Fig. 3c, d). The manipulated cells exhibit a mesenchymal phenotype, including protrusive activity, proliferation and long-range migration, which was not observed by the misexpression of other related genes (Supplementary Movie 1, Supplementary Fig. 6). Moreover, misexpression of Twist in the notochord and motor ganglion causes some disruptions in terminal differentiation, but does not transform these tissues into mesenchyme (Supplementary Fig. 7). The reprogrammed a9.49 cells exhibit expression of mesenchyme genes, including ERG (Supplementary Fig. 8), which is expressed in the ectomesenchyme of mouse embryos26. The affected lineage was visualized in juveniles using reporters for Tyr, a gene that is activated by Mitf in melanocytes14 (Fig. 3e,f). Normally the a9.49 derivatives are located solely in an anterior region of the CNS (Fig. 3e). In contrast, embryos expressing Mitf>Twist result in juveniles with ectopic a9.49 cells (Fig. 3f). The reprogrammed cells appear to produce mesodermal derivatives, such as tunic cells based on their location and their distinct, rounded morphology (Supplementary Fig. 9).
Additional evidence for the reprogramming of the a9.49 lineage was obtained with Kaede, a photoconvertible fluorescent protein that was previously used in Ciona to trace the formation of the CNS27. Here, embryos were co-electroporated with Mitf>Twist and Tyr>Kaede, which mediate expression in the a9.49 lineage (Fig. 4a). Tailbud embryos that were not exposed to UV light show no red fluorescence (Fig. 4b) and result in juveniles that have only green a9.49 descendants (Fig. 4c). In contrast, tailbud embryos treated with UV (Fig. 4d, e) develop into juveniles that display red cells throughout the body. Control juveniles lacking Mitf>Twist exhibit the expected expression solely in the CNS (Supplementary Fig. 10). Finally, time-lapse microscopy was used to examine the Mitf>Twist expressing cells in juveniles. Some of these cells migrate like the normal tunic cells derived from the mesenchyme (Fig. 4g, Supplementary Movie 2). Thus, the misexpression of Twist appears to be sufficient, in part, to reprogram the a9.49 lineage into ectomesenchyme.
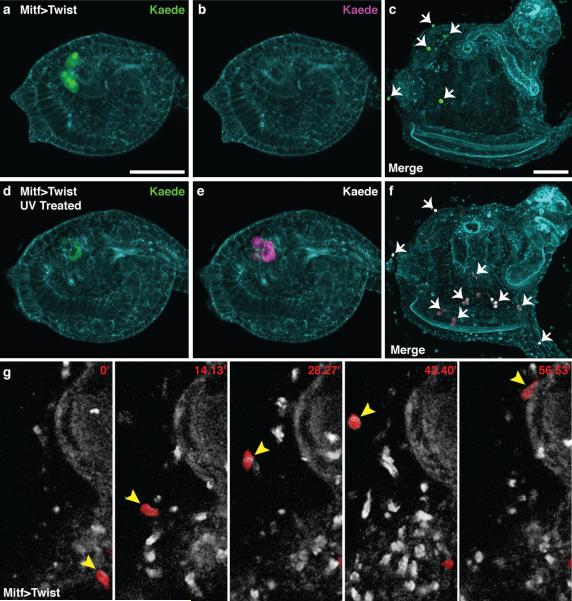
Figure 4. Lineage tracing of reprogrammed a9.49 cells.
a-f, Ciona electroporated with Mitf>Twist and Tyr>Kaede. a,b, Non-UV treated tailbud shows only green fluorescence c, Embryo never exposed to UV results in a juvenile with only green ectopic cells (arrows). d,e, UV treated tailbud shows green and red fluorescence respectively. f, UV treated embryo results in a juvenile with green and red ectopic cells (arrows). g, Time-lapse frames of Supplemental Movie 2 (minutes indicated) shows the migration of reprogrammed a9.49 cell labeled with Tyr>mCherry (arrowhead). Scale bars, 50 μm.
The mesenchymal properties of neural crest were proposed to be the last features to appear during its evolution3,28. Our studies of the non-vertebrate chordate Ciona intestinalis support this hypothesis. We propose that cephalic neural crest arose from the co-option of one or more mesenchyme determinants (e.g. Twist) in a rudimentary neural crest cell type. Thus, this enigmatic cell population should not be considered a vertebrate innovation but rather an elaboration of an ancestral chordate gene network.
Methods
Embryo preparation and imaging
Ciona intestinalis adults were obtained, in vitro fertilized, and electroporated for transient transgenesis as described29. For each electroporation, typically 70 μg of DNA was resuspended in 100 μl buffer. Embryos were fixed at the appropriate developmental stage for 15 minutes in 4% formaldehyde. The tissue was then cleared in a series of washes of 0.01% Triton-X in PBS. Actin was stained overnight with Alexa-647-conjugated phalloidin at a dilution of 1/500. Samples were mounted in 50% glycerol in PBS with 2% DABCO for microscopy. Differential interference contrast microscopy was used to obtain transmitted light micrographs with a Zeiss Axio Imager A2 using the 40× EC Plan Neofluar objective. Confocal images were acquired on a Zeiss LSM 700 microscope using a plan-apochromat 20× or 40× objective. Confocal stacks contained approximately 50 optical slices at a thickness of 1-2 μm each. Images were rendered in 3D using Volocity 6 with the 3D opacity visualization tool. For time-lapse microscopy, larvae and juveniles were anesthetized in artificial seawater supplemented with 0.04% tricaine mesylate in glass bottom dish. Time-lapse images were taken on a Zeiss LSM 700 microscope at intervals of 3-4 minutes.
Molecular cloning
The University of California Santa Cruz Genome Browser Gateway facilitated the identification of conserved non-coding sequences between Ciona intestinalis and Ciona savignyi. Primers (Supplementary Table 1) were used to PCR amplify these putative enhancer sequences which were cloned into a pCESA vector using either AscI/NotI restriction sites for Prop1, Twist, and βγ-crystallin or AscI/XhoI for Mitf. The Mitf enhancer sequence was cloned 5’ of a basal FOG promoter. The LacZ coding sequence of the pCESA vector was replaced with UNC-76 GFP, UNC-76 mCherry, H2B mCherry, eGFP, or Kaede. The ZicL, Msx, FoxD, Tyr, TRP, Brachyury, and DMBX enhancers have been previously described10,13,19,29,30. A similar cloning strategy was used to create misexpression vectors using Not1/EcoR1 sites for control group A bHLH genes, Wnt7, FoxD, and FoxD N-term or NotI/BlpI sites for stabilized β-catenin and Twist (Supplementary Table 1). The FoxD:DBD:WRPW coding sequence was made by amplifying the DBD of FoxD (Supplementary Table 1), which was subcloned into a pCESA vector containing an HA:NLS peptide and a WRPW repressor motif using NheI/SpeI sites. Additional coding sequences for Ets:VP16, Ets:WRPW, and dnTCF, were subcloned from existing expression vectors13.
In situ hybridization and immunohistochemistry
The double fluorescent in situ hybridizations and immunohistochemistry were performed as described29 using linearized cDNA clones for Wnt7 (cilv33g04), FoxD (citb8o13), ERG (cilv04i11), Mech2 (cicl04m09), and Twist (cicl20p07) from Nori Satoh's (OIST, Okinawa, Japan) cDNA library.
Kaede lineage tracing
Embryos electroporated with Tyr>Kaede and co-electroporated with Mitf>LacZ or Mitf>Twist were developed in the dark until the late tailbud stage. Tailbuds expressing Kaede were then photoconverted with UV using the DAPI filter on a Zeiss Stereo Lumar.V12 for 3 minutes. Embryos were then continuously reared in the dark to the juvenile stage and prepared for imaging.
Supplementary Material
Acknowledgements
We thank A. Stolfi for his continued support and guidance, Y. Satou for isolating the Twist enhancer, N. Ellis for cloning DMBX>Twist, and B. Gainous for critical reading of the manuscript. P.B.A is supported by a graduate fellowship from the NSF. This work was supported by a grant from the NIH (NS 076542).
Footnotes
Author Contributions P.B.A designed and performed most experiments in consultation with M.L. E.W. isolated the cis regulatory element for the βγ-crystallin reporter and made the stable β-catenin transgene. I.A.N. examined Mech2 and ERG expression in wild-type and reprogrammed tailbuds. P.B.A., M.L., and E.W. wrote the manuscript.
The authors declare no competing financial interests.
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Bronner ME, Le Douarin NM. Evolution and development of the neural crest: An overview. Dev. Biol. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Douarin NM, et al. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- 3.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 4.Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008;18:1127–1132. doi: 10.1101/gr.076208.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 6.Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–699. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- 7.Jeffery WR. Ascidian neural crest-like cells: phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J. Exp. Zool. B Mol. Dev. Evol. 2006;306:470–480. doi: 10.1002/jez.b.21109. [DOI] [PubMed] [Google Scholar]
- 8.Jeffery WR, et al. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: Insights into the ancestry and evolution of the neural crest. Dev. Biol. 2008;324:152–160. doi: 10.1016/j.ydbio.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Tassy O, et al. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 2010;20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo MT, et al. Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 2004;267:517–528. doi: 10.1016/j.ydbio.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 12.Wada H, Makabe K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int. J. Biol. Sci. 2006;2:133–141. doi: 10.7150/ijbs.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squarzoni P, Parveen F, Zanetti L, Ristoratore F, Spagnuolo A. FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development. 2011;138:1421–1432. doi: 10.1242/dev.057323. [DOI] [PubMed] [Google Scholar]
- 14.Curran K, et al. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yajima I, et al. Cloning and functional analysis of ascidian Mitf in vivo: insights into the origin of vertebrate pigment cells. Mech. Dev. 2003;120:1489–1504. doi: 10.1016/j.mod.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Nishida H, Satoh N. Determination and regulation in the pigment cell lineage of the ascidian embryo. Dev. Biol. 1989;132:355–367. doi: 10.1016/0012-1606(89)90232-7. [DOI] [PubMed] [Google Scholar]
- 17.Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 18.Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–1858. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai KS, Satoh N, Satou Y. An essential role of a FoxD gene in notochord induction in Ciona embryos. Development. 2002;129:3441–3453. doi: 10.1242/dev.129.14.3441. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Bildsoe H, et al. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev. Biol. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 22.Soo K, et al. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev. Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- 23.Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- 24.Vincentz JW, et al. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev. Biol. 2008;320:131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokuoka M, Satoh N, Satou Y. A bHLH transcription factor gene, Twist-like 1, is essential for the formation of mesodermal tissues of Ciona juveniles. Dev. Biol. 2005;288:387–396. doi: 10.1016/j.ydbio.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Vlaeminck-Guillem V, et al. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech. Dev. 2000;91:331–335. doi: 10.1016/s0925-4773(99)00272-5. [DOI] [PubMed] [Google Scholar]
- 27.Horie T, et al. Ependymal cells of chordate larvae are stem-like cells that form the adult nervous system. Nature. 2011;469:525–528. doi: 10.1038/nature09631. [DOI] [PubMed] [Google Scholar]
- 28.Shimeld SM, Holland PWH. Vertebrate innovations. Proc. Natl. Acad. Sci. USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135:931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- 30.Stolfi A, Levine M. Neuronal subtype specification in the spinal cord of a protovertebrate. Development. 2011;138:995–1004. doi: 10.1242/dev.061507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.