Abstract
The textbook view of gene activation is that the rate-limiting step is the interaction of RNA polymerase II (Pol II) with the gene’s promoter. However, studies in a variety of systems, including human embryonic stem cells and the early Drosophila embryo, have begun to challenge this view. There is increasing evidence that differential gene expression often depends on the regulation of transcription elongation via the release of Pol II from the proximal promoter. I review the implications of this mechanism of gene activation with respect to the orderly unfolding of complex gene networks governing animal development.
Introduction
The first step in gene expression is the formation of the preinitiation complex at the promoter DNA (reviewed by Thomas and Chiang, 2006; Sikorski and Buratowski, 2009). The preinitiation complex consists of RNA polymerase II (Pol II) and associated general transcription factors (such as TFIID) that recognize specific DNA sequence elements within the core promoter, including TATA, initiator, and the downstream promoter element (DPE) (Figure 1). This complex is initially unstable given that the promoter DNA is in a closed, double-helix conformation. The next step in transcription initiation is the transition of this closed complex into an open complex, with local melting of the promoter DNA. This transition depends on the recruitment of TFIIH, a large multisubunit complex that is approximately the same size as Pol II itself (Kim et al., 2000). TFIIH binds downstream of Pol II and contains ATP-dependent helicases that open the double-stranded DNA. Pol II now initiates RNA synthesis at the +1 transcription start site. However, this synthesis is initially inefficient and often results in the production of short, aborted RNAs composed of less than 10 nucleotides. Only after Pol II succeeds in synthesizing RNAs that are more than 10 nucleotides in length is it competent to enter the main body of the gene and produce a full-length transcript. This promoter clearance (or escape) depends on the phosphorylation of the carboxyl-terminal region (CTD) of the large subunit of Pol II by TFIIH and other general transcription factors such as the Mediator complex (Hirose and Ohkuma, 2007; Boeing et al., 2010). As I discuss below, there is increasing evidence that Pol II often arrests just after promoter clearance, typically ~30–50 nucleotides downstream of +1 (reviewed by Gilmour, 2009). Release of this paused Pol II from the proximal promoter is emerging as a major mechanism of gene control in development.
Figure 1. The First Steps in Transcription Activation.
A transcription factor, such as TBP/TFIID, binds to specific promoter elements, including TATA. This leads to the recruitment of additional general transcription factors, including TFIIA, TFIIB, TFIIF, and ultimately, RNA polymerase II (Pol II). The initial binding is unstable, given that the promoter complex is in a closed conformation. Recruitment of TFIIH leads to the formation of an open complex and the onset of transcription. Stable transcription depends on the phosphorylation of the Pol II C-terminal region (CTD), which fosters promoter escape. Adapted from Gilmour (2009).
Pol II Occupancy prior to Gene Expression
Some of the first evidence that release of promoter-associated Pol II might be a global mechanism of gene control came from studies in mouse embryonic stem (ES) cells (Bernstein et al., 2006; Guenther et al., 2007; Bilodeau et al., 2009). In these studies, the distributions of two different isoforms of histone H3, trimethylated lysine 4 (H3K4) and trimethylated H3K27, were assessed genome-wide. Earlier studies suggested that the former histone modification is associated with the promoter regions of actively transcribed genes (reviewed by Mohan et al., 2010), whereas the latter modification correlates with inactive genes (as in Müller et al., 2002). Many inactive genes in mouse ES cells contain both modifications, the so-called “bivalent mark,” indicative of genes that are active and inactive.
The occurrence of trimethylated H3K4 in the promoter regions of nonexpressed genes was interpreted to suggest that Pol II had access to their promoter regions (Guenther et al., 2007). Thus, genes containing bivalent marks were seen as repressed but “poised” for rapid activation once the proper inducing signals become available. Such a state of repression might be important for the orderly differentiation of specific cell types derived from multipotent progenitor cells.
Interactions of Pol II with the promoter regions of nonexpressing genes have been inferred by the occurrence of trimethylated H3K4. More definitive evidence was obtained by analyzing the genome-wide distribution of Pol II (Guenther et al., 2007). Whole-genome ChIP-chip assays were done using antibodies directed against different Pol II isoforms. These studies confirmed Pol II occupancy at the promoter regions of many inactive genes in the early Drosophila embryo (Zeitlinger et al., 2007; see Figure 2).
Figure 2. RNA Polymerase II Binding at Developmental Patterning Genes.
RNA polymerase II (Pol II) binding at the proximal promoter regions of developmental patterning genes, including sog (A), which encodes a bone morphogenetic protein inhibitor, and sim (B), which specifies the ventral midline of the central nervous system.
(A) Pol II chromatin immunoprecipitation (ChIP)-chip, genome-wide run on assays (Gro-Seq) and ChIP-Seq assays using extracts from 2–4 hr mutant embryos that lack a dorsal nuclear gradient and contain only dorsal ectoderm (Zeitlinger et al., 2007). Although sog is silent in these embryos, Pol II is clearly bound to the promoter region of the gene.
(B) An ~10 kb region of the Drosophila genome, which contains the linked sim and timeout genes. ChIP-chip assays identified Pol II binding in the promoter region of sim, but not timeout, in early, 2–4 hr embryos. Pol II binding is seen at the sim promoter in pipe and rm9/rm10 mutants, which produce only dorsal ectoderm and neurogenic ectoderm, respectively. No binding is observed in Toll10b mutants, which produce only mesoderm (Zeitlinger et al., 2007).
Whole-genome binding assays suggested that 10%–30% of the inactive genes in any given cell type contain Pol II bound to their promoter regions. For example, Pol II ChIP-chip assays (combining chromatin immunoprecipitation and microarray analysis) were done with pipe mutant embryos in Drosophila (Zeitlinger et al., 2007). These mutants produce just a single cell type, the dorsal ectoderm (Stein et al., 1991). Lateral regions of the embryo that normally form neurogenic ectoderm and ventral regions that give rise to mesoderm are transformed into dorsal ectoderm. Approximately 1000 inactive protein-coding genes (~7% of all such genes in the genome) exhibit Pol II binding in these mutants (Figure 2A). These genes are normally expressed in the mesoderm or neurogenic ectoderm but are inactive in pipe mutants.
In Drosophila, inactive genes containing promoter-associated Pol II are significantly overrepresented by developmental control genes. Many Hox genes, tissue determinants (such as tinman, a heart specification gene, and sim, which specifies the ventral midline of the central nervous system), and components of cell signaling pathways (for instance, genes encoding the fibroblast growth factor receptor and bone morphogenetic protein inhibitors) contain Pol II in the early embryo, prior to their activation later in development (Zeitlinger et al., 2007; Muse et al., 2007; Chopra et al., 2009a; see Table 1). Indeed, the first evidence that the release of paused Pol II is a critical mechanism of gene regulation in Drosophila development came from the analysis of three critical segmentation genes, slp1, engrailed, and wingless (Wang et al., 2007). Why are such genes “juiced” by the preloading of Pol II? I will first consider the different forms of Pol II that might be represented by whole-genome ChIP assays.
Table 1.
Drosophila Dorsal-Ventral Patterning Genes
| Gene | Function | Developmental Role | Pause Status |
|---|---|---|---|
| twist | transcription factor | mesoderm determinant | not paused |
| snail | transcription factor | mesoderm determinant | paused |
| wntD | Wnt ligand | dorsal-ventral patterning | not paused |
| heartless | FGF receptor | mesoderm migration and patterning | paused |
| heartbroken | FGF signaling | mesoderm migration and patterning | paused |
| mdr49 | multidrug resistance | berm cell migration | paused |
| single-minded | transcription factor | CNS ventral midline | pauseda |
| vein | EGF ligand | CNS patterning | ambiguous |
| rhomboid | EGF signaling | CNS patterning | ambiguous |
| brinker | transcription factor | inhibits BMP (Dpp) signaling | paused |
| short gastrulation | BMP (Dpp) Inhibitor | dorsal ectoderm | paused |
| thisbe | FGF8 ligand | mesoderm migration and patterning | not paused |
| intermediate neurogenic defective (ind) | transcription factor | CNS patterning | paused |
| drop (msh) | transcription factor | CNS patterning | paused |
| soxN | transcription factor | CNS patterning | ambiguous |
| tinman | transcription factor | heart patterning | paused |
| bagpipe | transcription factor | heart patterning | pauseda |
| ladybird (early) | transcription factor | heart patterning | paused |
| branchless | FGF ligand | trachea development | ambiguous |
| breathless | FGF receptor | trachea development | not paused |
| treachealess | transcription factor | trachea development | paused |
Paused in some, but not all, tissues of the early embryo.
Paused Pol II at Drosophila Heat Shock Genes
It is conceivable that Pol II binds and dissociates from the template DNA in a dynamic manner so that ChIP assays represent average occupancy of an unstable Pol II (reviewed by Fuda et al., 2009; Gilchrist et al., 2009). Another possibility is that Pol II binds and engages the DNA template but dissociates after producing short, aborted transcripts due to a failure of promoter clearance (see above). Alternatively, Pol II might be fully engaged and stably bound to the template DNA. These different forms of Pol II are sometimes referred to as “stalled” Pol II, a generic term for polymerase that is transcriptionally engaged but prevented from elongating (see Arndt and Kane, 2003). One particular form of stalled Pol II, paused Pol II, is particularly interesting from the standpoint of rendering genes “poised” for rapid and efficient induction in development, as I discuss below.
Paused Pol II binds the DNA template, undergoes promoter escape or clearance, and then stably pauses downstream of the transcription start site after producing a short nascent transcript, typically 30–50 bp in length (e.g., Nechaev et al., 2010; Figure 3). Evidence for this form of stalled Pol II, “promoter-proximal” paused Pol II, was first obtained more than 20 years ago for the Drosophila heat shock genes (e.g., Gilmour and Lis, 1986; Rougvie and Lis, 1988; O’Brien and Lis, 1991). There was also evidence that other genes, including human c-Myc and HIV early genes, were similarly controlled by Pol II elongation rather than by initiation (Bentley and Groudine, 1986; Kao et al., 1987; Krumm et al., 1992; Strobl and Eick, 1992; Marshall and Price, 1992). However, this mode of gene control was generally regarded by the research community to represent a specialized mechanism, primarily employed by genes that are subject to regulation by stress such as heat shock. In fact, recent whole-genome methods suggest that paused Pol II is a common feature of gene regulation in development (Core et al., 2008; Nechaev et al., 2010).
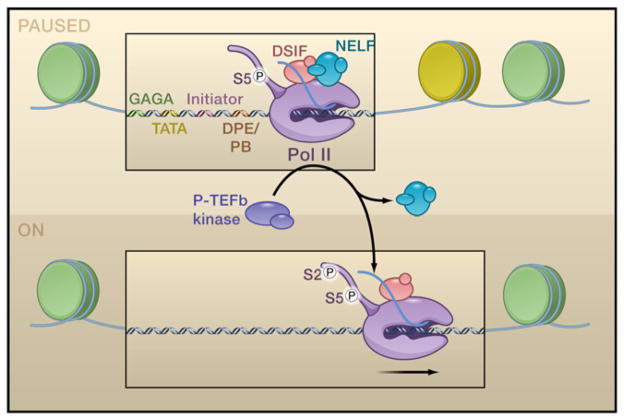
Figure 3. Paused Pol II and Its Release from the Proximal Promoter.
The top panel shows a promoter DNA template with paused RNA polymerse II (Pol II). The promoter region contains sequence elements that foster binding and activation of Pol II, including GAGA, TATA, initiator, and downstream promoter element/pause button (DPE/PB) motifs. Pol II is typically paused just downstream of the DPE region. It has undergone promoter escape and contains phosphorylation of serine 5 (Ser5) in the C-terminal domain (CTD). DSIF (5,6-dichloro-1-β-D-ribofuranoxylbenzimidazole sensivitiy-inducing factor) and NELF (negative elongation factor) help to arrest Pol II by binding to the nascent transcript (typically 30–50 nucleotides in length). Recruitment of P-TEFb (positive transcription elongation factor b) causes the release of NELF and the phosphorylation of Ser2 in the CTD, resulting in Pol II procession into the main body of the gene.
Systematic studies on the regulation of Drosophila heat shock genes, particularly hsp70, led to a detailed understanding of the nature of paused Pol II (reviewed by Fuda et al., 2009). There are a few key features that are relevant to the discussion of the importance of this mode of gene control in development.
First, Pol II is stably paused in promoter-proximal locations, typically +30 to +50 nucleotides downstream of the +1 transcription start site (see, for example, Nechaev et al., 2010). Recent studies suggest that Pol II often arrests downstream of one of the core promoter elements, the DPE, located between +28 bp and +32 bp downstream of the +1 transcription start site (Juven-Gershon et al., 2008). In some cases, the last nucleotides of the nascent transcript associated with paused Pol II correspond to the DPE or a specialized version of this motif called the PB (pause button), CGRWCG (Hendrix et al., 2008; Nechaev et al., 2010; Gilchrist et al., 2010; see Figure 3).
Second, it is estimated that there is just one stably bound Pol II complex bound per DNA template (see Lis, 2007). The short nascent transcripts that are associated with paused Pol II often contain the 5′ CAP modification that is typical of mature mRNAs. In addition, paused Pol II is sufficiently stable to permit detection of an associated “transcription bubble,” single-stranded DNA arising from the action of Pol II (Giardina et al., 1992; Krumm et al., 1992; Li et al., 1996; see below).
Finally, paused Pol II tends to be phosphorylated on serine-5 (Ser5P) of the heptad repeat comprising the C terminus of the large subunit of the Pol II complex (as in Boehm et al., 2003). Ser5P modification is mediated by the Cdk7 kinase located within the TFIIH complex (see Lu et al., 1992; Glover-Cutter et al., 2009; Akhtar et al., 2009).
Release of Pol II from the proximal promoter is associated with the phosphorylation of serine-2 (Ser2P) within the heptad repeats (Boehm et al., 2003; Keogh et al., 2003). This release depends on positive transcription elongation factor b (P-TEFb) (Boehm et al., 2003), which consists of the Cdk9 kinase and its regulator, cyclin T (Marshall and Price, 1995; Peng et al., 1998). Cdk9 either directly phosphorylates Pol II Ser2 or indirectly facilitates this phosphorylation (see below). Recruitment of Cdk9 is often sufficient to activate gene expression (reviewed by Peterlin and Price, 2006).
The recruitment of Pol II to the DNA template emerged as the dominant form of gene regulation based on in vitro transcription assays conducted in the 1980s and 1990s (see, for example, Stargell and Struhl, 1996; Levine and Tjian, 2003). There is little doubt that this mode of regulation is an important mechanism of gene activation. However, recent whole-genome assays suggest that the release of paused Pol II is also crucial for developmental gene control and may be employed by about one-third of the genes in a typical metazoan genome.
Identification of Paused Pol II Genome-wide
As discussed earlier, Pol II ChIP assays do not distinguish among the different forms of promoter-associated Pol II. Additional methods are required to identify paused Pol II. For example, a “transcription bubble,” a 15–20 bp region of single-standed DNA, arises in the immediate “wake” of the advancing Pol II complex during transcription (Figure 1). The bubble is stabilized by paused Pol II and can be identified based on the hypersensitivity of exposed A and T residues to oxidizing agents such as potassium permanganate (see Gilmour and Fan, 2009). The combination of Pol II ChIP binding assays and permanganate protection assays can provide compelling evidence for engaged Pol II (Zeitlinger et al., 2007; Muse et al., 2007). However, it is not yet possible to perform permanganate protection on a whole-genome scale, and consequently, this method can only be used to support the occurrence of pausing at select loci.
Recently, two methods have been developed for the whole-genome identification of paused Pol II, direct sequencing of the 30–50 nucleotide nascent transcripts that are associated with paused Pol II and genome-wide nuclear run-on assays (Gro-Seq) (Core et al., 2008; Nechaev et al., 2010). Both methods have been used with considerable success to identify paused Pol II in cultured cells, including Drosophila S2 and GL3 cells, as well as mammalian ES cells and human lung fibroblasts.
The former method relies on the deep sequencing of short nuclear RNAs. A critical trick for the successful application of this method is the enrichment of RNAs containing 5′ CAP modification (Nechaev et al., 2010). Once isolated, these transcripts are subject to extensive sequencing. An advantage of this technique is the identification of the exact location of the paused Pol II. As discussed earlier, in Drosophila, Pol II often arrests immediately after transcription of the first ~30 nucleotides of the transcription unit, just beyond the DPE (or PB). However, deep sequencing of small nuclear RNAs does not demonstrate the occurrence of paused Pol II, given that short RNAs can arise from termination or RNA-processing events.
A more reliable method for the genome-wide identification of paused Pol II is Gro-Seq. This method depends on the brief labeling (for instance, with biotin-tagged uracil) of nascent RNAs produced solely by engaged Pol II that are already present on the DNA template (Core et al., 2008). The isolated RNAs are subsequently sequenced and mapped onto whole-genome assemblies. As seen for Pol II ChIP-Seq assays in Drosophila embryos and mammalian ES cells, Gro-Seq assays suggest that ~30% of all protein-coding genes contain paused Pol II in cultured human lung fibroblasts.
The Gro-Seq method provides very high-resolution mapping of paused Pol II, given that nascent transcripts are subject to short pulses of labeling (~50–100 nucleotides). The enhanced resolution of this method, as compared with traditional ChIP-chip and ChIP-Seq methods, revealed an unexpected finding. Many, and perhaps most, paused mammalian promoters are associated with closely linked upstream, divergent transcripts units, which produce short, apparently unstable RNAs (Core et al., 2008; see also Seila et al., 2008; Preker et al., 2008). The function of these divergent transcription units is unknown, but it has recently been suggested that they might serve as docking sites for Polycomb repressors (Guenther and Young, 2010). Thus far, invertebrates such as Drosophila might contain a few such antisense transcription units, but not nearly on the scale seen for mammalian tissues.
How Many Paused Genes?
This question of the number of paused genes in a typical genome is somewhat controversial, partly due to inconsistencies in definition. Whole-genome ChIP assays suggest that Pol II has access to many inactive genes prior to the onset of de novo transcription (see Guenther et al., 2007). However, it is conceivable that just a subset of these genes contains stably paused Pol II with all of the features documented for hsp70. A stringent definition of pausing is the association of activated Pol II (marked by Ser5 phosphorylation) between 30 and 50 bp downstream of the +1 transcription start site, containing a nascent transcript with the 5′ CAP modification. Moreover, there is at least a 10-fold increase in the levels of Pol II in promoter-proximal regions as compared with the main body of the gene (a “stalling index” of 10 or more; see Zeitlinger et al., 2007; Muse et al., 2007). As discussed earlier, perhaps as many as one-third of all genes in a typical genome fulfill criteria for pausing and exhibit regulation via Pol II elongation at some point in the organism’s life cycle (Core et al., 2008).
For the Drosophila embryo, I consider a subset of paused genes, containing activated Pol II in promoter-proximal regions of tissues where the gene is known to be genetically inactive. For example, the snail gene is selectively activated in the presumptive mesoderm of the early Drosophila embryo (Zeitlinger et al., 2007). It is not significantly expressed in the dorsal ectoderm even though conventional Pol II ChIP-chip and Gro-Seq assays indicate Pol II occupancy and pausing at the snail promoter in this tissue (Zeitlinger et al., 2007). In these assays, it is possible to obtain homogenous, mutant embryos that produce just a single tissue, dorsal ectoderm. When these mutants are used for ChIP-chip, ChIP-Seq, and Gro-Seq assays, it is possible to identify all of the genes containing paused Pol II that are known to be silent in this tissue (see Figure 2A).
A reasonable guess is that there are several hundred genes selectively expressed in each of the primary presumptive tissues of the early Drosophila embryo, namely, gut, mesoderm, neurogenic ectoderm, and dorsal ectoderm. Thus, ~1000 genes are subject to stringent, tissue-specific de novo transcription. Conservatively, at least half of these genes contain paused Pol II in tissues where they are inactive (see Table 1). That is, at least 100 of the ~200 mesoderm-specific genes contain paused Pol II in the presumptive gut and/or ectoderm. Therefore, the release of paused Pol II is a crucial mechanism of gene control in Drosophila development. A current estimate is that at least half of all developmental control genes (such as Hox genes and components of cell signaling pathways, etc.) contain paused Pol II and are primarily regulated by release and elongation rather than binding and activation of Pol II at the core promoter (Table 1).
Mechanisms of Pol II Pausing
How does Pol II come to be paused in the proximal promoter? As mentioned earlier, the DPE located at +28 bp downstream of the +1 transcription start site, and a related sequence motif, the PB, might contribute to the pausing of activated Pol II within proximal promoters (Hendrix et al., 2008; Gilchrist et al., 2010). Another short sequence motif, the GAGA element, has also been implicated in promoter-proximal pausing (Lee et al., 1992; Wilkins and Lis, 1997) (Figure 3). GAGA is the binding site for the Trithorax-like (Trl) protein complex, which can decondense chromatin through the recruitment of NURF, an ATP-dependent enzymatic complex that displaces nucleosomes (Tsukiyama and Wu, 1995; Okada and Hirose, 1998). A GAGA element in the immediate 5′ flanking region of the Drosophila hsp70 promoter was shown to be important for paused Pol II (Wilkins and Lis, 1997). Its removal does not block induction of the modified hsp70 promoter, but it no longer contains paused Pol II. The mechanism by which Trl fosters pausing is uncertain. Perhaps GAGA helps to shift the balance between Pol II binding and nucleosome assembly at the core promoter in favor of Pol II (see Gilchrist et al., 2010).
The promoter regions of many paused genes contain a number of GC-rich sequence motifs, including the PB (as in Hendrix et al., 2008; Nechaev et al., 2010; Figure 3). Perhaps these sequences confer an energy barrier for the melting of the double helix by elongating Pol II and thereby function as “speed bumps” to impede Pol II movement. The resulting “dwelling” of Pol II might permit binding of DSIF (5,6-dichloro-1-β-D-ribofuranoxyl-benzimidazole sensivitiy-inducing factor) and NELF (negative elongation factor) to the nascent transcript, resulting in its stable association in the proximal promoter (as seen by Wu et al., 2005; Peterlin and Price, 2006; Missra and Gilmour, 2010). Recent studies suggest that the Spt5 subunit of DSIF binds nascent transcripts as they emerge from elongating Pol II (Winston, 2001; Missra and Gilmour, 2010). DSIF subsequently recruits NELF, and the two complexes appear to serve as a sort of brake to block procession of Pol II beyond the pause site (Wu et al., 2005; Gilchrist et al., 2008; Rahl et al., 2010).
Regulation of Pol II Pausing
It is conceivable that a variety of sequence-specific regulatory factors also contribute to the establishment of paused Pol II. Perhaps transcription factors such as Zelda, which has been implicated in the maternal/zygotic transition in the early Drosophila embryo (Liang et al., 2008), are important for pausing but are not sufficient to trigger Pol II release and gene activation. Zelda binds to a specific sequence motif, CAGGTAG, detected in a variety of developmental control genes, including those engaged in segmentation and dorsal-ventral patterning (ten Bosch et al., 2006). A purely speculative suggestion is that Zelda helps to prepare embryos for zygotic gene expression by recruiting Pol II to paused promoters. The main idea here is that some transcription factors might be essential for recruitment of Pol II, whereas others work subsequently to release paused Pol II.
It is currently unclear whether recruitment of paused Pol II is used as a regulatory strategy in development. For example, genes that are selectively activated in muscle cells may be paused throughout the early embryonic mesoderm, but not in tissues where the genes remain silent, such as the ectoderm. That is, Pol II pausing can be used to restrict the developmental potential of embryonic tissues. There are some cases of tissue-specific pausing. For example, the sim gene is the major determinant of a specialized region of the Drosophila cental nervous system, the ventral midline (Nambu et al., 1991; Wheeler et al., 2009). It is activated in the ventral-most regions of the neurogenic ectoderm by a Notch signal emanating from the adjacent presumptive mesoderm (Morel and Schweisguth, 2000; Cowden and Levine, 2002). sim contains paused Pol II in the ectoderm, but not mesoderm where the gene is never expressed (Figure 2B).
Does paused Pol II anticipate the activation of gene expression? Perhaps the induction of tissue-specific gene expression is a two-step process: recruitment and then release of Pol II from the proximal promoter. As mentioned earlier, there are ~1000 paused genes containing a stalling index of 10 or more in the early Drosophila embryo (Zeitlinger et al., 2007). Most of these genes will be activated within the next several hours of development. It does not make sense to establish paused Pol II in the early embryo for genes that are activated far later in the life cycle, such as the adult brain. Perhaps such genes become paused during larval or pupal development in preparation for efficient activation at the next phase of the life cycle.
Mechanisms of Pol II Release
As discussed earlier, P-TEFb is essential for the release of paused Pol II from the proximal promoter (reviewed by Peterlin and Price, 2006; Price, 2008). It either phosphorylates serine-2 (Ser2P) on the heptad repeats contained in the C-terminal region of the large subunit of Pol II or facilitates this phosphorylation by modifying NELF and DSIF (see Figure 3). Recent studies suggest that P-TEFb leads to the loss of NELF from the Pol II pausing complex, thereby permitting release of Pol II via Ser2 phosphorylation by another kinase, Cdk12 (Bartkowiak et al., 2010). Regardless of the detailed mechanism, at least a subset of developmental transcription factors, such as Dorsal, might activate gene expression by recruiting P-TEFb. This is a conceptually simpler mode of gene activation than Pol II recruitment, which depends on several large multisubunit complexes, including Mediator and TFIID (e.g., Levine and Tjian, 2003).
P-TEFb is a component of several multisubunit complexes, including SEC (super-elongation complex) (Lin et al., 2010; Mohan et al., 2010). In addition to P-TEFb, SEC contains AFF4, ELL, and several additional protein subunits. Each of these SEC subunits has been implicated in severe human leukemias arising from their fusion with MLL, a component of the trithorax H3K4 methyltransferase complex (Mohan et al., 2010). It has been proposed that these MLL-SEC fusion proteins result in “runaway” transcription of genes that are normally subject to stringent control in developing lymphocytes. SEC is a prime candidate for the regulated release of paused Pol II and subsequent processivity of Pol II through the DNA template during gene activation.
P-TEFb and other components of the SEC have also been implicated in the expression of HIV genes (see, for instance, Zhang et al., 2007; He et al., 2010). The LTR promoter of HIV contains paused Pol II, which is released by the interaction of the Tat activator with the nascent RNA associated with this pausing complex (Canduri et al., 2008). Tat has been shown to recruit P-TEFb, leading to the release of paused Pol II in the HIV LTR and efficient expression of viral genes (Zhu et al., 1997; He et al., 2010).
A link between the SEC and Pol II elongation in development is suggested by genetic studies in Drosophila. The SEC component AFF4 corresponds to lilliputian (lilli), which is essential for embryonic patterning; lilli mutants exhibit a variety of developmental defects, including a severe pair-rule phenotype (Wittwer et al., 2001; Vanderzwan-Butler et al., 2007). Moreover, another component of the SEC, ELL, exhibits a strong genetic interaction with the homeotic gene, Ubx, which contains stalled Pol II in both embryos and wing imaginal disks (Smith et al., 2008; Chopra et al., 2009a).
Speed of Induction
As discussed earlier, there are at least several hundred paused genes in the early Drosophila embryo (Zeitlinger et al., 2007; see Table 1). These include Hox genes, tissue determinants such as snail and sim, and genes encoding components of cell signaling pathways such as Heartless (a fibroblast growth factor receptor) (Leptin and Affolter, 2004) and Sog (a BMP inhibitor) (Wang and Ferguson, 2005). Why do such genes contain paused Pol II?
Classical studies of the heat shock genes suggest that paused Pol II significantly accelerates the timing of induction in response to stress as compared with comparable promoters lacking Pol II (see Lee et al., 1992; Wilkins and Lis, 1997). In the case of heat shock genes, this makes sense. Either these genes are rapidly induced to confer resistance to stress or the organism is dead. Perhaps it is similarly advantageous for developmental control genes. Either these genes are quickly and efficiently activated in response to appropriate inducing signals or the embryo is dead. Many of these signals—for example, a localized fibroblast growth factor or epidermal growth factor ligand—are only transiently expressed during development.
Paused Genes Are “Open” for Business
The preceding arguments apply to the release of the single promoter-associated Pol II in promoter-proximal regions of paused genes. Is the release of this one Pol II really sufficient to influence the dynamics of gene activation during development? In other words, it makes sense that a single primary transcript is produced more rapidly from a paused gene as compared with a nonpaused gene that must recruit Pol II de novo. However, it certainly takes more than a single primary transcript to produce meaningful levels of gene expression. It is reasonable to assume that at least several tens of transcripts must be produced to achieve sufficient levels of the corresponding protein to execute cellular functions such as transcriptional activation or repression of subordinate target genes.
How can the release of one paused Pol II facilitate another 10–30 rounds of transcription to achieve meaningful gene expression? Different types of mechanisms can be envisioned. For example, release and procession of the lone Pol II complex might lead to the decondensation of the associated transcription unit, thereby facilitating subsequent rounds of transcription (see Saunders et al., 2006). According to this view, paused Pol II “lubricates” the gene for more efficient transcription due to the intrinsic decondensing activities of the Pol II complex, such as histone acetyltransferases (see Timmers and Tora, 2005). As discussed below, paused Pol II might be more effective at rendering chromatin accessible for subsequent rounds of transcription than constitutive Pol II.
This lubricating action of paused Pol II might transcend procession of the lone Pol II that is released from the promoter upon induction. A recent study suggests that induction of the paused hsp70 heat shock locus leads to the rapid decondensation of the hsp70 transcription unit before Pol II reaches the end of the gene (Petesch and Lis, 2008). Indeed, this remodeling occurs even when transcription is inhibited. These observations raise the possibility that paused genes contain a chromatin remodeling complex, which is also poised for rapid deployment upon induction.
Reduced levels of NELF often result in the loss of paused Pol II and silencing of the associated promoters (Gilchrist et al., 2008). This observation provides clear-cut evidence that paused Pol II keeps the promoter in an open conformation. Indeed, it has been suggested that paused Pol II serves as a “bookmark” for the future expression of a gene (Nechaev et al., 2010; Gilchrist et al., 2010). In particular, Pol II is thought to preclude the binding of positioned nucleosomes at the core promoter, rendering the gene in an open conformation. In contrast, genes lacking paused Pol II might instead contain such positioned nucleosomes, resulting in a relatively closed conformation (Gilchrist et al., 2010). According to this view, it is easier to recruit Pol II to the open form of the gene for subsequent rounds of transcription as compared with genes in a closed conformation due to the absence of Pol II and the presence of positioned nucleosomes.
Another mechanism that might facilitate transcription of paused genes is the recycling of Pol II. Following a transcription cycle, tagged Pol II complexes were found to reassociate with the promoter regions of the genes that they had transcribed rather than being released into the nuclear “void” (Yao et al., 2007). This recycling might help to ensure efficient occupancy of active Pol II at paused loci.
The underlying mechanism of Pol II recycling is unknown, but it might be related to the observation that paused promoters contain an insulator activity (Chopra et al., 2009b). When a gene containing paused Pol II is placed between a 5′ enhancer and downstream reporter gene, the reporter gene is inactive. In contrast, the downstream reporter gene is activated when the intervening paused gene is replaced by a nonpaused gene lacking Pol II. It is possible that this insulator activity is a manifestation of nuclear compartmentalization of paused genes and recycling of Pol II.
Paused Pol II and Transcriptional Noise
It has been proposed that paused Pol II suppresses transcriptional noise arising from the stochastic recruitment of Pol II to the core promoter (Boettiger and Levine, 2009). In vitro transcription assays and single-molecule studies using atomic force microscopy suggest that Pol II recruitment is somewhat variable and inefficient (reviewed by Herbert et al., 2008). About less than one in ten Pol II interaction events leads to productive transcription of the target gene. This mechanism of gene activation would not permit coordinate expression among groups of cells. Even if such cells received an inducing signal at the same time, there would be cell-to-cell variability in the onset of transcription due to the inherently stochastic nature of Pol II recruitment and engagement at the core promoter.
In contrast, genes containing paused Pol II should exhibit coordinate patterns of activation. Such genes have skipped the stochastic step of Pol II recruitment and contain paused Pol II in an active and readied state. If a group of cells receives the inducing signal at the same time, then they should release paused Pol II from promoter-proximal regions and thereby launch de novo transcription in a synchronous fashion. This idea was tested by visualizing the first nascent transcripts produced from paused and nonpaused genes in the early Drosophila embryo (Boettiger and Levine, 2009). In situ hybridization assays were done using intronic probes located within the first several hundred base pairs of the transcription start site. Genes containing paused Pol II tend to exhibit synchronous patterns of activation, whereas nonpaused genes display more variable or stochastic patterns of activation (see Figure 4).
Figure 4. Activation of Paused and Nonpaused Genes.
(Left) The Drosophila thisbe gene lacks binding of RNA polymerase II (Pol II), based on both chromatin immunoprecipitation assays (not shown) and Gro-Seq assays (shown). The gene exhibits a stochastic pattern of activation in the early embryo, with nascent transcripts detected in only about half of all nuclei that will ultimately express the gene. (Right) In contrast, the sog gene contains paused Pol II and exhibits a synchronous pattern of activation.
It is possible that the synchronous patterns of activation observed for paused genes represent a manifestation of rapid transcription. After all, paused genes are on a “hair trigger” and are poised for rapid induction. Rapid activation would give the appearance of synchronous induction. Moreover, it is unlikely that synchrony of expression is maintained over many transcription cycles, at least not for large genes. After Pol II is released from the pause site, it does not appear to progress through the transcription unit at a uniform pace. Rather, Pol II undergoes irregular fits and starts of procession and dwelling across the DNA template (Herbert et al., 2008; Galburt et al., 2009). Such irregularities in the rate of Pol II movement might desynchronize gene expression among neighboring cells. Nonetheless, it is certainly possible that synchrony is maintained for the expression of small genes during “bursts” of gene activation. Moreover, it is likely that paused Pol II fosters homogenous patterns of gene expression within coordinate groups of cells, even if there is a loss in transcriptional synchrony.
In addition to promoting synchronous patterns of gene activation among groups of cells, it is possible that paused Pol II helps to ensure stoichiometric expression of gene batteries that are engaged in a common developmental process. That is, paused Pol II might help to produce equivalent transcript levels for genes encoding different subunits of multicomponent protein complexes. For example, both tinman and bagpipe are paused in the early Drosophila embryo (L. Core and V. Chopra, personal communication). The encoded NK homeodomain proteins interact to form heterodimeric transcription complexes that are essential for heart development (Zaffran and Frasch, 2005). Perhaps pausing helps to ensure coordinate levels of Tinman and Bagpipe products.
Another potential mechanism for the suppression of transcriptional noise is repression by the paused Pol II complex. The complex is quite large, and consequently, successive Pol II complexes cannot be loaded more than once every ~75 bp along an active template (Kornberg, 2007). Given that paused Pol II is mainly found in promoter-proximal locations within the first 30–50 bp of the transcription unit, it might block the binding of additional Pol II complexes prior to the induction of gene expression. The consequence of such “steric repression” is a lower “background” of expression as compared with genes lacking paused Pol II (see Chopra et al., 2009a). According to this view, paused Pol II might produce a sharp on/off switch in transcription upon induction.
In summary, there are several possible advantages for the occurrence of paused Pol II at developmental control genes: rapid activation of gene expression, reliability of expression due to Pol II recycling, synchronous activation among groups of cells undergoing coordinate development, and suppression of “basal” or adventitious transcription prior to induction.
Pausing as a Pol II Checkpoint
Pausing might ensure “fitness” of the Pol II complex before it embarks on its journey through the gene. Consider the case of large genes, frequently encountered in vertebrate systems and occasionally seen in Drosophila. The Drosophila Antennapedia locus is ~103 Kb in length (Laughon et al., 1986). It contains several promoters, and most of these contain paused Pol II (Chopra et al., 2009a, 2009b). At the established rates of Pol II procession (see Saunders et al., 2006), it should take more than an hour for a single Pol II complex to reach the end of the gene after it is released from the distal-most promoter. Even infrequent errors could lead to a catastrophic breakdown in gene expression. For example, suppose that Pol II backtracked, slipped out of frame, or was released from the template at an average of once every 10–25 Kb. The probability of producing meaningful levels of Antp products in a given cell at a given time becomes quite low.
Pausing might provide an opportunity to modify Pol II so that it is less likely to make mistakes during the transcription of essential genes. Certain modifications, such as phosphorylation of Pol II subunits and association of essential subunits, might occur only at the promoter. This is the location of many critical general transcription factors and complexes such as Mediator and TFIID (see Figure 1 and Levine and Tjian, 2003). Whatever maturation of Pol II that is imposed by these factors cannot occur after Pol II is released from the promoter.
Pol II might continue to “dwell in” promoter-proximal regions of paused genes even after they are activated (Zeitlinger et al., 2007; Muse et al., 2007). This dwelling might last for several seconds or up to a minute during each round of transcription—long enough so that the Pol II released from the promoter is fully mature and competent to complete transcription. According to this view, the key step in gene activation continues to be release of Pol II from the pause site in the proximal promoter. As discussed earlier, it is not possible to load more than one complex every 75 bp on the DNA template. Thus, paused Pol II at the typical proximal pause site, ~30–50 bp downstream of the transcription start site, will preclude the binding of another Pol II complex. Thus, only after the paused Pol II is fully released is it possible for another Pol II complex to enter the promoter region and dwell at the pause site. After receiving the appropriate maturation signals, this Pol II will then be released from the pause site, clearing the way for yet another Pol II complex.
According to this scenario, pausing represents a transcriptional checkpoint by ensuring the orderly flow and release of fully activated and mature Pol II through the promoter region (see Gilchrist et al., 2010). The Pol II that is released from the pause site is “fit” and is likely to complete the transcription of even large genes. In contrast, Pol II complexes that bind the DNA template and immediately transcribe the gene without dwelling at a proximal pause site might be more prone to error due to insufficient maturation at the checkpoint.
In summary, I have reviewed two basic mechanisms of paused Pol II in development, enhanced speed of gene activation and suppression of transcriptional noise. Speed of induction is influenced by chromatin decondensation, recycling of Pol II, and ease of Pol II re-engagement due to the open (that is, nucleosome-free) status of the paused promoter region. The suppression of transcriptional noise stems from bypassing the initial recruitment of Pol II to the core promoter, which is an inherently noisy, or stochastic, process. In addition, paused Pol II appears to foster synchronous and homogenous patterns of gene expression within and among coordinate groups of cells. Altogether, pausing appears to serve as a critical checkpoint to ensure the precise and reliable action of Pol II during development.
Acknowledgments
I thank the NIGMS for grant support, members of my lab for their enthusiasm and insights, and the anonymous referees for their guidance in polishing this Review.
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt KM, Kane CM. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. J Biol Chem. 2010;285:188–196. doi: 10.1074/jbc.M109.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canduri F, Perez PC, Caceres RA, de Azevedo WF., Jr CDK9 a potential target for drug development. Med Chem. 2008;4:210–218. doi: 10.2174/157340608784325205. [DOI] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol. 2009a;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009b;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden J, Levine M. The Snail repressor positions Notch signaling in the Drosophila embryo. Development. 2002;129:1785–1793. doi: 10.1242/dev.129.7.1785. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt EA, Grill SW, Bustamante C. Single molecule transcription elongation. Methods. 2009;48:323–332. doi: 10.1016/j.ymeth.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Pérez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fargo DC, Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Fan R. Detecting transcriptionally engaged RNA polymerase in eukaryotic cells with permanganate genomic footprinting. Methods. 2009;48:368–374. doi: 10.1016/j.ymeth.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Young RA. Transcription. Repressive transcription. Science. 2010;329:150–151. doi: 10.1126/science.1193995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci USA. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Laughon A, Boulet AM, Bermingham JR, Jr, Laymon RA, Scott MP. Structure of transcripts from the homeotic Antennapedia gene of Drosophila melanogaster: two promoters control the major protein-coding region. Mol Cell Biol. 1986;6:4676–4689. doi: 10.1128/mcb.6.12.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Leptin M, Affolter M. Drosophila gastrulation: identification of a missing link. Curr Biol. 2004;14:R480–R482. doi: 10.1016/j.cub.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Li B, Weber JA, Chen Y, Greenleaf AL, Gilmour DS. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci USA. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyl-transferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T, Lis JT. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Hirose S. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol Cell Biol. 1998;18:2455–2461. doi: 10.1128/mcb.18.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark ..get set...go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci USA. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Stein D, Roth S, Vogelsang E, Nüsslein-Volhard C. The polarity of the dorsoventral axis in the Drosophila embryo is defined by an extracellular signal. Cell. 1991;65:725–735. doi: 10.1016/0092-8674(91)90381-8. [DOI] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Vanderzwan-Butler CJ, Prazak LM, Gergen JP. The HMG-box protein Lilliputian is required for Runt-dependent activation of the pair-rule gene fushi-tarazu. Dev Biol. 2007;301:350–360. doi: 10.1016/j.ydbio.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Ferguson EL. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature. 2005;434:229–234. doi: 10.1038/nature03318. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. MidExDB: a database of Drosophila CNS midline cell gene expression. BMC Dev Biol. 2009;9:56. doi: 10.1186/1471-213X-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2:S1006. doi: 10.1186/gb-2001-2-2-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer F, van der Straten A, Keleman K, Dickson BJ, Hafen E. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development. 2001;128:791–800. doi: 10.1242/dev.128.5.791. [DOI] [PubMed] [Google Scholar]
- Wu CH, Lee C, Fan R, Smith MJ, Yamaguchi Y, Handa H, Gilmour DS. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Frasch M. The homeodomain of Tinman mediates homo- and heterodimerization of NK proteins. Biochem Biophys Res Commun. 2005;334:361–369. doi: 10.1016/j.bbrc.2005.06.090. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Gilmour DS, Henderson AJ. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem. 2007;282:16981–16988. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]