Abstract
We review recently identified mechanisms of transcriptional control that ensure reliable and reproducible patterns of gene expression in natural populations of developing embryos, despite inherent fluctuations in gene regulatory processes, variations in genetic backgrounds and exposure to diverse environmental conditions. These mechanisms are not responsible for switching genes on and off. Instead, they control the fine-tuning of gene expression and ensure regulatory precision. Several such mechanisms are discussed, including redundant binding sites within transcriptional enhancers, shadow enhancers, and ‘poised’ enhancers and promoters, as well as the role of ‘redundant’ gene interactions within regulatory networks. We propose that such regulatory mechanisms provide population fitness and ‘fine-tune’ the spatial and temporal control of gene expression.
Keywords: enhancer, paused polymerase, pioneer factors, robustness, gene regulatory networks
Transcriptional precision
The basic mechanisms for switching genes on and off during development were intensively studied in the 1980s and 1990s. The enhancer was shown to play a key role in integrating complex regulatory information to generate cell-specific patterns of gene expression [1]. However, in natural populations enhancer–promoter interactions can be affected by changes in temperature and variations in genetic background, but the developmental program remains unperturbed. What is the basis for this stability in developmental programming?
Our central premise is that the mechanisms used to provide stability in gene expression in natural populations also produce greater precision in developmental patterning mechanisms. By transcriptional precision we refer to the formation of sharp borders of gene expression, the exact timing of gene activation, coordinate expression of groups of genes within a developing tissue, and homogenous expression of a given gene across a field of coordinately developing cells. The advent of whole-genome technologies and improved imaging methods has provided recent insights into more subtle aspects of differential gene activity, namely the reproducible deployment of developmental programs in natural populations.
Redundant genetic interactions
Genetic analysis of Drosophila embryogenesis led to a conceptual breakthrough in our understanding of animal development [2]. The subdivision of the embryo into a series of body segments was first envisaged to be a regulatory cascade or genetic pathway, with maternal determinants, such as Bicoid, that establish sequential patterns of gap gene expression, pair-rule stripes and ultimately, segment-polarity stripes of gene expression (e.g. [3]). This view of a sequential pathway gave way to one of gene networks, whereby both maternal and zygotic activators and repressors interact with complex enhancers to produce localized stripes of gene expression [4]. In recent years, gene networks have been visualized as complex wiring-diagrams [5].
Such networks often contain seemingly redundant interactions. Moreover, two related transcription factors are sometimes seen to activate the expression of downstream target genes in the same cells at the same time. Removal of one copy of the regulatory gene often fails to produce an obvious or fully penetrant phenotype. Nonetheless, the gene might augment population fitness, which is what natural selection ultimately acts on.
Redundant interactions in gene regulatory networks have been suggested to provide stability and precision in metazoan development [5]. An illustrative example is seen for gut specification in Caennorhabditis elegans [6]. The intestine is composed of 20 cells that arise from a single progenitor in the early embryo. Intestinal identity is specified by a simple regulatory network, beginning with the maternal deposition of skn-1 transcripts and culminating in the expression of elt-2, which activates hundreds of ‘target’ genes required for gut differentiation (Figure 1a).
Figure 1.
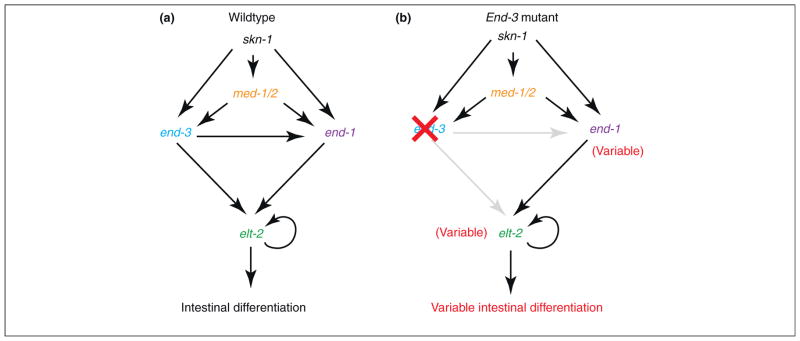
Redundant interactions in gene regulatory networks. Summary of the genetic cascade governing intestinal cell specification in C. elegans (see ref. [6]). (a) Wild-type network. skn-1 is maternally deposited and, in concert with other maternal and zygotic factors, activates the expression of transcription factors end-3 and end-1, both of which activate elt-2, the key regulator of intestine differentiation. (b) In end-3 mutants, end-1 can compensate and intestine differentiation is essentially normal. However, end-1 expression becomes significantly more variable, resulting in erratic expression of elt-2 and abnormal intestine differentiation in some individuals.
The activation of elt-2 depends on two related transcription factors, end-1 and end-3, which function in a largely redundant fashion. The consequences of disrupting either end-1 or end-2 gene activity have been examined, and evidence was obtained for increased ‘noise’ in gut specification from measurements of single mRNAs in individual embryos [6]. In particular, end-3 mutants show variability in both the timing and levels of elt-2 expression (Figure 1b), which might explain why 5% of end-3 mutants lack intestinal cells. Similarly, the overlapping activities of two T-box transcription factors, tbx-8 and tbx-9, appear to buffer stochastic variations in muscle differentiation [7].
These results suggest that redundant gene interactions within developmental networks can stabilize gene expression in natural populations. Such redundancy might also play a key role in ensuring transcriptional precision. That is, the combination of end-1 plus end-3 might ensure the precise timing and exact levels of elt-2 expression. There are numerous examples of potential redundancies in gene networks (e.g. [5]). Are these required for developmental patterning, or do they represent a means to stabilize complex processes despite genetic and environmental variations? These are not mutually exclusive concepts.
Intra-enhancer redundancy
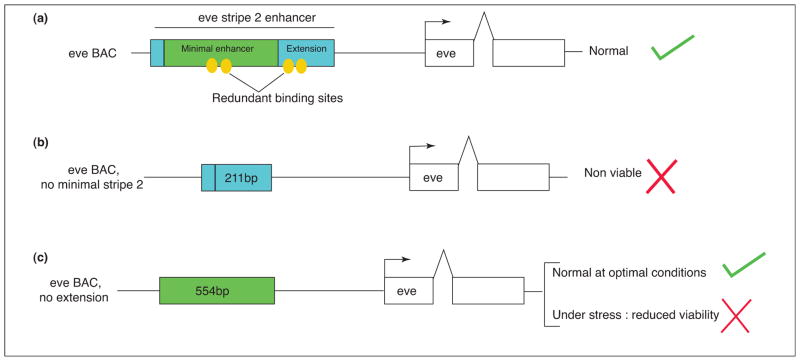
A typical developmental enhancer is several hundred bp in length and contains multiple binding sites for two or more sequence-specific transcription factors (reviewed in [1]). Some of the binding sites appear to be redundant, in that mutations in a subset of the sites do not qualitatively alter the expression patterns produced by the modified enhancers (e.g. [8]). What is the purpose of these ‘extra’ sites, which are often highly conserved? Evidence is gathering that in some cases they ensure robustness or stability in response to genetic and environmental variation. A recent analysis of the eve stripe 2 enhancer provides a particularly compelling example [9] (Figure 2).
Figure 2.
Importance of redundant binding sites for robustness. (a) Diagram of a BAC transgene containing the entire eve locus, including 5′ and 3′ stripe enhancers. Only the stripe 2 regulatory region is shown. The ‘full-length’ enhancer contains both the minimal ~500 bp enhancer (green) and ~200 bp 3′ extension (blue). The yellow ovals represent a subset of the TF binding sites in the stripe 2 regulatory DNA. (b) Removal of the minimal eve stripe 2 enhancer results in lethality, and embryos die with defects in the thorax (derived from the region of stripe 2 expression). (c) Removal of the 3′ extension does not impair embryogenesis under optimal culturing conditions, and normal adult flies are obtained. However, under genetic stress, only 5% of the flies survive. Thus, ‘redundant’ binding sites in the 3′ extension are required for robustness.
The full-length eve stripe 2 enhancer is over 700 bp in length and contains several binding sites for each of four key regulators: Bicoid, Hunchback, Krüppel, and Giant [10,11]. It produces a robust and authentic stripe 2 pattern when attached to a reporter gene and expressed in transgenic Drosophila embryos. Removal of ~200 bp from the 3′ end of the enhancer, which contains several TF binding sites, diminishes the levels of expression, but the resulting ~500 bp minimal enhancer produces an essentially normal pattern of expression [11]. BAC transgenesis and genetic complementation assays have been used to examine the contributions of the minimal enhancer and 3′ ‘extension’ [9].
Removal of the ~500 bp minimal enhancer from a ‘rescuing’ BAC transgene results in lethality due to a severely diminished stripe 2 pattern. Mutant eve−/eve− embryos carrying this BAC fail to hatch due to defects in the first thoracic segment. Interestingly, removal of the ~200 bp 3′ extension does not cause lethality under optimal culture conditions, and the viability of these flies is comparable to that of wild-type flies. These results suggest that the minimal ~500 bp eve stripe 2 enhancer is sufficient for segmentation, at least in the absence of environmental stress. However, there is a breakdown in the function of the minimal enhancer at elevated temperatures and in ‘sensitized’ genetic backgrounds. Thus, binding sites in the 3′ extension are not redundant under all conditions, but instead they appear to ensure reliable expression of eve stripe 2 under stress (Figure 2). This is likely to be a general mechanism of robustness or ‘canalization’ in development ([9,12,13] for a definition of canalization). So-called redundant binding sites in developmental enhancers are probably used in natural populations to cope with variability.
Shadow enhancers
A related mechanism for ensuring robustness is the use of multiple enhancers for a single pattern of gene expression. A variety of recently developed whole-genome assays (Box 1) permit the systematic identification of developmental enhancers (e.g. [14–16]). Such approaches suggest that many of the crucial developmental patterning genes in Drosophila are regulated by multiple enhancers that direct extensively overlapping patterns of gene expression and employ a similar regulatory ‘logic’ (e.g. [17]). The newly identified enhancers are sometimes termed ‘shadow enhancers’ because they map to more remote locations than the ‘classical’ or primary enhancers situated close to the gene [18,19]. Several examples are discussed below.
Box 1. Whole-genome identification of enhancers.
During the past 10 years a variety of ‘post-genome’ methods have been devised for the systematic identification of enhancers (which can exist both 5′ or 3′ of the gene or within the transcription unit). Transgenic assays are required to confirm their identities. Putative enhancers are attached to a minimal promoter and reporter gene, and introduced (via injection or electroporation) into a developing embryo. Either stable or transient transgenic embryos are assayed for reporter gene expression. Below we provide a brief review of some post-genome methods for identifying putative enhancers.
Computational methods: enhancers often contain a high density of transcription factor binding sites, typically one for every 30–50 bp across the length of the enhancer (200–300 bp or more). Algorithms have been developed for identifying high-density clusters of putative binding sites [58,59]. These methods work, but typically only 10–30% of ‘hits’ represent authentic enhancers when tested in transgenic embryos.
ChIP-Seq: permits the genome-wide identification of binding sites for sequence-specific transcription factors, or histone modifications (e.g. [40,41]). ChIP-Seq using antibodies against early Drosophila patterning determinants (e.g. Dorsal, Twist and Snail) led to the identification of shadow enhancers for a number of genes engaged in dorsal–ventral patterning [17]. In some systems it has been possible to identify active enhancers on a genome-wide scale for a given tissue by identifying particular histone modifications, or the enzymes responsible for these modifications (e.g. [16]).
Chromosome conformation capture (3C) assays: can identify the sequences in a genome that interact with specific promoters. It relies on the stabilization of transient ‘loops’ of distal enhancers to target promoters using formaldehyde cross-linking, similar to the chromatin cross-linking used for ChIP-Seq assays. 4C (chromosome conformation capture-on-chip) methods were used to identify multiple and overlapping enhancers for the regulation of Hoxd genes in the mouse limb bud [25]. 3C and 4C assays provide an estimate of the overall interactions that occur in vivo but do not reveal the dynamics of these long-range interactions.
MNase-Seq and FAIRE assays: micrococcal nuclease (MNase) induces double-strand breaks within nucleosome linker regions and single-strand nicks within the nucleosome and can be used to identify ‘nucleosome-free’ regions. In some cases, these regions coincide with ‘poised’ enhancers due to the binding of pioneer transcription factors (e.g. [60]). FAIRE (formaldehyde-assisted isolation of regulatory elements) also identifies nucleosome-free regions, or ‘open’ chromatin [61].
The shavenbaby locus (also known as ovo) is important for the specification of dorsal hairs in the cuticle of embryos and larvae [20]. It is regulated by a complex array of enhancers with extensively overlapping activities. It is possible to remove some of these enhancers and still obtain essentially normal cuticle patterns at optimal temperatures. However, these patterns are disrupted when the embryos are grown at either low (15 °C) or elevated (30 °C) temperatures. Moreover, normal embryos are resilient to genetic changes, such as reductions in the levels of Wingless, but produce abnormal cuticles upon removal of shavenbaby ‘shadow’ enhancers. Thus, the shadow enhancers ensure reliable expression when embryos are subject to genetic and environmental variation.
A similar situation is seen for the regulation of snail, which encodes a zinc finger transcription factor that establishes the boundary between the presumptive mesoderm and neurogenic ectoderm [12]. The snail gene is regulated by a proximal enhancer located near the transcription start site, as well as by a recently identified shadow enhancer located 5 kb upstream of the start site within the first intron of a neighboring gene. Quantitative imaging assays and genetic complementation experiments suggest that the two enhancers ensure reliable and uniform activation of snail expression in embryos containing only one maternal dose of Dorsal, or when subject to high temperatures (30 °C) [12,21]. Removal of either enhancer, particularly the distal shadow enhancer [21], causes defects in gastrulation under adverse conditions.
Shadow enhancers have also been implicated in vertebrate developmental processes. For example, the neurogenic regulatory gene, ATOH7 (Math5), is essential for the development of the mammalian retina [22]. A genetic disease causing blindness at birth (nonsyndromic congenital retinal nonattachment) results from the deletion of a remote ‘shadow’ enhancer located more than 20 kb away from the ATOH7 transcription unit [23]. The shadow enhancer directs a very similar spatiotemporal pattern of gene expression as the ‘primary’ proximal enhancer in the developing retina of a mouse. This result suggests that the primary enhancer alone cannot sustain sufficient levels of ATOH7 expression for normal development in the absence of the shadow enhancer. Thus, the two enhancers seem to be redundant in terms of the location and timing of the expression patterns they direct, but both are required to reinforce ATOH7 expression and achieve correct levels of expression during crucial stages of eye development.
There are additional examples of multiple enhancers for key vertebrate patterning genes. For example, deletion of a limb enhancer of the paired-box homeodomain transcription factor Prx has no obvious effect on Prx expression levels or on limb development in mice [24], suggesting the existence of additional, shadow enhancers. More recently, 4C assays (Box 1) identified multiple putative enhancers for Hoxd13 expression within a distal gene ‘desert’ that contains known regulatory elements, GCR and Prox [25]. Deletions of GCR and Prox have little effect on Hoxd13 expression in digits, thereby suggesting the occurrence of redundant regulatory elements. Indeed, complete abolition of Hoxd13 expression in digits is achieved only when the gene desert, together with the GCR and Prox regions, are completely deleted (830 kb deletion).
The preceding examples suggest that multiple enhancers represent a simple means for improving the reliability of gene expression. The underlying mechanism is uncertain, but they might increase the probability of gene activation at any given time during critical windows of development and make it more robust to perturbation. For example, if a typical enhancer has a 10% failure rate to loop and engage its target promoter, and if the proximal and distal enhancers function more or less independently of one another, then there is a combined failure rate of only 1% (e.g. [12]). That is, two enhancers function in an inherently multiplicative manner to activate gene expression (Figure 3). Such a mechanism also provides robustness. For example, if the failure rate of each individual enhancer increases to 30% due to stress, then the combined failure rate is only 9%.
Figure 3.
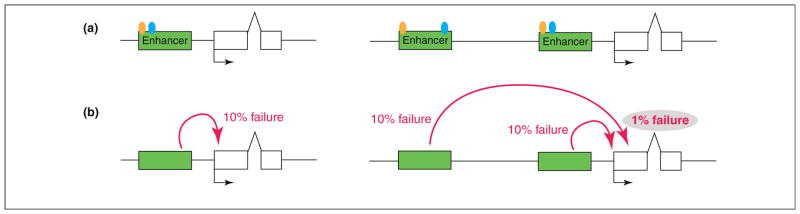
Model for enhancer synergy. (a) Schematic showing that the primary and shadow enhancers (green boxes) possess the same regulatory logic (TF binding sites are illustrated by colored circles). (b) To activate transcription, an enhancer loops to its cognate promoter. This interaction has a typical failure rate of 10%. In the presence of two enhancers regulating the same gene at the same time (primary and shadow), the combined failure rate is 1% (10% × 10% = 1%). This assumes that the two enhancers work independently of one another.
An alternative explanation is that multiple enhancers ensure high levels of expression above a minimal threshold required for genetic function (as suggested in the case of ATOH7 regulation). In reality, multiple enhancers could be important both for the reliable activation of gene expression and for maintaining high levels of expression. We still do not understand the details of how an enhancer switches on a gene and affects levels of expression, and therefore this is very much an open question. The source of shadow enhancers is uncertain, but it has been proposed that they might arise from ‘cryptic’ duplication events [18].
Rendering genes ‘poised’ for activation
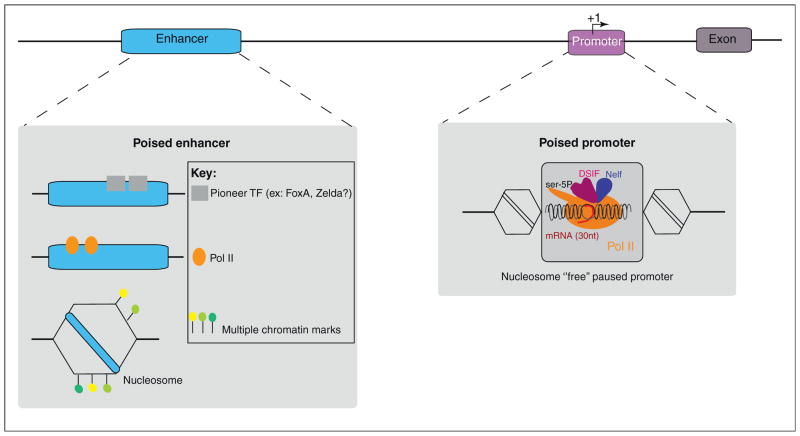
Timing is crucial in development, and recent studies have identified several mechanisms that ensure faithful activation of gene expression upon receipt of key inducing signals. We consider mechanisms that optimize induction of distal enhancers and the core promoter (Figure 4). In some cases, both are ‘primed’ for efficient activation.
Figure 4.
Summary of mechanisms of transcriptional priming. Gene transcription depends on enhancers (blue) and promoters (purple). The transcription start site (TSS) is indicated by an arrow labeled +1. The promoter can be primed or ‘poised’ for transcription by the recruitment of Pol II before gene expression. This ‘promoter pausing’ generates a small mRNA (around 30–50 nt) and then elongation is blocked by the binding of negative elongation factors such as Nelf and DSIF. The enhancer can be ‘prepared’ for activation by the binding of pioneer factors (represented by gray boxes), by recruitment of Pol II, or by the modification of the chromatin landscape (positioned nucleosomes and associated histone marks). These three features at enhancers may be linked, but for simplicity we illustrate them sequentially. Nucleosomes are represented by hexagons and histone marks with colored flags. A simplified scheme of a paused promoter is represented in the gray box.
Paused promoters
Many metazoan genes contain paused RNA polymerase II (Pol II) prior to their activation [26–28]. This paused Pol II is an active form of the enzyme that halts ~30–50 bp downstream of the +1 transcription start site (Figure 4; Box 2). It is present in ~30% of all genes in embryonic stem cells and about 15% of genes in the early Drosophila embryo [26,29,30]. The purpose is uncertain, but many developmental patterning genes contain paused Pol II. It has been suggested that it fosters rapid and synchronous activation of gene expression [31]. The idea is that regulating Pol II release, rather than recruitment, permits rapid induction of gene expression. This hypothesis has been explored using detailed mathematical modeling of transcription [32], but it still remains to be tested experimentally.
Box 2. Methods for identifying paused promoters.
Many developmental patterning genes contain paused Pol II before their activation during Drosophila embryogenesis (reviewed in [27]). There is also evidence that a significant number of inactive or weakly expressed genes contain paused Pol II in mammalian tissues, including embryonic stem cells. Several different methods have been used to identify paused genes, as summarized below.
Pol II ChIP-Seq assays: the simplest method is the genome-wide identification of Pol II binding. This is typically done with a mixture of antibodies recognizing different isoforms of Pol II (e.g. nonphosphorylated, ser-5P, ser-2P). Active genes contain Pol II extending signals across the length of their transcription units. Inactive genes fall into two classes: those completely lacking Pol II and those containing Pol II near the +1 transcription start site (e.g. [26]). These latter genes can be regarded as stalled or ‘provisionally’ paused. However, it is unclear whether Pol II has engaged the DNA template and undergone promoter escape, or if the signals detected in the promoter region represent an equilibrium of unstable Pol II associating and dissociating from the template. Additional methods are required to determine whether Pol II is truly paused, that is, activated polymerase containing a capped nascent transcript and arresting ~30–50 bp downstream of +1.
Permanganate protection assays: stably paused Pol II is associated with a ‘transcription bubble’ of ~20 bp due to the local denaturation of the double helix by the active polymerase. It is possible to detect the bubble by the modification of exposed, single-stranded thymidine residues with potassium permanganate. This method has been used to identify transcription bubbles for a number of genes containing stalled Pol II in Drosophila embryos and cultured S2 cells [62].
Direct sequencing: small nuclear RNAs containing 5′ caps are isolated, cloned, and then subjected to deep sequencing [63]. This method identified +34 as a common site of paused Pol II, with the DPE (downstream promoter element) or PB (pause button) motifs being the last nucleotides transcribed before arrest. A significant fraction of paused genes contain GAGA, INR, and DPE/PB motifs within or near their core promoters.
Gro-Seq assays: this has emerged as the method of choice for the systematic identification of paused Pol II [30,64]. However, it is not for the faint of heart. The method is a whole-genome nuclear run-on assay. Nuclei are harvested from embryos, tissues, or cultured cells, and treated with Sarkosyl to block de novo binding of Pol II. A modified nucleotide (e.g. bromouridine) is added along with a mixture of ATP and other agents to permit the elongation of pre-existing polymerases already engaged on DNA templates. These polymerases are allowed to extend ~50–100 nucleotides; the RNAs are then isolated using anti-bromo antibodies and subjected to deep sequencing. The resulting sequence information provides the exact locations of paused Pol II.
A nonexclusive alternative view is that paused Pol II is involved in recruiting chromatin-modifying enzymes that expedite transcription. For example, the chromatin landscape of the Hsp70 locus (the prototypic paused gene in Drosophila) is rapidly altered following heat shock, through a mechanism independent of transcription [33]. This rapid change is key to the effective activation of Hsp70 expression upon heat shock. Moreover, there is an inverse correlation between paused Pol II and positioned nucleosomes at the core promoter [34,35]. An increase in positioned nucleosomes has been observed upon destabilization of paused Pol II (e.g. NelfE knockdown in S2 cells) [34]. Conversely, diminished levels of the Polycomb repressor (in esc mutant embryos) correlates with augmented levels of paused Pol II [35]. It would appear that the promoter regions of developmentally regulated genes contain either paused Pol II or positioned nucleosomes, but the basis for this regulatory switch is uncertain.
These studies raise the possibility that paused Pol II might prepare genes for activation by establishing an ‘open’ configuration at the promoter. However, this possibility has not yet been critically tested.
Poised enhancers
There is also evidence that enhancers can be prepared for rapid deployment before gene activation (Figure 4). For example, the forkhead transcription factor FoxA binds to the Albumin enhancer in the primitive endoderm of mouse embryos where it is inactive (reviewed in [36]). FoxA is an example of a ‘pioneer’ factor [37]; it binds to inactive enhancers and renders them ‘poised’ for rapid induction upon the appearance of key activators, such as those mediating cell signaling.
To bind inactive enhancers, pioneer factors have the defining property of binding to nucleosomal DNA and compact chromatin, and remain bound even during mitosis. Since the initial discovery of FoxA and GATA factors as pioneer factors in the liver differentiation program, additional examples have been described [38,39].
Zelda is a maternal zinc finger transcription factor that is essential for the activation of ~100 genes 2–3 h after fertilization during Drosophila embryogenesis (maternal to zygotic transition) [40–42]. It binds to the enhancer regions of many or most developmental control genes before their activation. Disrupting Zelda binding sites can delay the onset of expression, or cause sporadic patterns of activation [40,41]. Thus, Zelda renders developmental enhancers poised for activation by maternal determinants such as Bicoid and Dorsal, and may function as a pioneer factor. It might also help ensure reliable patterns of gene activation in natural populations under stress, but this idea has not yet been tested.
The mechanisms by which pioneer factors prepare enhancers for efficient activation are not known. It has been suggested that they can displace nucleosomes and thereby render adjacent binding sites available for occupancy [36,38]. A nonexclusive possibility is that pioneer factors recruit chromatin-modifying enzymes that ‘mark’ enhancers for rapid deployment. For example, inactive liver and pancreas enhancers exhibit ‘active’ chromatin modifications in the mouse foregut endoderm where they are inactive [36]. This suggests ‘pre-patterning’ of the enhancers in progenitor tissues before their induction in the liver and pancreas. The P300 histone acetyltransferase and the EZH2 histone methyltransferase have been implicated in these modifications [43]. It is conceivable that such modifications are not strictly required for gene expression, but might improve the precision and stability of gene expression in natural populations.
More recently it has been suggested that histone modifications and Pol II help to prime distal enhancers [44] (Figure 4). In this study, whole-genome Chip-Seq assays were performed on isolated tissues obtained from staged Drosophila embryos. The timing of gene expression correlated with Pol II binding and two types of chromatin marks in enhancers. Pol II occupancy at enhancers is counterintuitive, but multiple studies, in human ES cells [45] and mice [45,46], suggest that enhancers can be bound by Pol II and are sometimes transcribed. Additional members of the general transcription machinery, such as the TATA binding protein TAF3 [47], are also seen at particular enhancers. It was suggested that these factors might foster looping interactions between distal enhancers and promoters, but it is currently unclear how Pol II and associated factors might render enhancers poised for activation. It is possible that they are recruited to enhancers by pioneer TFs, but this idea awaits further studies.
When stochastic expression is ‘purposeful’
Many developmental patterning genes in Drosophila contain paused Pol II, shadow enhancers, or both. We have discussed how these mechanisms might foster the precision and stability of gene expression in development. However, there are examples of developmental control genes that exhibit sporadic or stochastic patterns of expression. Some might exhibit such expression because there is no selective pressure for them to be expressed in a precise and synchronous manner. However, there are cases where stochastic expression is used as a purposeful strategy for generating regulatory diversity among the cells of a population [48]. One of the most striking examples is seen in the eye of the adult fly [49–51].
Color vision depends on the differential expression of rhodopsin-3 (Rh3) and Rh4 in the R7 photoreceptor cells and the differential expression of Rh5 and Rh6 in the R8 photoreceptor cells. These differential patterns depend on stochastic expression of spineless, which encodes a homeobox transcription factor that activates Rh4 in R7 [52]. Approximately 70% of the ommatidia express spineless, but the patterns of activation differ among adult flies. When spineless is expressed, Rh4 is activated in R7; if not, Rh3 is expressed instead. The identity of these distinct classes of R7 cells dictates the identities of the underlying R8 cells. When spineless and Rh4 are expressed in R7, then Rh6 is expressed in the associated R8 cell. Conversely, when spineless is absent and Rh3 is expressed in R7, then Rh5 is expressed in the associated R8 cell. Thus, diverse patterns of rhodopsin expression are achieved by the stochastic expression of spineless. The underlying mechanism is uncertain.
There are other examples of the imporatance of stochastic expression in the control of developmental genes. Notably, Nanog, one of the key determinants of pluripotent stem cells, exhibits stochastic expression in cultured ES cells and in early mouse embryos [53,54]. There is a correlation between elevated levels of Nanog expression and self-renewal of pluripotent stem cells in culture [55,56]. By contrast, low levels correlate with a propensity for the cells to differentiate.
Concluding remarks
The preceding examples are probably exceptional. We believe that most regulatory genes are ‘primed’ for rapid and precise deployment during development. Several mechanisms were discussed, including redundancies in gene networks and developmental enhancers, shadow enhancers, and primed promoters and enhancers (via paused Pol II and pioneer TFs). There is little doubt that additional mechanisms await discovery (Box 3).
Box 3. Outstanding questions.
How do multiple enhancers provide precision in gene expression: do they increase the levels or probability of expression?
Are genes with multiple enhancers more or less ‘evolvable’? Do shadow enhancers increase the probability of evolving novel gene activities?
How do pioneer factors prime enhancers?
How does paused Pol II prime the promoter?
When are imprecise, stochastic modes of gene activation advantageous in development?
There is something of a chicken and egg issue that we have skirted. Namely, what is the source of these mechanisms of developmental precision? It is conceivable that they arose from the demands of natural populations, namely, to stabilize complex developmental processes in response to inherent (genetic) and extrinsic (environmental) fluctuations. Alternatively, they might have arisen from the demands of the embryo, to produce timely and dynamic on/off patterns of gene expression underlying cell specification processes. These are not mutually exclusive concepts. A regulatory mechanism selected to provide stability in natural populations (e.g. shadow enhancer) might be incorporated into the core patterning process to produce sharper borders of gene expression [12] or homogenous patterns of activation [57]. Conversely, a mechanism selected for developmental precision (e.g. paused Pol II) might foster robustness of expression in natural populations. We suggest that the dynamic interplay between the demands of natural populations and the embryo has produced the exquisite patterning processes that underlie animal development.
Glossary
- Canalization
a measure of the ability of a population to produce the same phenotype regardless of fluctuations in its environment, genotype or other sources of variability. Our use of the term ‘robustness’ conveys the same essential meaning
- Enhancer
the predominant regulatory DNA for controlling gene expression. It has the defining property of driving reporter expression in transgenic assays from a heterologous promoter
- Gene regulatory network
interacting genes and their associated regulatory DNAs that are responsible for a specific developmental process such as the specification of gut or muscle
- Paused polymerase
RNA Pol II that has initiated transcription, but arrests after producing a small nascent RNA of 30–50 nt. The Pol II is ‘ready to go’ but needs additional regulators to undergo elongation
- Pioneer factor
a specialized TF (sequence-specific) that binds to nucleosomal DNA and prepares enhancers for rapid and timely deployment
- Poising
preparing genes for rapid and timely transcription. This can be achieved by priming the promoter, the enhancer, or both
- Redundancy
two genes are considered to be redundant if they play similar functions and are able to replace one another. This can be extended to a genetic interaction or enhancers or binding sites within enhancers. However, we do not believe in true redundancy. Instead, genes or regulatory DNAs might appear to possess redundant, or overlapping, activities in the laboratory, but not in natural populations subject to stress
- Shadow enhancer
an enhancer that is sometimes located far from the gene it regulates. The term ‘shadow’ is a metaphor which reflects that, historically, these distal enhancers tended to be discovered after the proximal/primary enhancer and in unexpected locations such as in the introns of neighboring genes
References
- 1.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Nüsslein-Volhard C, Roth S. Axis determination in insect embryos. Ciba Found Symp. 1989;144:37–55. doi: 10.1002/9780470513798.ch4. [DOI] [PubMed] [Google Scholar]
- 4.Ip YT, et al. The bicoid and dorsal morphogens use a similar strategy to make stripes in the Drosophila embryo. J Cell Sci. 1992;16 (Suppl):33–38. doi: 10.1242/jcs.1992.supplement_16.5. [DOI] [PubMed] [Google Scholar]
- 5.Davidson EH. Network design principles from the sea urchin embryo. Curr Opin Genet Dev. 2009;19:535–540. doi: 10.1016/j.gde.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj A, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burga A, et al. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 8.Arnosti DN, et al. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig MZ, et al. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 2011;7:e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanojevic D, et al. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 11.Small S, et al. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry MW, et al. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 14.Zinzen RP, et al. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 15.He Q, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 2011;43:414–420. doi: 10.1038/ng.808. [DOI] [PubMed] [Google Scholar]
- 16.May D, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2011;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Gene Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong JW, et al. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays. 2011;34:135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunipace L, et al. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;4084:4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riesenberg AN, et al. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci. 2009;29:12865–12877. doi: 10.1523/JNEUROSCI.3382-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghiasvand NM, et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cretekos CJ, et al. Regulatory divergence modifies limb length between mammals. Gene Dev. 2008;22:141–151. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev. 2011;21:231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther MG, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Gene Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boettiger AN, et al. Transcriptional regulation: effects of promoter proximal pausing on speed, synchrony and reliability. PLoS Comput Biol. 2011;7:e1001136. doi: 10.1371/journal.pcbi.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chopra VS, et al. The Polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42:837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Gene Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watts JA, et al. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnani L, et al. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Fakhouri THI, et al. Dynamic chromatin organization during foregut development mediated by the organ selector gene pha-4/FoxA. PLoS Genet. 2010;6:e1001060. doi: 10.1371/journal.pgen.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nien CY, et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison MM, et al. Zelda Binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu CR, et al. Chromatin ‘prepattern’ and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 45.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, et al. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasiliauskas D, et al. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature. 2011;479:108–112. doi: 10.1038/nature10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston RJ, et al. Interlocked feedforward loops control cell-type-specific rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jukam D, Desplan C. Binary fate decisions in differentiating neurons. Curr Opin Neurobiol. 2010;20:6–13. doi: 10.1016/j.conb.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 54.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalmar T, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glauche I, et al. Nanog variability and pluripotency regulation of embryonic stem cells–insights from a mathematical model analysis. PLoS ONE. 2010;5:e11238. doi: 10.1371/journal.pone.0011238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry MW, et al. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci USA. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berman BP, et al. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Markstein M, et al. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilmour DS, Fan R. Detecting transcriptionally engaged RNA polymerase in eukaryotic cells with permanganate genomic footprinting. Methods. 2009;48:368–374. doi: 10.1016/j.ymeth.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 63.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]