SUMMARY
Transcription is commonly held to be a highly stochastic process, resulting in considerable heterogeneity of gene expression among the different cells in a population. Here, we employ quantitative in situ hybridization methods coupled to high-resolution imaging assays to measure the expression of snail, a developmental patterning gene necessary for coordinating the invagination of the mesoderm during gastrulation of the Drosophila embryo. Our measurements of steady-state mRNAs suggest that there is very little variation in snail expression across the different cells comprising the mesoderm, and that synthesis approaches the kinetic limits of Pol II processivity. We propose that rapid transcription kinetics and negative autoregulation are responsible for the remarkable homogeneity of snail expression and the coordination of mesoderm invagination.
INTRODUCTION
Recent studies suggest that inherent stochastic processes underlying transcription are a significant source of cell-cell variation in gene expression. Such variation can be explained by theoretical considerations of diffusion-driven processes involving small numbers of molecules (Munsky et al., 2012). Stochastic variation has been substantiated by a growing number of experimental studies that employ single-molecule imaging methods to measure variation in mRNA in individual cells (Golding et al., 2005; Raj et al., 2006, 2008; Zenklusen et al., 2008; To and Maheshri, 2010; So et al., 2011; Itzkovitz et al., 2011). However, many developmental processes, such as coordinated cell movements during gastrulation, are dependent upon uniform expression of key patterning genes. It remains to be understood what mechanisms exist to either compensate or correct the variation often observed during transcription to allow for the coordination of cell behavior within an embryonic tissue.
Drosophila gastrulation provides an excellent model for investigating the mechanisms of coordinate gene expression. snail is a key regulator of epithelial-mesenchyme transitions in a variety of animal embryos. It exhibits sharp spatial limits of expression within the presumptive mesoderm, and is required for coordinating mesoderm invagination during Drosophila gastrulation (Leptin, 1991; Kosman et al., 1991). It exhibits sharp spatial limits of expression within the presumptive mesoderm. snail contains paused RNA polymerase, a feature that correlates with synchronously expressed genes (Zeitlinger et al., 2007a; Boettiger and Levine, 2009). It has also two enhancers, each independently capable of driving expression in the presumptive mesoderm – an apparent redundancy that has been shown to ensure a more uniform distribution of nascent transcription (Perry et al., 2010, 2011). Theoretical arguments and mathematical modeling have suggested that these features may increase the average synthesis rate and decrease cell-cell variations in mRNA expression (Perry et al., 2011; Boettiger et al., 2011) although these hypotheses have not been critically tested. In order to measure the homogeneity of expression, we developed a single molecule fluorescent in situ hybridization (smFISH) technique for detection of snail transcripts in early Drosophila embryos. The approach is similar to that described by Paré et al (2009), but extended to permit the analysis of many more cells. We further developed this method to allow measurement of the rates of snail RNA synthesis, in order to better understand the relationship between Pol II kinetics and transcriptional precision.
RESULTS AND DISCUSSION
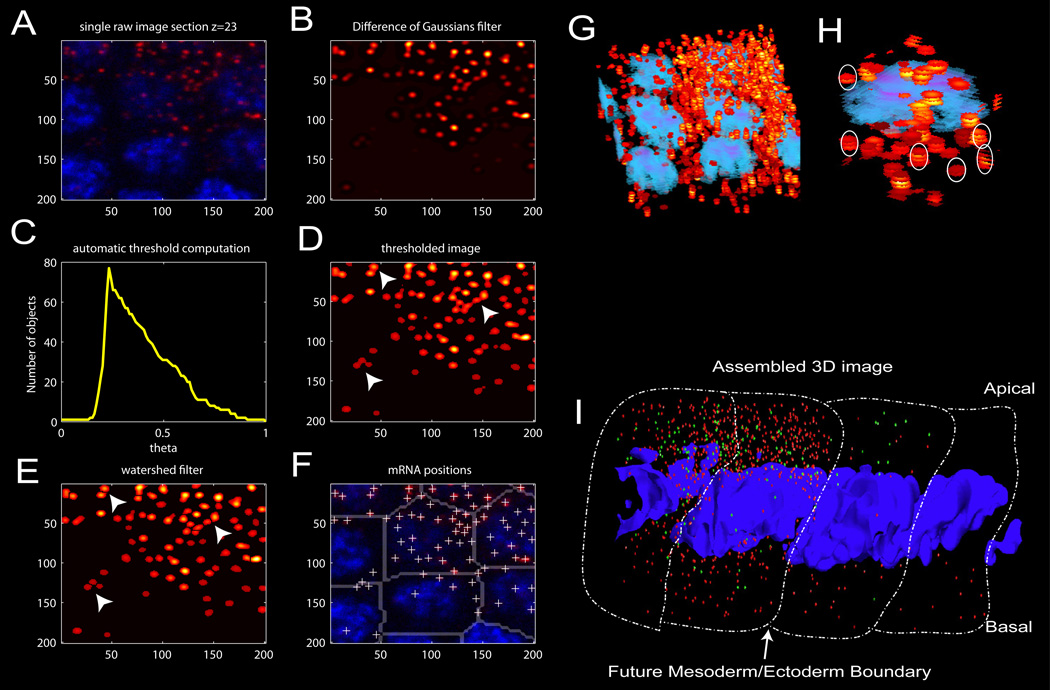
Staged embryos were hybridized with snail antisense RNA probes, and mRNAs can be resolved into individual hybridization “dots” with a high numerical-aperture objective and confocal microscope (Figure 1A and Figure S2A). These dots were computationally detected and traced through consecutive Z-sections to determine total counts per “cell” (Extended Experimental Procedures and Figure 1B–H). We define a cell as the cytoplasm in closest proximity to a given nucleus in syncitial embryos. Control experiments with equal molar levels of “red” and “green” snail antisense hybridization probes suggest that each dot represents a single mRNA (Figure S1).
Figure 1. single molecule mRNA counting.
(A) Representative individual confocal section of several adjacent nuclei, showing bright, diffraction limited spots. (B) Gaussian filtered version of the image in (A). (C) Plot of the number of separate objects identified vs. intensity threshold applied. The script selects the intensity threshold, theta, which maximizes the number of separate objects. (D) Resulting image after threshold determined in (C) is applied. Note under dense conditions, several spots remain fused (white arrows indicate examples). (E) Segmented image after a watershed algorithm is applied to unlink spots joined by the threshold in (D) (see white arrows). (F) White crosses mark the centroids of all the mRNA molecules identified in the image, which are assigned to parent nuclei using the computed nucleoid region map, indicated by gray partitions (see Methods). (G) Three dimensional-projection of 'stacked disks' (red/yellow) identified in each image plane in the previous steps. Projection of volume reconstructions of nuclei are shown in blue. (H) These disks are clustered along z to identify which dots correspond to different focus planes from the same molecule. White ovals indicate some examples. (I) Three-dimensional reassembly of individual snail mRNA transcripts (denoted by small red spheres), yellow mRNA driven from a single-copy snail BAC transgene (green spheres) and nuclei (Draq5 labeled DNA, blue). Approximate cellular boundaries have been outlined.
An example of a processed image is shown in Figure 1I. It displays four consecutive nuclei in a late-stage cell-cycle 14 (cc14) embryo. The sharp border between the future mesoderm and lateral ectoderm is clearly evident. The red hybridization dots correspond to endogenous snail mRNAs, while the green dots correspond to yellow mRNAs encoded by a BAC transgene containing the snail 3’ UTR and regulatory DNAs. There are lower levels of yellow RNAs than snail since these embryos contain just one copy of the BAC transgene (see Figure S2). There is an asymmetric distribution of the hybridization dots, with higher levels in the apical cytoplasm (near the surface of the embryo) as compared with more basal regions (Figure 1I). Indeed, many mRNAs encoded by developmental patterning genes exhibit such localization (e.g., Davis and Ish-Horowicz, 1991).
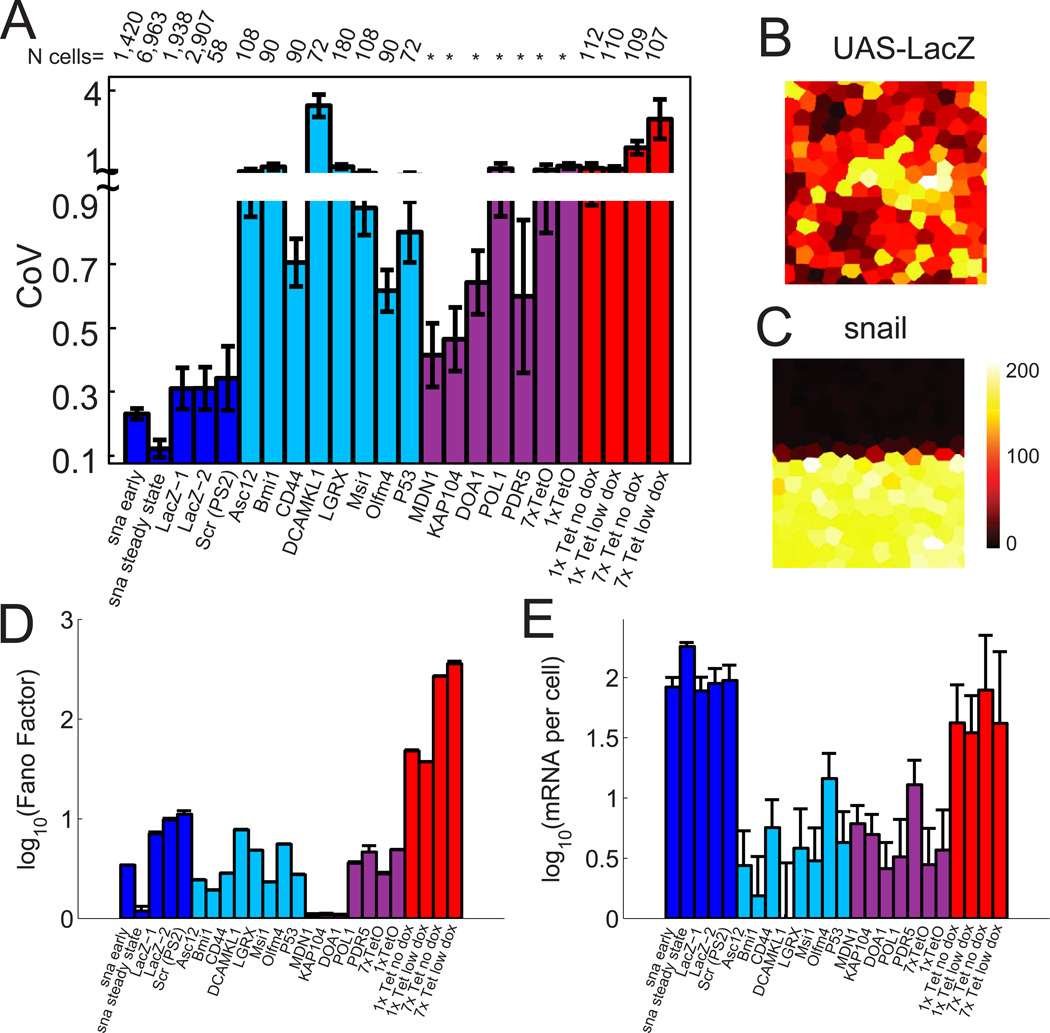
Using this approach we analyzed nearly 7,000 cells from 22 embryos in mid cell cycle 14 (cc14) when snail expression has reached its steady-state. There is remarkable homogeneity of snail mRNA levels between cells, just 10–12% variation, close to the limits of experimental error (Figure 2A and 2C). In contrast, previous smFISH studies in mice (Itzkovitz et al., 2011), yeast (Zenklusen et al., 2008; To and Maheshri, 2010), and cultured mammalian cells (Raj et al., 2006), reported 60%–300% variation (Figure 2A). We sought to identify the distinctive features of snail regulation that might be responsible for its homogeneous expression.
Figure 2. Homogenous expression of snail in mesodermal cells.
(A) Comparison of the coefficient of variation (CoV) for genes expression in D. melanogaster (blue) M. mus. (cyan)(Itzkovitz et al., 2011), S. cerevisiae (magenta) (Zenklusen et al., 2008; To and Maheshri, 2010) and mammalian cultured CHO cells (red) (Raj et al., 2006). 'Snail early' refers to CoV measured at the onset of cycle 14 (telophase of cycle 13). 'Snail steady state' is the mid cycle 14 stable levels. lacZ-1 and lacZ-2 are from ubiquitous induction of two different UAS-lacZ lines with different maternal drivers. Scr measurements from Paré et al (Paré et al., 2009). Error-bars indicate standard deviation in CoV between embryos (for Drosophila) or by bootstrapping the population of single-cell measurements. (B) Spatial distribution of mRNA counts per cell for ubiquitously induced lacZ expression during cycle 14. The colored tiles outlines the “nucleoid region” around each nucleus. The color of the tile indicates the number of mRNA molecules counted within (see colorbar next to (C)). (C) Spatial distribution snail mRNA counts per cell during cycle 14. (D) Fano factor comparison for lacZ, snail and all previously published data shown in (A). (E) Comparison of median mRNA counts per cell over all cycle 14 embryos for snail, lacZ and previously published data.
To determine whether this homogeneity arises from extrinsic factors such as synchronized cell cycles, shared pools of general transcription factors within syncitial embryos, the uniformity of nuclear volumes (etc.), we examined the variability of lacZ transcripts driven by a ubiquitous maternal Gal4 driver in transgenic embryos (Figure 2B). We chose this comparison since lacZ transcription is first detected at a similar stage as snail (shortly after nuclei reach the periphery), and like snail, lacZ shows little systematic spatial variation within the expressed domain of cc14 embryos (unlike many other zygotic genes). Moreover, the 5 kb lacZ mRNA provides a strong signal for smFISH assays, and the potent Gal4 UAS allows it to reach comparable levels as snail.
lacZ transcripts exhibit nearly three-fold greater variability than snail, though generally less than has been reported in C. elegans embryos (Raj et al., 2010), mouse embryonic gut cells (Itzkovitz et al., 2011) or cell culture (Raj et al., 2006; Zenklusen et al., 2008; To and Maheshri, 2010). Even greater variation in lacZ expression is observed using an alternative measure of noise, the Fano factor, which gives deviations from Poisson type noise. A Fano factor of ~10 is calculated for lacZ, nearly 5-fold greater than the variability observed for snail at the same stage (Figure 2D). This analysis also shows that a reasonable fraction of the variation measured in other species comes from Poisson noise, as expected for infrequently transcribed genes which have low average mRNA counts (Figure 2E). The striking differences between snail and the lacZ controls suggest that a substantial degree of the homogeneity of snail transcription is not due to extrinsic factors, but instead depends on distinctive features of its regulation. We also observed that lacZ accumulates to somewhat lower levels than snail (Figure 2E). We therefore asked whether the kinetics of snail transcription might serve to limit cell-cell variation in expression.
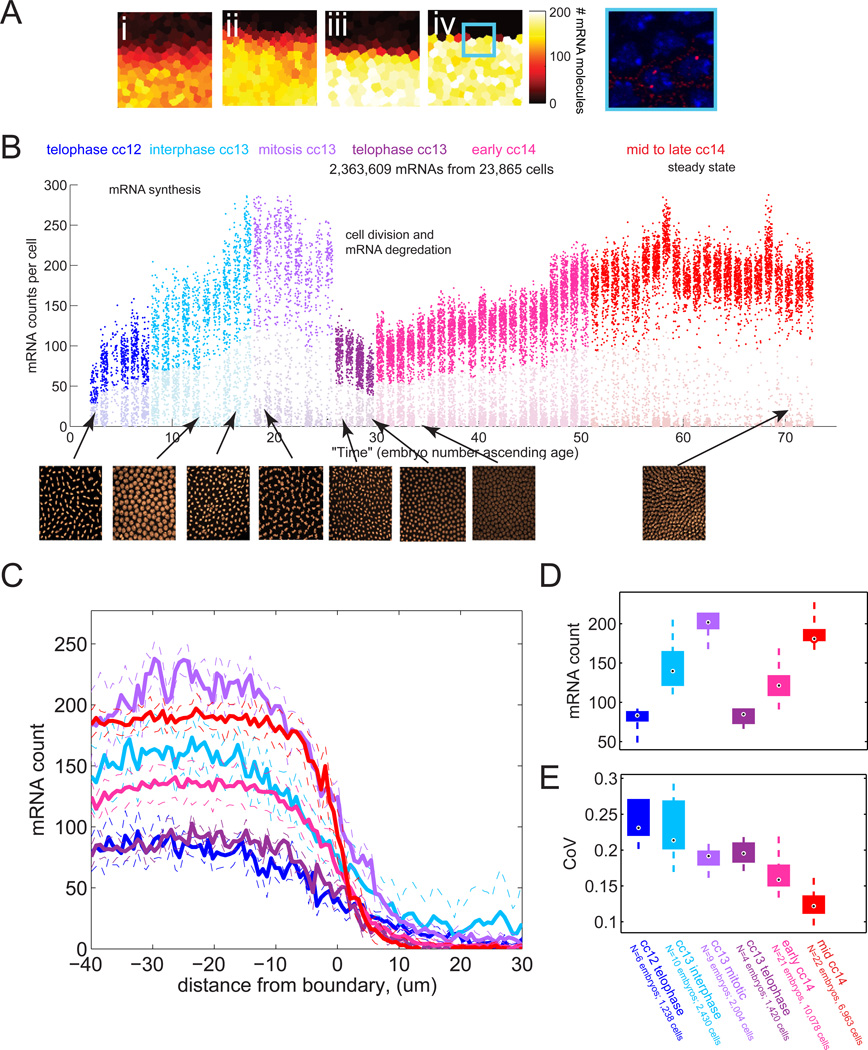
To assay the kinetics of transcription we first analyzed the dynamics of snail mRNA accumulation from the onset of cc13 to the end of cc14. Representative patches of cells from early, mid and late cycle 14 embryos are shown in Figure 3A. The complete data-set is shown in Figure 3B, which plots the mRNA counts in all cells from a given embryo as a cluster of dots, with embryos sorted temporally based on nuclear morphology. During the ~15 minute interphase of cc13, the hybridization dots increase from an average of ~80 per cell to ~200 per cell (Figure 3D). After mitosis, there is an average of ~80 mRNAs per cell and this increases to a steady state of ~180 per cell by the midpoint of cc14 (Figure 3B–D). These data also revealed an interesting dynamic trend in snail cell-cell variation (Figure 3E), which we will return to later.
Figure 3. Dynamics of mRNA expression at the single cell level.
(A) Heat map representation of the number of mRNA in each cell for progressively older embryos (i)–(iv) in cycle 14. The colored tiles outlines the “nucleoid region” around each nucleus. The color of the tile indicates the number of mRNA molecules counted within (see colorbar after (iv)). A single confocal slice from the box in panel (iv) is shown at right. (B) mRNA counting results. Each column represents a single embryo, each dot a single cell, the y position indicates the number of mRNA found in that cell. Embryos are sorted approximately by age. The color of the dot indicates the age class as determined by nuclear density and nuclear morphology. Representative nuclei images for each class are shown in the insets below. (C) Average mRNA per cell for embryos in each age class as a function of cell distance from the snail boundary. Dashed lines represent +/− standard deviation. Color code as in (B). (D) Distribution of average mRNA counts per mesodermal cell, for embryos in each age class. The mesodermal boundary is defined as the area where the mRNA count, averaged across a line perpendicular to the the boundary, drops below half its maximal value. The box spans from the lower to upper quartile. The median is indicated by the black dot. Whiskers extend to greatest and smallest data point. (E) Box plots of coefficient of variation for embryos in each age class.
In order to calculate the rate of synthesis it is essential to determine the half-life of the snail mRNA. Towards this end, we measured the reduction in the number of snail hybridization dots during mitosis, when de novo transcription is arrested. By late prophase of cc13, (identifiable by the disappearance of nascent transcripts and more compact, intense DAPI staining in nuclei), there is an average of ~210 transcripts per nucleus. By telophase, ~5 min later (estimated from live imaging of RFP-histone tagged embryos), we observed 80 transcripts per nucleus (and twice as many nuclei). This ~25% drop suggests a snail mRNA half-life of ~13 min (lower quartile uncertainty bound: 7.2 min, upper uncertainty 22 min, details see Figure S3A and Extended Results).
With this information it is possible to estimate the rate of synthesis of snail mRNAs (Figure S3B and Extended Results). For this calculation we used the mRNA counts at telophase of cc12 (~80 transcripts), the counts at prophase at the end of cc13 (~210 transcripts), and the degradation rate calculated above. We also need to know the number of snail templates per nucleus, which depends on the timing of DNA replication. This was estimated by close inspection of transcription foci (see Figure S3C) in embryos throughout cc13. Half of the embryos exhibit twin-spots of nascent transcription, suggesting that they have undergone DNA replication at the snail locus. Consequently we infer that snail is replicated at the midpoint of cc13. Our measurements suggest that each template synthesizes one full-length snail mRNA every ~10 seconds.
This rate approaches the speed limit set by the intrinsic kinetics of Pol II. Measured elongation rates in Drosophila are ~20 nt per second at room temperature (Ardehali and Lis, 2009). Because of the large Stoke’s radius of the Pol II complex, it engages the gene at a density of one per ~80 nt, or one per 4 seconds assuming maximum packing and maximum rates of Pol II elongation can be simultaneously achieved (bumper to bumper traffic moving at the speed limit). This is likely an over-estimate of the true physical limit, since intrinsic Pol II kinetics, such as variations in the speed of transcription (Neuman et al., 2003), the frequency of pausing during mRNA synthesis (Neuman et al., 2003), and the stochastic loading of Pol II (Darzacq et al., 2007) will further limit Pol II packing. Moreover, recent research suggests that the rate of Pol II movement (and hence mRNA synthesis) may decrease significantly at high densities due to the effects of transcription traffic (Klumpp and Hwa, 2008), see also Extended Results. Thus, it would appear that snail RNA synthesis is within a factor of 2 of the upper limit of transcription kinetics. This rapid rate of synthesis is likely to be a major factor in the low cell-cell variability of snail expression. Moreover, this continuous transcription throughout the interphase cycle substantially limits the effects of transcriptional bursts on expression noise (by minimizing promoter 'off' time), which is believed to be a prime contributor to cell-cell variation in gene expression (Munsky et al., 2012; Raser and O’Shea, 2005; Raj et al., 2006).
Since we measured snail mRNA degradation only at the onset of cc14, it is possible that different kinetics apply to the previous 20 min (cc13) or ensuing 45 min (onset of gastrulation). If snail transcripts degrade slower than we have measured for a substantial portion of the cell cycle, mRNA synthesis rates will be lower than 1 every 10s per template. In the extreme case of no degradation, mRNA synthesis rates could be as low as 1 mRNA every 17s. If snail mRNAs degrade faster than our estimate, Pol II packing densities would be even higher than one every ~200 bp. Thus, even the most dramatic changes in degradation rates would not affect our computed synthesis rates by more than a factor of two.
Surprisingly, the steady-state mRNA levels observed in cycle 14 (~180 transcripts/cell) are substantially lower than expected. Specifically, they are lower than those achieved in the much shorter cycle 13 (~210 transcripts), and substantially lower than predicted by extrapolation of the synthesis and degradation rates we measured in cycle 13 to cycle 14 (~400 transcripts/cell). This effect could be due to global changes associated with the maternal-zygotic transition (such as an enhanced degradation rate for mRNAs in cycle 14) or indicate a negative autoregulatory effect of Snail protein on its own production. In the latter case, the lag in protein synthesis would explain why slightly higher levels are reached in cycle 13 (before negative feedback has reached full strength) than in cycle 14, and the subsequent repressive effect would account for the lower final steady-state levels during cc14.
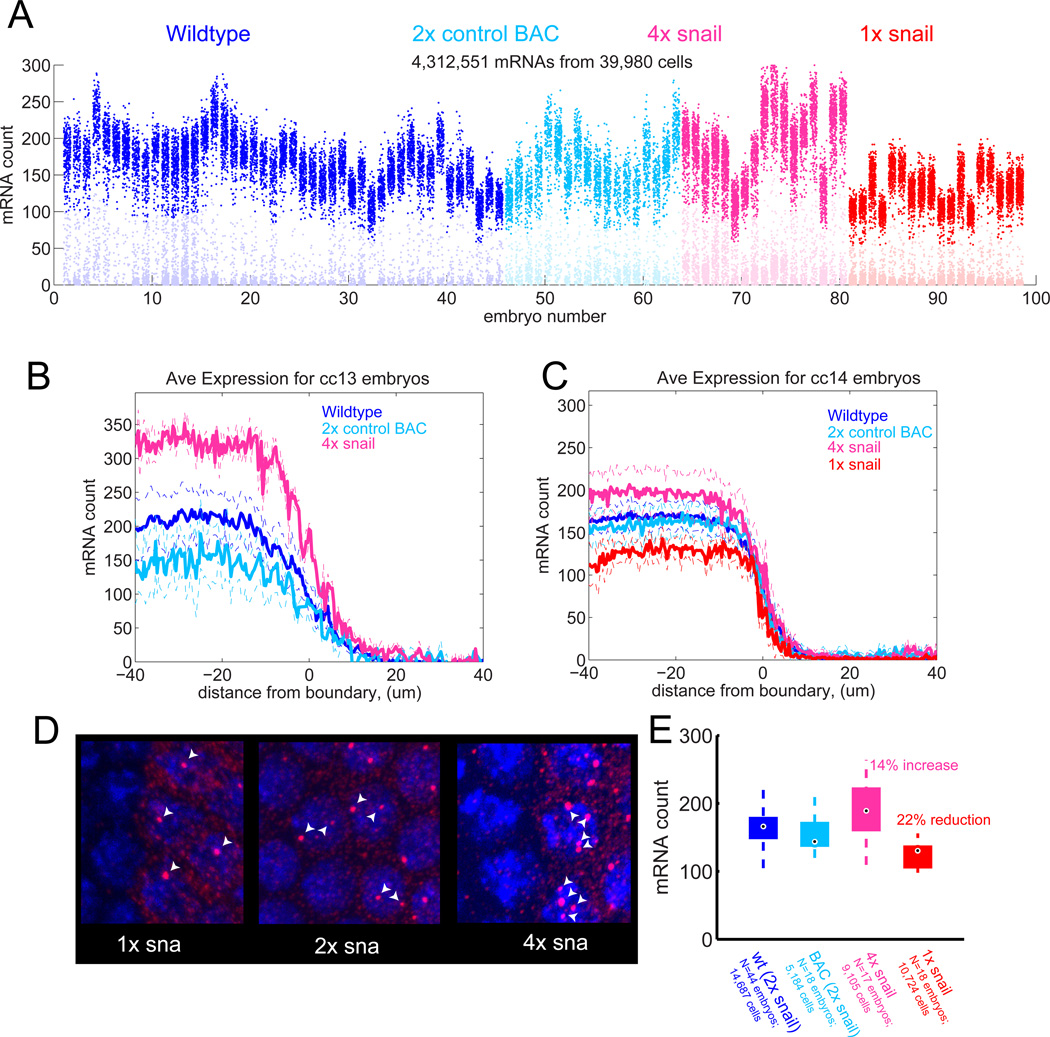
To test for the existence of such negative autoregulation, we measured the effect on snail mRNA levels of adding two additional copies of the snail locus via BAC transgenesis (Figure 4A). If the observed changes in snail resulted from cycle 14 global differences rather than specific autoregulation, doubling the number of snail alleles should double the steady state number of transcripts. If instead feedback exists, such that high levels of Snail result in down-regulation of snail production, then the increase will be significantly less than double. By the end of cc13, mRNA counts are almost twice wild-type levels, equal to the sum of the counts observed with just the 2 endogenous copies or the 2 added, transgenic copies, suggesting an absence of feedback at this stage (Figure 4B). However, by mid-cc14 this over-expression has largely been suppressed, and the average snail mRNA counts are only 14% higher in the 4X background as compared with wild-type embryos (Figure 4C). It would appear that higher levels of Snail protein reduce snail mRNA accumulation. This could be through direct autorepressive effects of Snail protein binding to snail regulatory regions. Consistent with an auto-regulatory role for snail, whole-genome ChIP binding data (Zeitlinger et al., 2007b) reveal substantial peaks of Snail binding at both the proximal and distal (shadow) enhancers (Figure S4).
Figure 4. Dosage compensation by weak negative feedback.
(A) Counting results from 98 cycle 14 embryos with wildtype snail locus (blue), 2 copies of a snail BAC in a snail deletion background (cyan), this BAC in a wildtype background providing 4 copies of snail (pink), single copy of snail (red). (B) Average spatial profiles of mRNA expression from embryos at mitosis of cycle 13: endogenous snail (blue), BAC transgene (cyan) or both (pink). 1X snail embryos can not be identified at mitotic stages due to the absence of nascent transcripts, see (D). (C) As in (B) but for cycle 14. (D) Identification of 1× snail embryos by counting nascent transcripts. Note the median number of detectable nascent transcripts per cell provides a reliable indication of the copy number for snail. (E) Box-and-whisker plot summarizing effect of copy number on mRNA levels. Whiskers mark the positions of the highest and lowest count in sample. Boxes indicate inter-quartile range and median.
To further test the ability of Snail to compensate for changes in production rate, we examined snail/+ heterozygotes (Figure 4C–E). By mid-cc14, these embryos show only 22% lower levels of snail mRNAs as compared with wild-type levels (Figure 4C). As a control we compared these results to measurements from embryos where the snail coding sequence was replaced with the yellow reporter gene within the BAC transgene. In these embryos we see essentially equal levels of yellow and snail transcripts at cc14 (attesting to the full functionality of the transgenes). Embryos containing a single copy of the snail/yellow BAC transgene exhibit the expected 50% reduction in transcript levels (Figure S2C–F). This suggests that snail is de-repressed in snail/+ heterozygotes as compared with +/+ wild-type embryos at cc14.
Weak autorepression also helps explain the progressive decrease in cell-cell variability of snail expression over time (Figure 3E). Once engaged, cells with too much mRNA experience reduced transcription because of negative feedback, while cells with too little mRNA are derepressed, experiencing enhanced transcription until they approach normal expression levels.
The preceding findings significantly extend our understanding of transcription variability in the Drosophila embryo. Previous studies suggest that shadow enhancers and paused Pol II might help suppress cell-cell variations of gene expression within an embryo and among different embryos. However, these earlier studies examined nascent transcripts within nuclei and did not characterize variation in the accumulation of cytoplasmic mRNAs (e.g. Perry et al., 2010). Consequently, this study provides the first estimate of transcription kinetics in the Drosophila embryo. We estimate that snail transcription is quite rapid and might serve to reduce variability in gene expression. We have also presented the first evidence that Snail autorepression dampens transcriptional noise and buffers fluctuations in snail gene dose.
In summary, we have presented evidence that snail exhibits remarkably homogenous expression in the presumptive mesoderm of early embryos. There is only ~10–12% variation in the levels of snail transcripts by the time that steady-state expression is established in cc14 embryos. This homogeneity contrasts with the heterogeneous expression observed for maternally-driven lacZ transcripts, induced expression of select genes in mouse or C. elegans embryos (Raj et al., 2010) or cultured cells (Raj et al., 2006; Zenklusen et al., 2008; To and Maheshri, 2010). During the early phases of snail expression, rapid rates of RNA synthesis minimize noise by driving transcription towards the saturation limit set by the intrinsic properties of Pol II processivity. Once sufficient protein has accumulated to allow transcriptional feedback, weak auto-regulation dampens the transcription rate and further contributes towards uniform expression. Only 30 min after steady-state expression is achieved during cc14, the snail-expressing cells undergo coordinated invagination at the onset of gastrulation. We propose that the exquisite homogeneity of snail expression helps coordinate this complex morphogenetic process.
Materials and Methods
Fly crosses and embryo selection
Flies containing the snail-BAC transgene on chromosome III (with a miniwhite casette) were crossed and back-crossed to y,w; wg[Sp]/CyO-hb-lacZ ; Pr,Dr/TM3,Ser,Sb to generate flies of genotype: y,w; wg[Sp]/CyO-hb-lacZ ; BAC-transgene/TM3,Ser,Sb. Flies of genotype: y,w; osp[29]/CyO, containing an allele which deletes esg and snail (Gift from Tony Ip) were also crossed and back-crossed to y,w; wg[Sp]/CyO-hb-lacZ; Pr,Dr/TM3,Ser,Sb to generate y,w; osp[29]/CyO; Pr,Dr/ TM3,Ser,Sb. These flies were crossed with y,w; wg[Sp]/CyO-hb-lacZ ; BAC-transgene/TM3 and progeny with CyO and miniwhite (linked to BAC construct), that lacked wg[Sp] were selected. The BAC was then homozygoused by selection against TM3, Sb. Rescue embryos were identified by selecting against expression of esg and lacZ. Embryos from y,w; osp[29]/CyO parents were analyzed for the 1× snail measurements, and identified by the predominance of single sites of nascent transcription and lack of multiple sites within a nucleus.
Probe synthesis
Probes were synthesized by in vitro transcription with digoxigenin or biotin RNA labeling mix (Roche Applied Sciences) from PCR linearized DNA templates. Long probes were fractionated into 100–500 bp fragments by short treatment with carbonate buffer and fractionation was checked on a gel. For competition assays short ~500 bp template sequences were used and probe length validated without carbonate treatment. Isolated probe RNA was stored in fresh hybridization buffer (50% formamide (Amresco), 5× SSC, 100 ug/mL sonicated salmon sperm DNA (Sigma-Aldrich), 50 ug/mL heparin (Sigma-Aldrich), 0.1% Tween-20 (Fisher Sciences)) at −20C.
In situ hybridization for mRNA counting
Embryos were dechorinated in bleach and washed with 0.1% Triton-X and water, before being fixed in 8% formaldehyde (Polysciences Inc) and heptane for 25 min. Embryos were then devitelinized by shaking in heptane and methanol and stored in methanol. Prior to labeling embryos were gradually moved to ethanol, treated in xylenes for 1 hr, post-fixed with 4% formaldehyde for 25 min, washed and incubated in hybridization buffer at 55C. Probes (1–3 ng/uL in hybridization buffer) were then added for 15 to 18 hours at 55C, and were rinsed out with heated hybridization buffer, washed, blocked, and incubated in primary antibodies (shp a-dig at 1:400 dilution, (Roche 11333089001), m a-bio at 1:400 (Invitrogen 03-3700)) overnight at 4C. Samples were washed and blocked again, incubated in secondaries antibodies (Alexa488 a-m and/or Alexa555 a-shp at 1:500, (Invitrogen A-21436,A-21202)) for 1.5 hours at 22C, washed again, and treated with Draq5 (BioStatus) to label nuclei. All wash steps are for a minimum of 1 to 3 hours and involve a minimum of five fluid changes. After a brief rinse whole embryos were mounted in Prolong Gold (Invitrogen) and cured overnight prior to imaging.
Image Acquisition
Images were acquired using a 63× oil immersion objective on a Zeiss 700 laser scanning confocal microscope at slow scan speed. The objective was allowed an hour of warm up scanning prior to imaging to avoid changes in the focal plane which occur during this period. Images were acquired at 2048×2048 pixels in 50 z-sections and were taken spanning the epithelial layer in 330 nm steps.
Image Processing
Zeiss .lsm files were converted to .tif files using custom Matlab scripts available in our online software toolbox through Github (https://github.com/AlistairBoettiger/Image_Analysis). Nuclear images were smoothed with a difference of Gaussians filter and segmented using a watershed algorithm. The resulting area map was used to assign mRNA to parent nuclei. Since a substantial portion of the data was acquired prior to cellularization cell membrane labeling could not be used for assignment. The mRNA counts are then corrected for differences in cell areas. This prevents brighter nuclei which get slightly larger regions during segmentation from having inflated counts. The algorithm for detecting and counting mRNA is illustrated schematically in Figure 1 and described in detail in the Extended Experimental Procedures. Briefly, in each frame, bright spots are identified by a difference of Gaussians filter followed by an adaptive, image dependent threshold and then a watershed algorithm to split neighboring fused spots. Absolute spot intensity varies in images depending on probe length, depth of the section, and small displacements out of the focal plane. The adaptive threshold is based on maximizing the number of objects in the field, which balances intensity variation and minimizes fusing of adjacent diffraction separated regions. Next, spots in consecutive z-planes are stitched together based on centroid alignment. Most molecules appear in at least two consecutive planes and often three. Localizations not traceable through at least two frames are not counted.
Supplementary Material
Highlights.
The gene snail is expressed with unprecedented low cellular variation.
The rapid snail mRNA synthesis approaches the kinetic limits of RNA Pol II processivity.
Continual, rapid transcription reduces transcription variation in early expression phases.
Negative auto-regulation reduces transcription variation in later expression phases.
Acknowledgments
The authors thank Shalev Itzkovitz and Arjun Raj, for generously providing original data from their respective publications (Itzkovitz et al., 2011; Raj et al., 2006, 2010), Tony Ip for the gift of osp[29] flies, the Kunes lab and Zhuang lab for use of facilities, Chiahao Tsui for technical support, and Jacques Bothma, Mounia Lagha, Mike Perry and other members of the Levine lab for helpful discussions. A.N.B. was supported by an N.S.F. graduate research fellowship and a postdoctoral fellowship grant from the Damon Runyon Foundation. This work was funded by a grant from the National Institute of Health (GM46638) to M.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nature structural & molecular biology. 2009;16:1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Ralph PL, Evans SN. Transcriptional regulation: effects of promoter proximal pausing on speed, synchrony and reliability. PLoS computational biology. 2011;7:e1001136. doi: 10.1371/journal.pcbi.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, De Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nature structural & molecular biology. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3’ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Itzkovitz S, Lyubimova A, Blat IC, Maynard M, Van EsJ, Lees J, Jacks T, Clevers H, Van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nature Cell Biology. 2011;14:1–10. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18159–18164. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Ip YT, Levine M, Arora K. Establishment of the Mesoderm-Neuroectoderm Boundary in the Drosophila Embryo. Science. 1991;254:118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes & Development. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- Munsky B, Neuert G, Van Oudenaarden A. Using Gene Expression Noise to Understand Gene Regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri Ea, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Paré A, Lemons D, Kotake Y, Beaver W, Freund Y, McGinnis W. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Current Biology. 2009;19:2037–2042. doi: 10.1016/j.cub.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Current Biology. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2011;108 doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Van Den Bogaard P, Rifkin S, Van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biology. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, Van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So L, Ghosh A, Zong C, Sepúlveda La, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nature genetics. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T-L, Maheshri N. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science. 2010;327:1142–1145. doi: 10.1126/science.1178962. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007a;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young Ra, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes & Development. 2007b;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature structural & molecular biology. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.