Abstract
The plant-specific NAC transcription factors (TFs) play important roles in regulation of diverse biological processes, including development, growth, cell division and responses to environmental stimuli. In this study, we identified the members of the NAC TF family of chickpea (Cicer arietinum) and assess their expression profiles during plant development and under dehydration and abscisic acid (ABA) treatments in a systematic manner. Seventy-one CaNAC genes were detected from the chickpea genome, including 8 membrane-bound members of which many might be involved in dehydration responses as judged from published literature. Phylogenetic analysis of the chickpea and well-known stress-related Arabidopsis and rice NACs enabled us to predict several putative stress-related CaNACs. By exploring available transcriptome data, we provided a comprehensive expression atlas of CaNACs in various tissues at different developmental stages. With the highest interest in dehydration responses, we examined the expression of the predicted stress-related and membrane-bound CaNACs in roots and leaves of chickpea seedlings, subjected to well-watered (control), dehydration and ABA treatments, using real-time quantitative PCR (RT-qPCR). Nine-teen of the 23 CaNACs examined were found to be dehydration-responsive in chickpea roots and/or leaves in either ABA-dependent or -independent pathway. Our results have provided a solid foundation for selection of promising tissue-specific and/or dehydration-responsive CaNAC candidates for detailed in planta functional analyses, leading to development of transgenic chickpea varieties with improved productivity under drought.
Introduction
Chickpea (Cicer arietinum L.) is one of the major legume crops cultivated throughout the world, especially in the Afro-Asian countries, providing great supplies of protein-, carbohydrate-, mineral-, vitamin-, and health-promoting fatty acid-rich food for human consumption [1]. A number of chickpea by-products, such as low-grade and culled chickpeas, chickpea hay and straw and chickpea pod husks are also widely used for animal feeding [2]–[4]. However, chickpea productivity is severely affected by drought which has made development of drought-tolerant chickpea cultivars is the most important goal in many chickpea research programs [5]–[7].
To cope with drought stress, intensive research has been conducted in recent years in both model and crop plants to discover and elucidate genes and molecular mechanisms that regulate drought responses [8]–[11]. Within the regulatory networks that control the signal transduction from stress signal perception to stress-responsive gene expression, various transcription factors (TFs) and their DNA binding sites, the so-called cis-acting elements, act as molecular switches for stress-responsive gene expression, enabling plants adapt better to the adverse stressor [12], [13]. Discovery and genetic engineering of genes encoding novel TFs have the potential to develop transgenic crop plants with superior yield under stress conditions.
The plant-specific NAC (NAM - no apical meristem, ATAF - Arabidopsis transcription activation factor, and CUC- cup-shaped cotyledon) TF family was discovered in Petunia more than 18 years ago [14]. Since then an amazingly large number of studies have provided evidence for the functions of NAC members in almost every biological process in plants, ranging from lateral root formation [15], embryo development [16], flowering [17], regulation of secondary cell wall synthesis, cell division [18], to biotic and abiotic stress responses [19]–[22]. A typical NAC TF contains a highly conserved N-terminal DNA-binding NAC domain and a variable C-terminal transcriptional regulatory region (TRR) that can serve as either a transcriptional activator or a repressor [19], [21]. Several cis-motifs have been identified as DNA-binding sites for the NAC TFs, including the drought-responsive NAC recognition sequence (NACRS) [23], the iron deficiency-responsive IDE2 motif [24], the calmodulin-binding CBNAC [25], the secondary wall NAC binding element (SNBE) [26], and the 21-bp sequence motif (−83 to −63) in the 35S promoter [15]. Being multiple functional proteins, NAC TFs are also able to mediate protein-protein interactions through their DNA-binding NAC domains [15], [19]. A number of the NAC TFs contain transmembrane (TM) helices (TMHs) in their C-terminal region that are responsible for the anchoring to the plasma membrane [27]. These NAC members are classified as membrane-associated, designated as NTL (NTM1-Like or “NAC with Transmembrane Motif 1”-Like) TFs and are grouped in NTL subfamily. Studies of NTL members in Arabidopsis and rice (Oryza sativa) indicated that the majority of the NTL genes are stress-responsive [27], [28].
The advance in genomic sequencing has allowed research community to identify the NAC family members in many sequenced species at genome-wide scale, such as 117 genes in Arabidopsis, 151 in rice [29], 163 in poplar (Populus trichocarpa) [30], 152 in tobacco (Nicotiana tabacum) [31], 152 in maize (Zea mays) [32], [33], 147 in foxtail millet (Setaria italica L.) [34], 110 in potato (Solanum tuberosum) [35], 74 in tomato (Solanum lycopersicum) [36], 204 in Chinese cabbage (Brassica rapa) [37], 88 in pigeonpea (Cajanus cajan) [38] and approximately 200 in soybean (Glycine max) [39] of which 152 members were identified with full-length open reading frame (ORF) [40]. Taking the advantage of the available genomic sequence of chickpea [41], [42], in this study we have identified CaNAC genes in annotated chickpea genome and provided a nomenclature for all the identified CaNAC members. We also carried out sequence alignment and phylogenetic analyses to classify the CaNACs according to their phylogeny. Additionally, we studied the expression patterns of the CaNAC genes in various organs under different developmental stages using available transcriptome data. Furthermore, to identify CaNAC candidate genes responsive to dehydration/drought in either ABA (abscisic acid)-dependent or independent manner for in planta functional studies, we characterized the expression profiles of phylogenetically predicted stress-related CaNAC and membrane-bound CaNAC/CaNTL genes in leaves and roots of chickpea plants treated with dehydration or ABA using real-time quantitative PCR (RT-qPCR). Our results have provided an insight into the regulatory functions of the CaNACs in chickpea, and laid a foundation for in-depth in planta functional characterization of selected CaNAC genes with the final aim to use them for the improvement of drought tolerance in chickpea by genetic engineering.
Materials and Methods
Plant growth, treatments and collection of tissues
Seeds of chickpea (Cicer arietinum L.) Hashem “kabuli” cultivar [43] were germinated in pots containing vermiculite and were well-watered and grown under greenhouse conditions (continuous 30°C temperature, photoperiod of 12 h/12 h, 150 µmol m−2 s−1 photon flux density and 60% relative humidity). For expression profiling of CaNAC genes under normal and dehydration stress conditions, 9-day-old chickpea plants were subjected to dehydration, ABA and water (control) treatments for 2 and 5 h as previously described [44]. The relative water content of chickpea seedlings was 55% at 2 h and 33% at 5 h after dehydration. Leaf and root tissues of treated plants were separately collected for expression analysis.
Identification of the CaNAC genes in chickpea
All CaNACs annotated in genotype CDC Frontier, a larger-seeded chickpea “kabuli” cultivar (Ca v1.0) [41] were first collected from PlantTFDB (http://planttfdb.cbi.pku.edu.cn/index.php) [45] and from iTAK (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) for manual analysis. The sequences of the CaNAC genes and encoded proteins were then individually checked at NCBI (Bioproject: PRJNA175619) [41], using both blastN and blastP to identify the chromosomal location of each CaNAC gene. Sequences of CaNAC genes were also collected from the genome of the small-seeded “desi” chickpea ICC4958 cultivar (Ca v1.0) (Bioproject: PRJNA78951) available at http://nipgr.res.in/CGAP/home.php for comparison [42]. TMHHM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was applied for prediction of the membrane-bound CaNAC/CaNTL members.
Phylogenetic analysis
Sequence alignments of NAC proteins from chickpea, Arabidopsis and rice were performed with a gap open penalty of 10 and a gap extension penalty of 0.2 to construct the unrooted phylogenetic trees by the neighbor-joining method using MEGA (V6.0) software (http://www.megasoftware.net/) [46]. The confidence level of monophyletic groups was estimated using a bootstrap analysis of 10 000 replicates. Bootstrap values are displayed next to the branch nodes. The alignments were subsequently visualized using GeneDoc (http://www.nrbsc.org/gfx/genedoc/) as presented in Figure S1.
In silicon expression analysis of CaNAC genes
Expression data available for each putative CaNAC gene were retrieved from the Chickpea Transcriptome Database (CTDB) (http://www.nipgr.res.in/ctdb.html) [47], and used for expression analysis of the CaNACs in different tissues and organs of chickpea during development. Detailed information about sample collections for transcriptome analyses was provided in references [47]–[49]. Briefly, shoots and roots were collected from 15-day-old plants [48], and shoot apical meristem (SAM) was dissected from 21-day-old plants [49] grown in pots containing autoclaved mixture (1∶1) of agropeat and vermiculite in culture room (22±1°C, photoperiod of 14 h). Young leaves, mature leaves, flower buds (FB1-FB4, where FB1, FB2, FB3 and FB4 were 4 mm, 6 mm, 8 mm and 8–10 mm in size, respectively), flowers (FL1-FL4, where FL1 was young flower with closed petals, FL2 was mature flower with partially opened petals, FL3 was mature flower with opened and faded petals and FL4 was drooped flower with senescing petals) and young pods were harvested from field-grown plants [47], [49]. Germinating seedlings (GS) were 5-day-old plants grown on wet Whatman papers in Petri dishes [47].
RNA isolation, DNaseI treatment, cDNA synthesis
Collected chickpea leaf and root samples were ground into a fine powder using Retsch MM300 shaker and mortar and pestle, respectively. Total RNA was isolated using RNeasy Plant Mini Kit and QIAcube system (Qiagen). Measurement of RNA concentration, DNaseI digestion and cDNA synthesis were performed according to methods published earlier [44].
RT-qPCR and statistical analyses
Gene-specific primers for selected CaNAC genes were designed using the Primer 3 [50] (Table S1). RT-qPCR reactions and data analyses were performed as previously described [51]. The IF4a gene was used as a reference gene [52], and the ΔCT method was used to calculate initial amount of target genes [53]. Statistical significance of the differential expression patterns between treatments was determined using the Student's t-test (one tail, unpaired, equal variance). For considering a gene as dehydration- or ABA-induced or -repressed, the criterion of minimum 2-fold expression change (at least at one time point) with P-value <0.05 was applied.
Results and Discussion
Identification and nomenclature of the CaNAC genes in chickpea
To identify all the CaNAC genes annotated in the chickpea genome, we first collected all the predicted CaNAC genes from PlantTFDB and iTAK. These two databases collected the TF sequences from the annotated genomic sequence (Ca v1.0) of the genotype CDC Frontier, a chickpea “kabuli” cultivar [41]. Next, all the CaNAC gene sequences were subjected to a sequence comparison to remove all the overlapped genes to build a list of 71 potential CaNAC genes in chickpea (Table S2). Subsequently, the identified CaNAC genes were blasted against the assembled "kabuli" genome (Ca v1.0) available at NCBI (Bioproject: PRJNA175619) to identify their chromosomal location of each CaNAC gene (Table S2). If the gene annotation predicted several splice variants for a given gene, splice variants encoding the longest open reading frames were selected as representative members as provided in Dataset S1 along with their respective protein sequence.
In addition to the genomic sequence of the larger-seeded chickpea “kabuli” CDC Frontier cultivar, the genomic sequence of the small-seeded “desi” ICC4958 chickpea cultivar was also available from another independent genome sequencing project (Bioproject: PRJNA78951) [42]. Thus, we also searched for the CaNAC genes annotated in the “desi” ICC4958 chickpea cultivar. Only 62 CaNACs were found in the “desi” ICC4958 annotated genome, with 9 members less as compared with those identified in “kabuli” CDC Frontier genome (Table S2). This may be due to the fact that a lower number of protein encoding genes (27,571 genes) were annotated in the “desi” chickpea genome [42], in comparison with the “kabuli” chickpea genome (28,269 annotated genes) [41]. Additionally, Jain et al. [42] estimated that chickpea genome might have around 32,000 genes, which is approximately 13–15% more than the number of currently annotated gene models. Therefore, we might expect to identify more CaNAC(s) in chickpea genome in future by fine-tuning of the annotation. We proposed a nomenclature to name the identified CaNAC members from CaNAC01 to CaNAC71 following their chromosomal localization in the “kabuli” chickpea genome and the chromosomal order starting with chromosome 1.
Chromosomal localization, gene duplication and phylogenetic analyses of CaNAC TFs
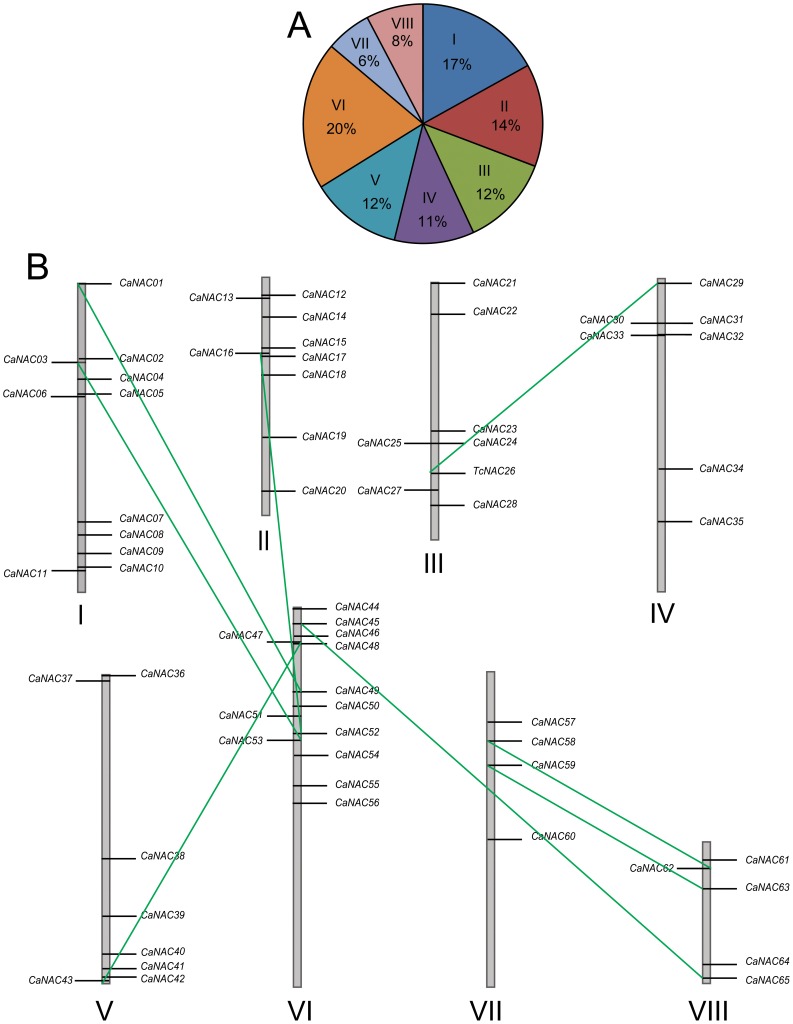
Out of 71 identified CaNAC genes, 65 members were able to be mapped to the 8 chickpea chromosomes according to the currently available sequence data [41], [42]. These 65 CaNACs are distributed on the 8 chromosomes with an uneven ratio (Figure 1A). The highest number of CaNACs was detected on chromosome VI, with 13 members representing ∼20% of the identified CaNACs, while the lowest number of CaNACs was found on chromosome VII, containing 4 out of 65 mapped CaNAC genes (∼6%) (Figure 1A). The exact location site of each CaNAC gene is shown in Table S2, and the relative locations of the CaNACs on their respective chromosome are illustrated in Figure 1B. Among the 65 CaNACs, using the criterion >60% homology at nucleotide level we found 8 duplicated pairs, none of which was tandemly duplicated pair (Figure 1B). In comparison with chickpea, in soybean 13 tandemly duplicated clusters of 2 or more GmNAC members were identified among 152 examined GmNACs [40]. This observation might suggest the absence of the recent whole genome duplication (WGD) in chickpea, as found in soybean ∼13 million years ago [54]. Indeed, analysis of the rates of synonymous substitution per synonymous site (Ks) within the paralogous gene pairs indicated that the latest WGD event in chickpea was ∼58–60 million years ago [41], [42].
Figure 1. Distribution percentage of 65 chickpea CaNAC genes identified in this study in 8 chickpea chromosomes.
(A) Chromosomal distribution of CaNAC genes with their percentage on each chromosome. (B) Graphical representation for chromosomal localization of CaNAC genes. Greek numbers indicate chromosome numbers. Green lines indicate duplicated gene pairs.
A multiple alignment indicated that all of the CaNACs shared a highly conserved N-terminal DNA binding NAC domain, which consists of five consensus subdomains (A–E), and a variable C-terminal transcriptional regulation domain. Additionally, a conserved bipartite nuclear localization signal was also found in the D subdomain of the majority of CaNACs, suggesting that these CaNACs may be localized to the nucleus (Figure S1) [55]. To examine the structure and phylogenic relationship between the CaNAC TFs and the ANACs of Arabidopsis, we constructed a unrooted phylogenic tree based on the alignment of their deduced protein sequences (Figure S2). On the basis of the phylogenetic analysis, we could classify the CaNACs into 12 subgroups together with their ANAC orthologs. This result suggests that the CaNACs are as diverse as the ANACs. On the other hand, increasing evidence has suggested that phylogenetic analysis can be used to predict the function of genes because genes with similar functions are phylogenetically related [32], [40], [56]–[58]. Thus, the phylogenetic analysis also allowed us to predict the function of the CaNACs as many ANACs have known functions [19]–[22], [59].
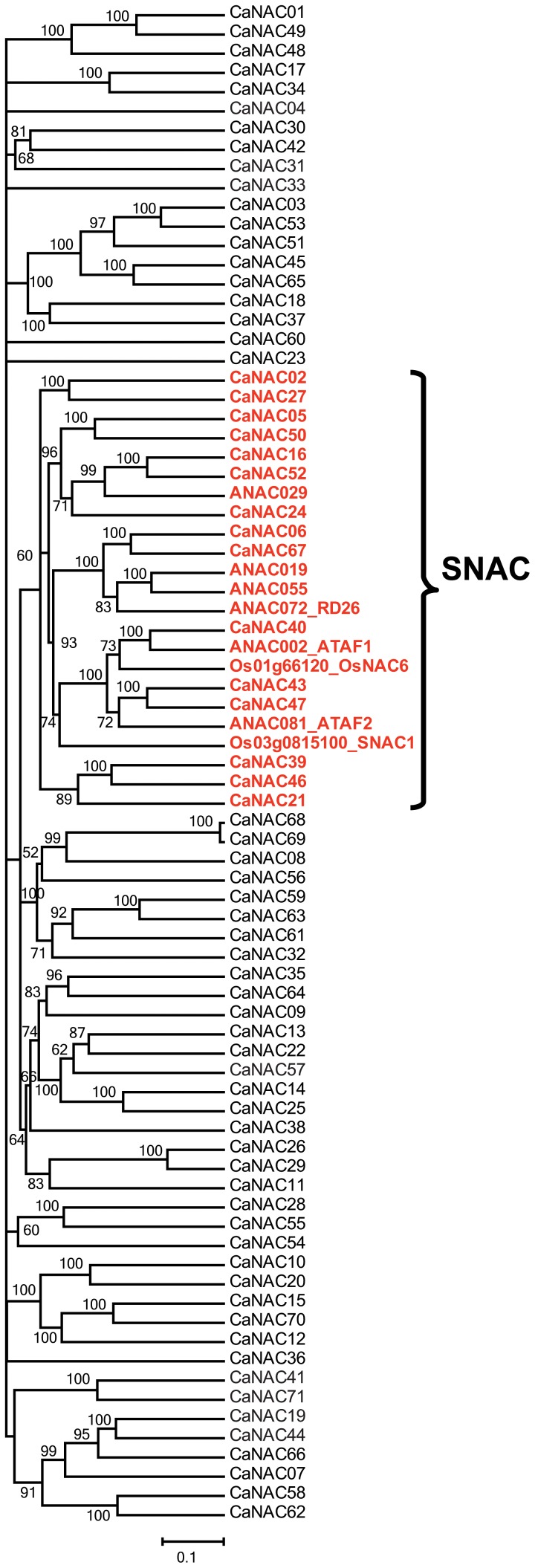
As we were interested in identifying abiotic stress-, especially drought-related, CaNAC genes through phylogenetic analysis for further studies, the most well-known stress-related ANACs and ONACs were selected and included into a phylogenetic analysis of identified CaNACs [23], [60]–[62]. According to the phylogenetic tree shown in Figure 2, by using these 6 stress-related Arabidopsis ANACs (ANAC002, 019, 029, 055, 072 and 081) and 2 stress-related rice ONACs (SNAC1/ONAC002 and OsNAC6/SNAC2/ONAC048), 15 CaNACs could be classified as stress-related TFs. Obviously, we cannot rule out that there would be more stress-related CaNAC genes, scattered on other branches of the tree, out of 71 identified CaNACs. As evidenced in soybean, when more stress-related ANAC and ONAC proteins were used in phylogenetic analysis-based prediction, more stress-related GmNAC genes, clustered into different clades, were predicted [40].
Figure 2. Prediction of stress-responsive CaNAC genes based on phylogenetic analysis.
The unrooted phylogenetic tree was constructed using the full NAC protein sequences of all 71 CaNAC and well-known stress-responsive NAC genes from Arabidopsis (ANAC002, 019, 029, 055, 072 and 081) and rice (SNAC1/ONAC002 and OsNAC6/SNAC2/ONAC048). Bootstrap values are displayed next to the branch nodes. SNAC, stress-related NAC group.
The membrane-associated CaNAC/CaNTL subfamily
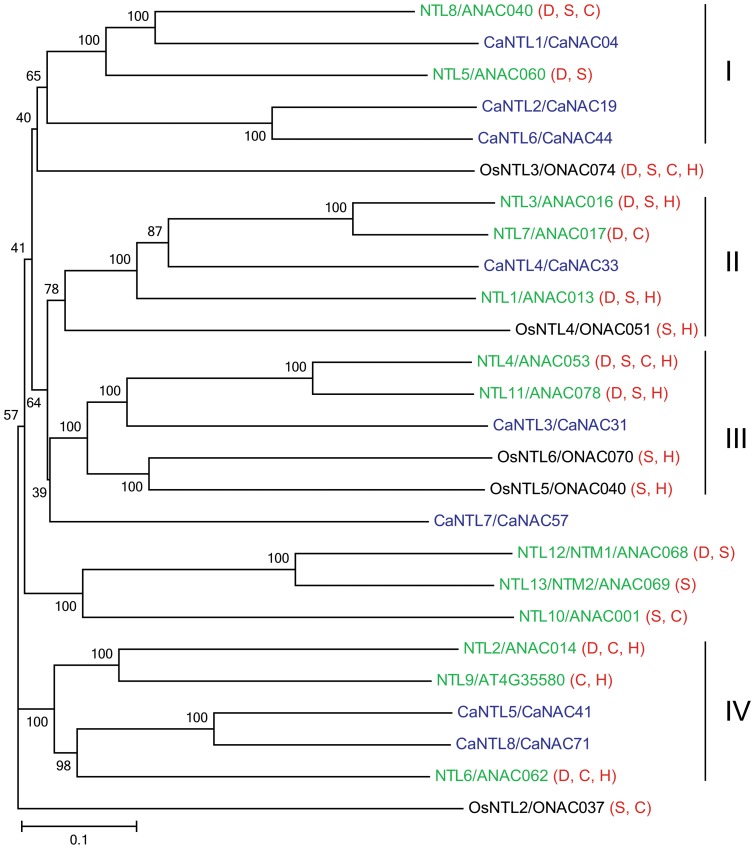
Membrane-associated NTL TFs are stored in their dormant form, and when required, their cytoplasmic anchors are degraded, resulting in activated TFs that will then enter the nucleus to regulate expression of target genes [27]. Among 71 CaNACs, 8 members (CaNAC04, 19, 31, 33, 41, 44, 57 and 71) were identified as membrane-associated CaNTLs using the TMHMM v2.0 (Table 1), of which 4 (CaNAC31, 33, 41 and 71) and 4 (CaNAC04, 19, 44 and 57) members contain one and two TMHs, respectively. When compared with NTLs identified in other recently studied plant species, out of 11 putative GmNTLs of soybean 2 members possess two TMHs [40], whereas all the NTLs predicted in Arabidopsis, rice, maize, potato, foxtail millet, Chinese cabbage and tomato contain only one TMH [27], [28], [33]–[37], suggesting that the existence of doubled TMHs might be specific to leguminous plants. Interestingly, among 8 CaNTLs, CaNAC57/CaNTL7 contains both of its two TMHs in its N-terminal region. With the exception of SlNAC65 of tomato, which has only a TMH in the N-terminus, none of the other NTLs identified in Arabidopsis, rice, maize, potato, foxtail millet, Chinese cabbage and tomato has TMH(s) located in the N-terminus [27], [28], [33]–[37]. As the so-called “Exp number, first 60 AAs” (“Exp” = “Expected”) of CaNTL7 provided by the TMHMM server 2.0 was 35.9 (Table 1), much higher than 10, the two predicted TMHs of CaNAC57 might be parts of a signal peptide. In agreement with the unique structure of CaNTL7, a phylogenetic tree constructed from the CaNTLs from chickpea (CaNTLs/CaNACs), Arabidopsis (NTLs/ANACs) and rice (OsNTLs/ONACs) indicated that the chickpea CaNTLs were scattered into 4 major groups, whereas the CaNTL7 stayed alone on a distinct branch (Figure 3).
Table 1. Putative membrane-bound chickpea CaNTLs.
| Gene name | Membrane-bound member | Length (aa) | Transmembrane sequences | Exp number of AAs in TMHs | Exp number, first 60 AAs | |
| CaNAC04 | CaNTL1 | 476 | 331…350 | 362…384 | 43.78637 | 0.00061 |
| CaNAC19 | CaNTL2 | 624 | 535…552 | 600…622 | 38.70603 | 0 |
| CaNAC31 | CaNTL3 | 578 | 553…575 | 20.90626 | 0.0002 | |
| CaNAC33 | CaNTL4 | 558 | 530…552 | 21.93664 | 0.00065 | |
| CaNAC41 | CaNTL5 | 612 | 585…607 | 22.28558 | 0.00171 | |
| CaNAC44 | CaNTL6 | 636 | 539…558 | 614…633 | 40.15937 | 0 |
| CaNAC57 | CaNTL7 | 430 | 15…32 | 37…59 | 39.78718 | 35.9005 |
| CaNAC71 | CaNTL8 | 610 | 582…604 | 22.60723 | 0 | |
AA, amino acid; Exp, expected; TMHs, transmembrane helices.
Figure 3. Phylogenetic tree of membrane-bound NACs from chickpea (CaNTLs/CaNACs), Arabidopsis (NTLs/ANACs) and rice (OsNTLs/ONACs).
The unrooted phylogenetic tree was constructed using the full protein sequences. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch. Stress-responsiveness of each NTL gene is shown next to its name in the parentheses. D, dehydration/drought; S, salt stress; C, cold stress; H, heat stress.
Expression patterns of CaNAC genes in various tissues during development
Tissue-specific expression profiles are helpful as these data enable us to determine whether a gene of interest plays a role in defining the precise nature and function of given tissue(s). CTDB (www.nipgr.res.in/ctdb.html) provided a comprehensive transcriptome atlas that was generalized for young chickpea seedlings and various types of chickpea tissues collected at various stages of development, including roots, shoots, shoot apical meristem, young leaves, mature leaves, flower buds, flowers and young pods, using either 454 pyrosequencing (Figure 4A) [47] or Illumina sequencing (Figure 4B) [49]. Overall, the expression data for 44 CaNAC genes in these tissues could be retrieved from the CTDB, which were presented in a heatmap representation shown in Figure 4. According to the data, the CaNACs possess highly variable transcript abundance. For example, CaNAC01, CaNAC49 and CaNAC63 exhibited a very weak expression in all the tissues as compared with other CaNACs. The putatively predicted stress-related CaNACs (SNACs) (Figure 2) and the membrane-bound CaNTLs (Table 1) are among those with high transcript abundance measured in the tissues. A number of CaNAC genes exhibited differential expression patterns being specific in some particular tissues, such as CaNAC16, CaNAC20 and CaNAC50 (Figure 4A), while many of them appeared to be ubiquitously expressed in the tissues examined across the developmental stages. This phenomenon was also observed for the NAC genes in other plants, such as Arabidopsis, rice and soybean, suggesting that the functions of the NACs are diversified both in monocotic and dicotic plants [20], [40], [59], [63]. Additionally, increasing evidence has suggested that overexpression of tissue-specifically expressed genes can promote the development of that particular tissue. Transgenic Arabidopsis with overexpressed NAC1 and AtNAC2 genes, which are preferentially expressed in roots, displayed enhanced lateral root development [15], [64]. Overexpression of the rice SNAC1 gene, which was induced mainly in guard cells by drought, resulted in an enhanced stomatal function under drought, leading to an increase in drought tolerance [60]. Thus, taken together our results provide a first insight for the readers to link the CaNAC genes to their putative in planta functions through their temporal and spatial expression patterns.
Figure 4. Heatmap representation for expression of CaNAC genes in different tissues.
(A) The expression data generated by 454 pyrosequencing of cDNA libraries prepared from shoots, roots, mature leaves, flower buds and young pods were obtained from CTDB. Elevated expression levels are indicated by increasing intensities of brown color expressed in RPM (reads per million) values. (B) The expression data generated by Illumina sequencing of RNA-seq libraries prepared from germinating seedling (GS), young leaf (YL), shoot apical meristem (SAM), flower bud stages (FB1-FB4) and flower stages (FL1-FL4) were obtained from CTDB. Blue and red color gradients indicate an increase or decrease, respectively, in transcript abundance represented in log2 values. NTLs, membrane-bound CaNACs; SNACs, stress-related CaNACs.
Expression patterns of predicted stress-related and membrane-bound CaNAC genes in chickpea roots and leaves during dehydration treatment
Previously, through phylogenetic analysis using several well-known stress-related ANAC and ONAC proteins, which are not membrane-bound NTLs, as seed sequences, 15 CaNAC genes were predicted to be stress-related (Figure 2). On the other hand, all of the NAC genes encoding membrane-bound NTLs in Arabidopsis and rice were reported to be induced by at least one type of environmental stresses, namely dehydration/drought (D), salt (S), cold (S) or heat (H) stress as summarized from published literature and visualized in Figure 3 [21], [65]. Thus, in order to identify dehydration-responsive genes for our follow-up in planta functional analyses, next we used RT-qPCR to examine the expression of 23 CaNAC genes, including all 15 stress-related CaNACs predicted by phylogenetic analysis and all 8 membrane-bound CaNTLs, in leaf and root tissues of dehydrated chickpea plants. The expression analyses separately performed with dehydrated chickpea leaves and roots might provide information on the tissue-specific mode of action of the tested CaNACs under dehydration.
Using the criterion of fold-change ≥2 and P<0.05, the majority of the tested CaNACs were found to be dehydration-responsive in leaves and/or roots of chickpea plants (Figure S3A). Among the 23 CaNACs, 14 genes, of which 3 CaNTLs (CaNTL2/CaNAC19, CaNTL5/CaNAC41 and CaNTL7/CaNAC57), were up-regulated, whereas only 4 genes, of which one CaNTL (CaNTL1/CaNAC04), were down-regulated by at least 2-fold in leaves after 2 and/or 5 h of dehydration (Figure 5, Figure S3A). CaNAC06 and CaNAC67 were the two most highly induced genes (over 200- and 300-fold, respectively), whereas CaNAC02 and CaNAC04 were the two most significantly repressed genes (23.8-fold and 28.6-fold, respectively after 5 h of dehydration) in chickpea leaves by dehydration. As for the roots, 12 genes, of which 2 CaNTLs (CaNTL2/CaNAC19 and CaNTL6/CaNAC44), were induced, whereas 3 genes, of which one CaNTL (CaNTL1/CaNAC04) were repressed by at least 2-fold after dehydration for 2 and/or 5 h (Figure 6, Figure S3A). In comparison with dehydrated leaves, the degree of induction in dehydrated roots was approximately 10-fold lower, with the highest induction of ∼23-fold recorded for CaNAC67 at 5 h of dehydration. Thus, CaNAC67 being induced the most highly in both tissues is a promising candidate gene which deserves further and in-depth in planta molecular and functional analyses under drought. A number of studies have indicated that TF encoding genes with high inducibility by stress are preferable for selections of further in planta functional studies as they might have potential for development of improved stress-tolerant transgenic plants by overexpression approach [58], [66]. Additionally, the repression degree of CaNACs in dehydrated roots versus dehydrated leaves was also lower by approximately 4-fold. For instance, CaNAC02 was the most highly down-regulated by dehydration in roots with a fold-change of only 6.2. In addition, CaNAC24 was deserved to be mentioned as this gene was induced (2.6-fold and 3.7-fold at 2 and 5 h after dehydration, respectively) in dehydrated roots but repressed (3-fold at 2 h of dehydration) in dehydrated leaves. It would be then interesting to study how CaNAC24 is involved in regulation of chickpea responses to drought. We hypothesize that under drought stress the up-regulation of CaNAC24 in roots might contribute to enhancement of root development, whereas its down-regulation in leaves might contribute to repression of leaf and/or shoot growth. These changes would enhance the adaptation of chickpea plants under limited water conditions.
Figure 5. Expression of selected CaNAC genes in chickpea leaves under dehydration and ABA treatments.
Expression data were obtained by RT-qPCR of treated (ABA or dehydration) and well-watered (WW) control leaf samples collected at indicated time points. Mean relative expression levels were normalized to a value of 1 in water-treated control leaf samples. Error bars = SE values of three biological replicates. Asterisks indicate significant differences as determined by a Student's t-test (*P<0.05; **P<0.01; ***P<0.001). Membrane-bold CaNACs are underlined.
Figure 6. Expression of selected CaNAC genes in chickpea roots under dehydration and ABA treatments.
Expression data were obtained by RT-qPCR of treated (ABA or dehydration) and well-watered (WW) control root samples collected at indicated time points. Expression data were obtained by RT-qPCR of collected root samples. Mean relative expression levels were normalized to a value of 1 in water-treated control root samples. Error bars = SE values of three biological replicates. Asterisks indicate significant differences as determined by a Student's t-test (*P<0.05; **P<0.01; ***P<0.001). Membrane-bold CaNACs are underlined.
Our data together indicated that out of 23 CaNACs examined, 19 genes were dehydration responsive in roots and/or leaves of young chickpea seedlings (Figure S3A). Of 8 membrane-bound CaNACs, 5 genes were determined as dehydration-responsive, representing 62.5% of the CaNTLs and almost reaching to the percentage of Arabdopsis NTL genes identified as responsive to dehydration/drought (10/13 genes, i.e. 76.9%) (Figure 3) [21], [65]. A Venn diagram analysis indicated that the majority of dehydration-responsive CaNACs are overlapped in roots and leaves, with 10 and 3 genes up-regulated and down-regulated in both two organs, respectively (Figure S3B, left panel). Three genes, CaNAC05, 21 and 57, were induced only in dehydrated leaves, while 2 gene (CaNAC24 and 44) were specifically up-regulated in dehydrated roots only under our experimental conditions. As for down-regulation, only CaNAC24 was found to be specifically down-regulated in leaf tissues by dehydration (Figures 5–6; Figure S3A). It should also be noticed that out of 15 phylogenetically predicted stress-related CaNACs (Figure 2), 14 genes are dehydration-responsive (Figure S3A), demonstrating that the phylogenetic analysis-based method has an accuracy rate of 93.33%; a quite good rate for a prediction.
Expression patterns of predicted stress-related and membrane-bound CaNAC genes in chickpea roots and leaves under ABA treatment
It is well-established that the NAC TFs can regulate plant responses to water stress through either ABA-dependent or -independent manner [19], [22]. Thus, it was of interest to examine the expression of the selected 23 CaNACs in both roots and leaves treated with ABA. Results indicated that a total of 12 of 23 examined CaNACs were responsive to ABA as their expression levels were altered by at least 2-fold at a P-value <0.05. Out of these, 7 and 8 CaNACs were up-regulated, whereas only 2 and 2 CaNACs were down-regulated in ABA-treated leaves and roots, respectively (Figures 5–6; Figure S3A). Similar to dehydration, ABA treatment also resulted in a significant overlap among the ABA-responsive CaNAC genes detected in roots and leaves. Out of 12 CaNAC genes responsive to ABA in leaves and/or roots, 5 and 2 were found to be ABA-induced and -repressed, respectively, in both organs (Figure S3B, right panel). Additionally, according to our data, the majority of dehydration-related CaNACs identified in this present study may regulate drought-responsive responses in chickpea in an ABA-dependent manner. Out of the 19 CaNACs responsive to dehydration in leaves and/or roots, 7 genes were recorded as ABA-independent, whereas 12 CaNACs were identified as ABA-dependent. Four CaNAC genes (CaNAC31, CaNAC33, CaNAC39 and CaNAC71) were not responsive to either ABA or dehydration (Figures 5–6, Figure S3A). Our data also suggested that CaNAC02 and CaNAC67, showing the highest down- and up-regulation, respectively, in both roots and leaves by dehydration, act in dehydration/drought responses in an ABA-dependent pathway. In addition to dehydration-inducible promoters [67], ABA-inducible promoters, when coupled with dehydration-inducible genes, are also useful in biotechnological applications for enhancing drought tolerance of transgenic plants [68].
Conclusions
Research efforts on identification and characterization of the NAC TFs using high-throughput genomic surveys and expression analyses will undoubtedly describe key features of the members of this novel plant-specific TF family. As a result, our current understandings of the regulatory functions of the NAC TFs in various plant species will be definitely accelerated. Our current study, which reported the comprehensive identification and characterization of the CaNAC family in chickpea, has provided an insight into the functional diversity of the CaNAC family. Furthermore, our expression analyses of a number of CaNAC genes during development, dehydration and ABA treatments have established a solid foundation for chickpea scientists to select candidate genes and their associated tissue-specific and/or dehydration- and/or ABA-responsive promoters for follow-up in planta functional analyses, leading to engineered chickpea cultivars with enhanced drought tolerance.
Supporting Information
Multiple alignment of 71 CaNACs of chickpea and well-known stress-responsive NACs from Arabidopsis (ATAF1/ANAC002, 019, 029, 055, 072 and ATAF2/081) and rice (SNAC1/ONAC002 and OsNAC6/SNAC2/ONAC048). Conserved NAC domain and subdomains (A–E) are indicated by thick blue line and black thin black lines, respectively, above the sequences. The putative nuclear localization signal (NLS) is shown by a blue double-headed arrow below the sequence. Putative stress-related NAC subgroup is highlighted in red-colored background, and membrane-bound CaNAC members are highlighted in turquoise-colored background.
(PDF)
Phylogenetic relationship of NAC proteins from chickpea (CaNACs) and Arabidopsis (ANACs). The unrooted phylogenetic tree was constructed using the full NAC protein sequences. The membrane-bound CaNAC and ANAC proteins are indicated in blue-colored and green-colored letters, respectively.
(PDF)
Expression of 23 selected CaNAC genes in chickpea roots and leaves under dehydration and ABA treatments. (A) Summary of the results of the expression data. (B) Venn diagram analysis of dehydration- and ABA-responsive CaNAC genes in roots and leaves of chickpea plants. The ABA- and/or dehydration-responsive genes were defined as those whose expression is altered by at least 2-fold (P<0.05) at 2 and/or 5 h of dehydration and/or ABA treatment.
(PDF)
Primers used in RT-qPCR analysis.
(XLS)
Putative CaNAC genes identified in this study and their major features.
(XLS)
Nucleotide and amino acid sequences of 71 CaNAC genes obtained from (Ca v1.0) (Bioproject: PRJNA175619).
(TXT)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
CVH is supported by a PhD fellowship from "International Program Associate" of Rikagaku Kenkyusho (Institute of Physical and Chemical Research, Japan) (http://www.riken.jp/en/careers/programs/ipa/). This work was also supported in part by a grant (Project Code 905/HĐ-KHCN-CNSH) from the Ministry of Agriculture and Rural Development of Vietnam and the Ministry of Science and Technology of Vietnam to the Research Group of DVN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jukantil AK, Gaur PM, Gowda CLL, Chibbar RN (2012) Nutritional quality and health benefits of chickpea (Cicer arietinum L.): A review. Br J Nutr 108:11–26. [DOI] [PubMed] [Google Scholar]
- 2. Rubio LA (2005) Ileal digestibility of defatted soybean, lupin and chickpea seed meals in cannulated Iberian pigs: I. Proteins. J Sci Food Agric 85:1313–1321. [Google Scholar]
- 3. Bampidis VA, Christodoulou V (2011) Chickpeas (Cicer arietinum L.) in animal nutrition: A review. Anim Feed Sci Technol 168:1–20. [Google Scholar]
- 4. Ngwe T, Nukui Y, Oyaizu S, Takamoto G, Koike S, et al. (2012) Bean husks as a supplemental fiber for ruminants: Potential use for activation of fibrolytic rumen bacteria to improve main forage digestion. Anim Sci J 83:43–49. [DOI] [PubMed] [Google Scholar]
- 5. Jain D, Chattopadhyay D (2010) Analysis of gene experession in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol 10:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Molina C, Rotter B, Horres R, Udupa SM, Besser B, et al. (2008) SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics 9:553–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nasr Esfahani M, Sulieman S, Schulze J, Yamaguchi-Shinozaki K, Shinozaki K, et al. (2014) Mechanisms of physiological adjustment of N2 fixation in Cicer arietinum L. (chickpea) during early stages of water deficit: single or multi-factor controls. Plant J 79:964–980. [DOI] [PubMed] [Google Scholar]
- 8. Hadiarto T, Tran LS (2011) Progress studies of drought-responsive genes in rice. Plant Cell Rep 30:297–310. [DOI] [PubMed] [Google Scholar]
- 9. Jogaiah S, Govind SR, Tran LS (2013) Systems biology-based approaches toward understanding drought tolerance in food crops. Crit Rev Biotechnol 33:23–39. [DOI] [PubMed] [Google Scholar]
- 10. Shanker AK, Maheswari M, Yadav SK, Desai S, Bhanu D, et al. (2014) Drought stress responses in crops. Funct Integr Genomics 14:11–22. [DOI] [PubMed] [Google Scholar]
- 11. Albacete AA, Martinez-Andujar C, Perez-Alfocea F (2014) Hormonal and metabolic regulation of source-sink relations under salinity and drought: From plant survival to crop yield stability. Biotechnol Adv 32:12–30. [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803. [DOI] [PubMed] [Google Scholar]
- 13. Tran LS, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K (2007) Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol 428:109–128. [DOI] [PubMed] [Google Scholar]
- 14. Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170. [DOI] [PubMed] [Google Scholar]
- 15. Xie Q, Frugis G, Colgan D, Chua N-H (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duval M, Hsieh T-F, Kim SY, Thomas TL (2002) Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248. [DOI] [PubMed] [Google Scholar]
- 17. Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis . PLoS One 2:e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong R, Richardson EA, Ye Z-H (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis . Planta 225:1603–1611. [DOI] [PubMed] [Google Scholar]
- 19. Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K (2010) Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1:32–39. [DOI] [PubMed] [Google Scholar]
- 20. Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87. [DOI] [PubMed] [Google Scholar]
- 21. Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381. [DOI] [PubMed] [Google Scholar]
- 22. Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103. [DOI] [PubMed] [Google Scholar]
- 23. Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, et al. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogo Y, Kobayashi T, Nakanishi Itai R, Nakanishi H, Kakei Y, et al. (2008) A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J Biol Chem 283:13407–13417. [DOI] [PubMed] [Google Scholar]
- 25. Kim HS, Park BO, Yoo JH, Jung MS, Lee SM, et al. (2007) Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis . J Biol Chem 282:36292–36302. [DOI] [PubMed] [Google Scholar]
- 26. Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis . Mol Plant 3:1087–1103. [DOI] [PubMed] [Google Scholar]
- 27. Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, et al. (2010) Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95:56–65. [DOI] [PubMed] [Google Scholar]
- 28. Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, et al. (2007) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, et al. (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44. [DOI] [PubMed] [Google Scholar]
- 30. Hu R, Qi G, Kong Y, Kong D, Gao Q, et al. (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa . BMC Plant Biol 10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rushton PJ, Bokowiec MT, Han S, Zhang H, Brannock JF, et al. (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol 147:280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voitsik AM, Muench S, Deising HB, Voll LM (2013) Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant Biol 13:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shiriga K, Sharma R, Kumar K, Yadav SK, Hossaina F, et al. (2014) Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puranik S, Sahu PP, Mandal SN, B VS, Parida SK, et al. (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS One 8:e64594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS (2013) Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res 20:403–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kou XH, Wang S, Wu MS, Guo RZ, Xue ZH, et al. (2014) Molecular characterization and expression analysis of NAC family transcription factors in tomato. Plant Mol Biol Rep 32:501–516. [Google Scholar]
- 37. Liu T, Song X, Duan W, Huang Z, Liu G, et al. (2014) Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in chinese cabbage. Plant Mol Biol Rep 32:1041–1056. [Google Scholar]
- 38. Satheesh V, Jagannadham PT, Chidambaranathan P, Jain PK, Srinivasan R (2014) NAC transcription factor genes: genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.). Mol Biol Rep DOI10.1007/s11033-014-3669-5 [DOI] [PubMed] [Google Scholar]
- 39. Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, et al. (2009) In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res 16:353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, et al. (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varshney RK, Song C, Saxena RK, Azam S, Yu S, et al. (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246. [DOI] [PubMed] [Google Scholar]
- 42. Jain M, Misra G, Patel RK, Priya P, Jhanwar S, et al. (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74:715–729. [DOI] [PubMed] [Google Scholar]
- 43. Shamsi K (2010) The effect of sowing date and row spacing on yield and yield components on Hashem chickpea variety under rainfed condition. Afr J Biotechnol 9:7–11. [Google Scholar]
- 44. Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, et al. (2011) Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res 18:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, et al. (2011) Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol 156:1661–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh VK, Garg R, Jain M (2013) A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol J 11:691–701. [DOI] [PubMed] [Google Scholar]
- 50. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 51. Ha CV, Le DT, Nishiyama R, Watanabe Y, Sulieman S, et al. (2013) The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Res 20:511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garg R, Sahoo A, Tyagi AK, Jain M (2010) Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem Biophys Res Commun 396:283–288. [DOI] [PubMed] [Google Scholar]
- 53. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. [DOI] [PubMed] [Google Scholar]
- 55. Greve K, La Cour T, Jensen MK, Poulsen FM, Skriver K (2003) Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem J 371:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–563. [DOI] [PubMed] [Google Scholar]
- 57. Zhang G, Chen M, Chen X, Xu Z, Guan S, et al. (2008) Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot 59:4095–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tran LS, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, et al. (2009) Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics 281:647–664. [DOI] [PubMed] [Google Scholar]
- 59. Demura T, Fukuda H (2007) Transcriptional regulation in wood formation. Trends Plant Sci 12:64–70. [DOI] [PubMed] [Google Scholar]
- 60. Hu H, Dai M, Yao J, Xiao B, Li X, et al. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, et al. (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630. [DOI] [PubMed] [Google Scholar]
- 62. Wu Y, Deng Z, Lai J, Zhang Y, Yang C, et al. (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290. [DOI] [PubMed] [Google Scholar]
- 63. Nuruzzaman M, Sharoni AM, Satoh K, Moumeni A, Venuprasad R, et al. (2012) Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday Selection (drought tolerant) and IR64. Mol Genet Genomics 287:389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, et al. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916. [DOI] [PubMed] [Google Scholar]
- 65. Lee S, Lee H-J, Huh SU, Paek K-H, Ha J-H, et al. (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227:76–83. [DOI] [PubMed] [Google Scholar]
- 66. Quach TN, Tran LS, Valliyodan B, Nguyen HT, Kumar R, et al. (2014) Functional analysis of water stress-responsive soybean GmNAC003 and GmNAC004 transcription factors in lateral root development in Arabidopsis . PLoS One 9:e84886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350. [DOI] [PubMed] [Google Scholar]
- 68. Guttikonda SK, Valliyodan B, Neelakandan AK, Tran LS, Kumar R, et al. (2014) Overexpression of AtDREB1D transcription factor improves drought tolerance in soybean. Mol Biol Rep DOI10.1007/s11033-014-3695-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of 71 CaNACs of chickpea and well-known stress-responsive NACs from Arabidopsis (ATAF1/ANAC002, 019, 029, 055, 072 and ATAF2/081) and rice (SNAC1/ONAC002 and OsNAC6/SNAC2/ONAC048). Conserved NAC domain and subdomains (A–E) are indicated by thick blue line and black thin black lines, respectively, above the sequences. The putative nuclear localization signal (NLS) is shown by a blue double-headed arrow below the sequence. Putative stress-related NAC subgroup is highlighted in red-colored background, and membrane-bound CaNAC members are highlighted in turquoise-colored background.
(PDF)
Phylogenetic relationship of NAC proteins from chickpea (CaNACs) and Arabidopsis (ANACs). The unrooted phylogenetic tree was constructed using the full NAC protein sequences. The membrane-bound CaNAC and ANAC proteins are indicated in blue-colored and green-colored letters, respectively.
(PDF)
Expression of 23 selected CaNAC genes in chickpea roots and leaves under dehydration and ABA treatments. (A) Summary of the results of the expression data. (B) Venn diagram analysis of dehydration- and ABA-responsive CaNAC genes in roots and leaves of chickpea plants. The ABA- and/or dehydration-responsive genes were defined as those whose expression is altered by at least 2-fold (P<0.05) at 2 and/or 5 h of dehydration and/or ABA treatment.
(PDF)
Primers used in RT-qPCR analysis.
(XLS)
Putative CaNAC genes identified in this study and their major features.
(XLS)
Nucleotide and amino acid sequences of 71 CaNAC genes obtained from (Ca v1.0) (Bioproject: PRJNA175619).
(TXT)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.