The structures of retinal-bound and retinol-bound hCRBPII are reported. The X-ray flux-dependent and wavelength-dependent rearrangement of retinol in the binding pocket of hCRBPII is demonstrated. The binding modes of retinol and retinal in CRBPI and CRBPII are contrasted.
Keywords: cellular retinol-binding protein II, retinol, retinal
Abstract
Cellular retinol-binding proteins (CRBPs) I and II, which are members of the intracellular lipid-binding protein (iLBP) family, are retinoid chaperones that are responsible for the intracellular transport and delivery of both retinol and retinal. Although structures of retinol-bound CRBPI and CRBPII are known, no structure of a retinal-bound CRBP has been reported. In addition, the retinol-bound human CRBPII (hCRBPII) structure shows partial occupancy of a noncanonical conformation of retinol in the binding pocket. Here, the structure of retinal-bound hCRBPII and the structure of retinol-bound hCRBPII with retinol fully occupying the binding pocket are reported. It is further shown that the retinoid derivative seen in both the zebrafish CRBP and the hCRBPII structures is likely to be the product of flux-dependent and wavelength-dependent X-ray damage during data collection. The structures of retinoid-bound CRBPs are compared and contrasted, and rationales for the differences in binding affinities for retinal and retinol are provided.
1. Introduction
Retinol, one of several forms of vitamin A, is essential for vision, cell growth, reproduction and metabolism. Because of its low solubility in aqueous solution and its relative instability, retinol requires carrier proteins for transport, bioavailability and stability. Different proteins in different tissues play important roles in the transport, storage and metabolism of vitamin A (Ong, 1984 ▶; Li & Norris, 1996 ▶). These are retinol-binding proteins (RBPs), cellular retinoic acid-binding proteins (CRABPs), cellular retinol-binding proteins (CRBPs), cellular retinal-binding protein (CRALBP) and interphotoreceptor retinoid-binding protein (IRBP). The retinoid ligand in all of these proteins binds noncovalently and with high affinity within a relatively deep binding pocket. CRBPs belong to the superfamily of intracellular lipid-binding proteins (iLBPs). This family also includes the cellular retinoic acid-binding proteins (CRABPs) and the fatty acid-binding proteins (FABPs) (Veerkamp et al., 1993 ▶; Banaszak et al., 1994 ▶). The structures of the proteins in this family are quite similar. They all share a ten-stranded β-sandwich domain (a–j) with two short α-helices near the mouth of the large binding cavity (Cowan et al., 1993 ▶; Franzoni et al., 2002 ▶; Winter et al., 1993 ▶; Vaezeslami et al., 2006 ▶). Although four isoforms of human cellular retinol-binding protein (hCRBP) have been identified (hCRBPI, hCRBPII, hCRBPIII and hCRBPIV; Folli et al., 2001 ▶, 2002 ▶), only hCRBPI and hCRBPII have so far been shown to be involved in the metabolism and transport of intracellular retinol. CRBPI is found predominantly in the liver and kidney, while hCRBPII is more prevalent in the small intestine (Crow & Ong, 1985 ▶; Ong, 1984 ▶; Chytil, 1984 ▶). It has also been reported that hCRBPII regulates retinoid metabolism in the intestine and facilitates the reduction of retinal to retinol and the subsequent esterification of retinal to retinyl ester (Vogel et al., 2001 ▶). CRBPI and CRBPII share approximately 56% sequence identity (Li et al., 1986 ▶) and both proteins can bind all-trans-retinol, all trans-retinal and 13-cis-retinol, while neither binds 9-cis-retinol (MacDonald & Ong, 1987 ▶; Ong, 1984 ▶), although CRBPII is found to be the most selective for all-trans-retinoids. The first crystal structures of CRBP were those of apo and all-trans-retinol-bound rat CRBPII published in 1993 (Winter et al., 1993 ▶). Comparison of the two structures showed no significant differences upon ligand binding. More recently, the crystal structures of both apo human CRBPII and zebrafish CRBP (PDB entries 2rcq and 1kqx, respectively) and all-trans-retinol-bound human CRBPII and zebrafish CRBP (PDB entries 2rct and 1kqw, respectively) have been reported (Tarter et al., 2008 ▶; Calderone et al., 2002 ▶). The ligand was modelled as a mixture of retinol and a retinol derivative in which C12—C13 is a single bond. In this paper, we report the first structure of an all-trans-retinal-bound CRBP (hCRBPII). We further show that a retinoid derivative virtually identical to that seen in the zebrafish CRBP and hCRBPII structures can be generated by X-ray radiation damage, and further show that this process is dependent on the wavelength of the X-ray radiation. We finally report the first bona fide structure of retinol-bound hCRBPII and compare it with the retinal-bound structure.
2. Materials and methods
hCRBPII was cloned, expressed and purified as previously described for other hCRBPII mutants, with the single exception that the induction time was 5 h (Wang et al., 2012 ▶). All-trans-retinol and all-trans-retinal were purchased from Sigma–Aldrich. The dissociation constant K d was determined by fluorescence titration as described previously (Vasileiou et al., 2007 ▶).
2.1. Crystallization of hCRBPII retinal-bound and retinol-bound complexes
The pure protein was concentrated to 5–10 mg ml−1. The complexes of protein with all-trans-retinal and all-trans-retinol were prepared by adding 3–4 equivalents of either retinal or retinol solution (30 mM retinal or retinol in ethanol). The final concentration of ethanol in the protein solution was no higher than 10%(v/v). The mixture of protein and ligand was incubated at room temperature in Black LiteSafe Microcentrifuge Tubes for 1 h. Crystals of both complexes were grown at room temperature by the hanging-drop vapour-diffusion method in Linbro plates wrapped in aluminium foil to prevent light-initiated degradation of the retinal and retinol. The best crystals were grown using crystallization screens from Hampton Research in 30–35% PEG 4000 (Sigma–Aldrich), 0.1 M sodium acetate (Columbus Chemical Industry) pH 4.6–4.8, 0.1 M ammonium acetate (J. T. Baker) with 1 µl hCRBPII–ligand complex. The crystals appeared after 3 d and reached maximum size in one week. The crystals were briefly soaked in a cryoprotectant solution (30% PEG 4000, 0.1 M sodium acetate pH 4.6–4.8, 0.1 M ammonium acetate, 10–20% glycerol) and flash-cooled in liquid nitrogen. The two crystal forms of retinol-bound hCRBPII were grown under identical conditions.
2.2. Data collection and refinement
All diffraction data were collected on beamline 21-ID-D, LS-CAT (Argonne National Laboratory, Advanced Photon Source, Chicago, Illinois, USA) using a MAR300 detector and radiation of wavelength 1.00 Å at 100 K. The diffraction data were indexed using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶). Initial phases were produced using MOLREP (as implemented in the CCP4 program package) using human apo CRBPII (PDB entry 2rcq) as a model (Tarter et al., 2008 ▶). The structures were refined using REFMAC5 in the CCP4 program suite (Vagin & Teplyakov, 2010 ▶; Winn et al., 2011 ▶). Model rebuilding and placement of ordered water molecules were performed manually using Coot (Emsley et al., 2010 ▶). The chromophore was created using JLigand and manually fitted into the electron density at the end of the refinement. All refinements were carried out using REFMAC5 and PHENIX (Murshudov et al., 2011 ▶; Adams et al., 2010 ▶). Ramachandran outliers were found in PDB entries 4qyn (0.83%), 4qyp (1.58%), 4qzt (2.12%) and 4qzu (2.49%). The higher number of outliers for the last three structures come from molecule D, where the density is poor in several regions.
2.3. HPLC analysis of retinol extracted from crystals
To verify the identity of all-trans-retinol in the crystals before data collection, we constructed a set of experiments in which retinol was extracted from the freshly grown wild-type (WT) hCRBPII crystals and the resulting sample was analyzed via high-performance liquid chromatography (HPLC) and UV–Vis spectroscopy. Both the HPLC chromatogram and the UV–Vis spectrum of all-trans-retinol were taken as references to compare with the extracted sample. All-trans-retinol-bound WT hCRBPII crystals, grown as described above, were dissolved in phosphate-buffered saline (PBS; 500 µl). The PBS solution was extracted with hexane (200 µl) by vortexing for 1 min and the organic layer was transfered to a 1 ml eppendorf tube and dried under a nitrogen stream. 30 µl ethyl acetate was then added and 15 µl of the resulting sample was purified by reverse-phase HPLC chromatography (silica column; Microsorb MV-86-100-C5, 5 mm, 100 A). The running solvent mixture was hexane/ethyl acetate (85:15) with a flow rate of 1 ml min−1. The detection wavelength was set to 325 nm. The retinoid extracted from the crystals showed a single peak in the HPLC trace with a retention time identical to that of all-trans-retinol. The fraction was also collected and a UV–Vis spectrum identical to that of retinol was observed. Notably, the extraction of retinol from the freshly grown retinol-bound WT hCRBPII crystals demonstrated that the bound chromophore was completely protected within the protein cavity during crystallization, with no evidence of degradation or rearrangement. We also performed an HPLC run for the co-sample, in which all-trans-retinol and the other half of the crystal extract were premixed together. As anticipated, co-injection resulted in a single peak, verifying that the extracted sample is all-trans-retinol.
3. Results and discussion
3.1. The affinity of hCRBPII for retinol and retinal
The affinity of rat CRBPII for both retinol and retinal has been measured using both a fluorescence quenching assay and fluorine NMR. Although earlier reports indicated an order of magnitude difference in binding between retinol and retinal, the most recent study showed rat CRBPII binding to retinol and retinal with similar affinity (K d of ∼10 nM; Li et al., 1991 ▶; Malpeli et al., 1995 ▶; Kane et al., 2011 ▶). Since, as far as we know, no affinity measurements for hCRBPII have been reported, we measured its affinity for both retinal and retinol using a tryptophan fluorescence quenching assay. While the binding of retinal was significantly weaker compared with that most recently reported for the rat protein, 130 ± 10 nM (Supplementary Fig. S11) compared with 10 nM for rat CRBPII (Kane et al., 2011 ▶), hCRBPII showed significantly higher affinity for retinol, with a lower limit of about 2 nM (Supplementary Fig. S1a). The binding constant appeared to be too high for accurate measurement using our fluorescence assay. This seems to indicate a potential difference in the interaction of rat versus human CRBPII with these ligands.
3.2. Structure of the retinal-bound hCRBPII complex
hCRBPII–retinoid complexes were crystallized in a variety of crystal forms, although none corresponded to the previously published crystal form of the hCRBPII complex (Wang et al., 2012 ▶). All of the crystal forms have multiple copies of the complex in the asymmetric unit. Retinal-bound hCRBPII crystallizes in space group P1 with four molecules per asymmetric unit (all of the crystallographic data are tabulated in Table 1 ▶). Electron density corresponding to the entire ligand was clear in only one of the four chains (chain B). In chain C density for retinal is well defined for all atoms apart from C15 and the hydroxyl group, where the density is relatively weak. In molecule A electron density is relatively well defined for the carbonyl group and C11–C15 but is weaker for the rest of the molecule. Scattered, uninterruptable electron density is seen in molecule D. The overall structure of the protein is little changed either in the apo state or when bound to either retinal or retinol (Figs. 1 ▶ a and 1 ▶ b). The retinal is essentially identical in all three molecules where the electron density is clear (Fig. 2 ▶ a). In fact, apart from the oxygen to be discussed below, the position of retinal is almost perfectly superimposable with that of retinol in the retinol-bound hCRBPII structure. In both ligands the β-ionone ring adopts the 6-s-trans conformation (Fig. 2 ▶ a). Two hydrophobic residues, Phe16 and Leu77, surround the ionone ring of the ligand, locking the chromophore in position and restricting the rotation of the ionone ring about the C6—C7 single bond (Figs. 2 ▶ b and 2 ▶c). The distances between the C5 methyl group of retinal and Leu77 and Phe16 are about 3.7 and 3.8 Å, respectively. It is clear that rotation of the ionone ring would lead to steric clashes between the gem-dimethyl group and these two residues.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the last resolution shell.
| WT hCRBPIIretinol | WT hCRBPIIretinal | WT hCRBPIIretinol, 7keV | WT hCRBPIIretinol, 11keV | |
|---|---|---|---|---|
| Data-collection statistics | ||||
| Space group | P21 | P1 | P1 | P1 |
| Unit-cell parameters | ||||
| a () | 34.65 | 35.44 | 36.41 | 36.48 |
| b () | 75.14 | 54.89 | 54.18 | 54.14 |
| c () | 54.65 | 68.71 | 68.44 | 68.30 |
| () | 90.00 | 107.75 | 107.72 | 107.64 |
| () | 100.78 | 97.66 | 97.19 | 96.94 |
| () | 90.00 | 103.08 | 103.59 | 103.71 |
| Resolution () | 50.001.19 (1.211.19) | 50.001.62 (1.651.62) | 29.791.89 (1.941.89) | 34.6611.496 (1.511.50) |
| Total reflections | 306503 | 241247 | 76284 | 358399 |
| Unique reflections | 87302 | 76879 | 34646 | 76284 |
| Completeness (%) | 99.8 (84.0) | 91.5 (93.8) | 92.36 (86.0) | 95.20 (88.0) |
| Molecules per asymmetric unit | 2 | 4 | 4 | 4 |
| Average I/(I) | 20.19 (2.04) | 51.85 (6.87) | 12.97 (6.23) | 26.45 (2.41) |
| R merge (%) | 9.1 (43.4) | 3.2 (32.1) | 7.8 (32.5) | 4.2 (59.8) |
| Mosaicity () | 0.486 | 0.806 | 0.58 | 0.58 |
| Refinement statistics | ||||
| R work/R free (%) | 18.71/21.48 | 20.43/25.24 | 17.98/24.42 | 16.15/21.85 |
| R.m.s.d., bond lengths () | 0.0297 | 0.025 | 0.008 | 0.007 |
| R.m.s.d., angles () | 2.381 | 2.200 | 1.079 | 1.066 |
| Average B, main chain (2) | 9.148 | 22.545 | 34.628 | 23.69 |
| Average B, water and side chain (2) | 16.410 | 26.406 | 45.016 | 39.549 |
| No. of water molecules | 507 | 323 | 331 | 468 |
| Ramachandran plot | ||||
| Most favoured (%) | 97.10 | 95.66 | 96.04 | 96.20 |
| Allowed (%) | 2.07 | 2.76 | 3.21 | 2.28 |
| Outliers (%) | 0.83 | 1.58 | 2.12 | 2.49 |
| PDB code | 4qyn | 4qyp | 4qzt | 4qzu |
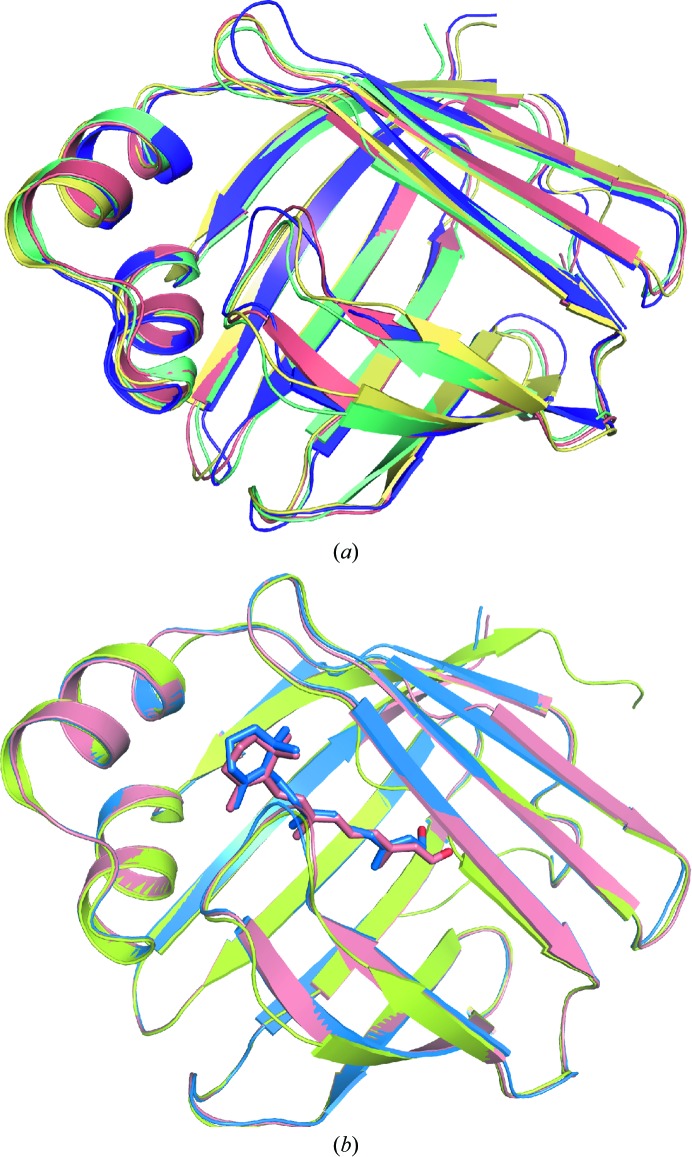
Figure 1.
(a) An overlay of the structures of hCRBPII (salmon), hCRBPIII (PDB entry 1ggl, yellow; Folli et al., 2001 ▶), hCRBPIV (PDB entry 1lpj, green; Folli et al., 2002 ▶) and hCRBPI (PDB entry 1kgl, purple; Franzoni et al., 2002 ▶). (b) An overlay of the structures of apo hCRBPII (PDB entry 2rcq, lime; Tarter et al., 2008 ▶) and of wild-type hCRBPII bound to all-trans-retinal (4qyp, salmon) and to all-trans-retinol (PDB entry 4qzt, blue)
Figure 2.
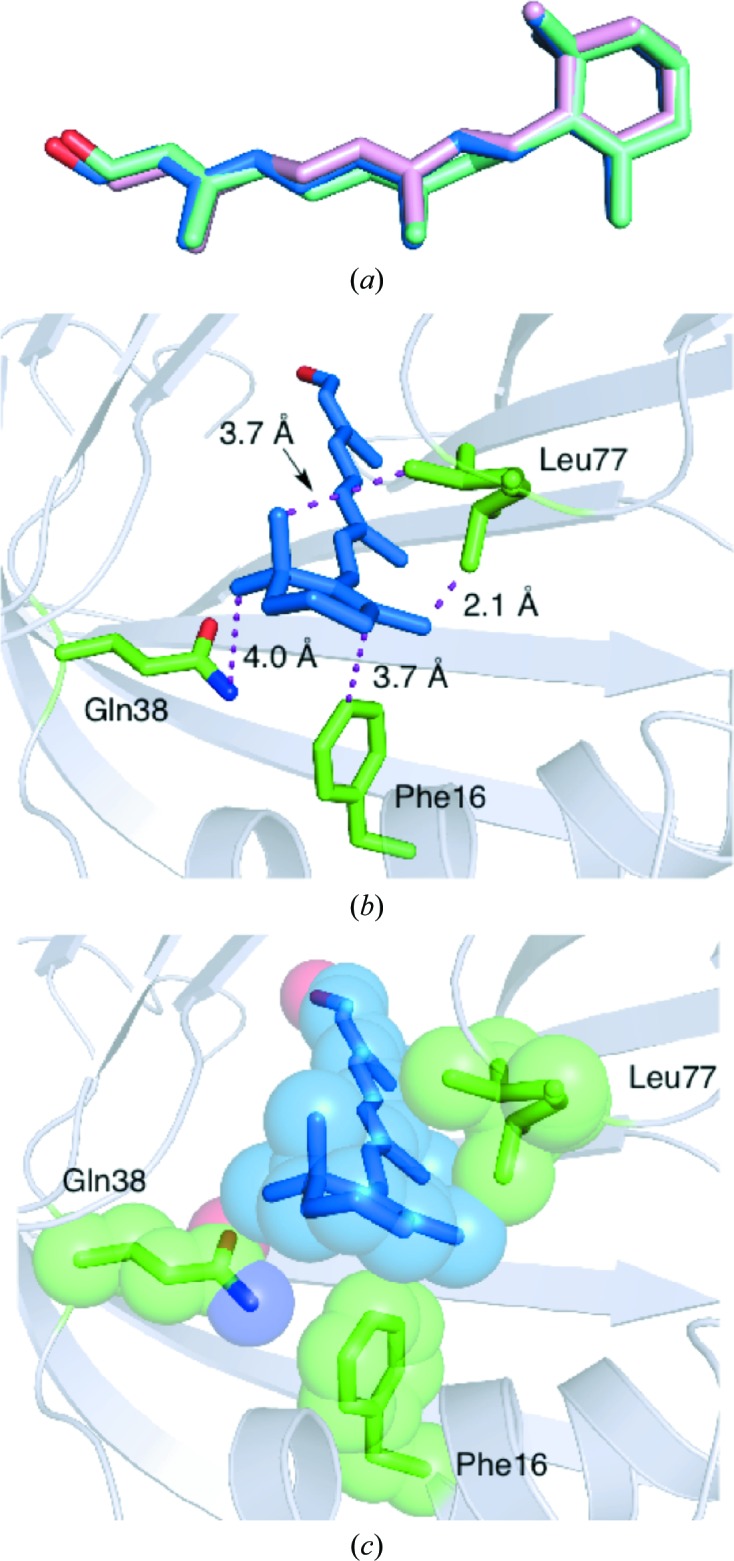
Retinal binding in hCRBPII (PDB entry 4qyp). (a) An overlay of retinal from molecule A (cyan), molecule B (blue) and molecule C (pink), showing the similarity in the binding in the three molecules. (b) Stick representation of the interaction between Phe16, Leu77 (both with green C atoms, blue N atoms and red O atoms ) and the retinal ionone ring (blue C atoms) (in molecule B). (c) Space-filling depiction of (b).
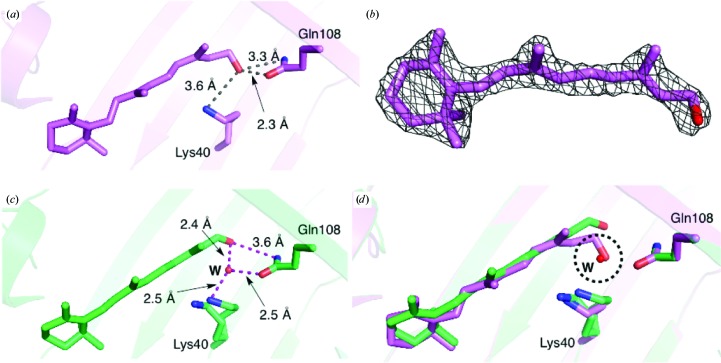
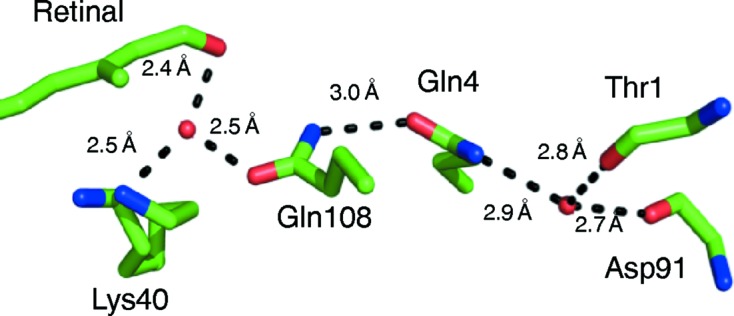
The carbonyl group of retinal makes a water-mediated interaction with Gln108 and Lys40 (Fig. 3 ▶). The side chain of Gln108 is fixed in position by a small network of water molecules and residues, including the main-chain carbonyls of Thr1 and Asp91 and the side chains of Gln4 and Gln108 (Fig. 3 ▶). This network is unique to hCRBPII and is not found in zebrafish CRBP (PDB entry 1kqw), rat CRBPII (PDB entry 1pob), rat CRBPI (PDB entry 1ocb) or hCRBPI (PDB entry 1kgl; Franzoni et al., 2002 ▶) (Supplementary Fig. S2).
Figure 3.
The water-mediated interaction of the retinal hydroxyl group. Atoms are coloured by type and hydrogen-bonded distances are shown.
The first water molecule in this network makes hydrogen bonds to the main-chain carbonyls of Thr1 and Asp91 and determines the orientation of Gln4 in hCRBPII (Fig. 3 ▶). Since water must act as hydrogen-bond donor to the two main-chain carbonyl groups, only the lone pairs on the O atom of water are left to make the hydrogen bond to the amide N atom of Gln4, defining its orientation. Therefore, it is the carbonyl O atom of Gln4 that makes a hydrogen bond, necessarily to the amide of Gln108, defining its orientation as well. The orientation of Gln108, with the carbonyl group pointing toward the carbonyl of the ligand, precludes a direct hydrogen bond to the retinal carbonyl O atom and necessitates the water-mediated hydrogen bond. The hydrogen bond between this water and the retinal carbonyl is 2.49 Å, which is an ideal distance for a strong low-barrier hydrogen bond (Fig. 3 ▶). This water makes an additional hydrogen bond to the ∊-amino group of Lys40, which is fixed in space by two hydrogen bonds in the binding pocket.
3.3. Wavelength-dependent damage of the ligand in retinol-bound hCRBPII
We next sought to compare our retinol-bound structure with those of retinol-bound CRBPII. Structures from rat, human and zebrafish have already been determined (Supplementary Fig. S3). In both structures the binding pocket is partially occupied by another retinoid species, postulated to be a degradation product of the original retinol (Tarter et al., 2008 ▶; Calderone et al., 2002 ▶). It is characterized by a torsion angle around the C13—C14 bond of around 60°, which is inconsistent with the trans double bond between these two C atoms expected for retinol. The hydroxyl group of this species makes hydrogen bonds to both Gln108 and Lys40, essentially occupying the same position as the water molecule responsible for the water-mediated interaction observed in our hCRBPII–retinal complex (Supplementary Fig. S4). In contrast, the rat CRBPII–retinol complex shows only retinol in the binding pocket, with the hydroxyl group in a different position to that seen in the other structures, allowing it to make hydrogen bonds to both the amide O and N atoms of Gln108. In an effort to produce a structure of human CRBPII bound only to retinol, we redetermined the structure of the hCRBPII–retinol complex in a new P21 crystal form with two complexes per asymmetric unit. To our surprise, we saw exclusively what appears to be exactly the same retinoid derivative bound to both molecules in the asymmetric unit (Supplementary Fig. S3a). The hydroxyl of the retinoid derivative makes hydrogen bonds to both Gln108 and Lys40, and occupies a position that is essentially identical to that of the water in the retinal complex (Supplementary Fig. S4d). No evidence for retinol is seen in our structures. Although the exact identity of the retinoid is unclear, a possible candidate that is consistent with the electron density is the rearranged retinol derivative shown in Supplementary Fig. S3(b). To determine whether the new molecule is an impurity, we took an NMR spectrum of the retinol sample. The NMR spectrum showed the retinol to be quite pure, with no significant extraneous peaks. Further, analytical HPLC of the sample also showed only a single peak consistent with retinol (Supplementary Fig. S5). Since our crystals were all grown at the relatively low pH of 4.6, we investigated the possibility that the low pH of the crystallization conditions could lead to the retinoid derivative by incubating retinol, both alone and in the presence of hCRBPII, overnight at low pH. In both cases a decomposition product was identified by HPLC. However, this product was much less polar as evident from the HPLC retention times, which is inconsistent with a hydroxyl-group containing compound. We surmised that the low-pH product was a dehydration product, similar to that seen previously, but it is structurally inconsistent with that observed in the crystal structures (McBee et al., 2000 ▶; Sorensen, 1965 ▶). We then grew over 30 crystals of the complex, dissolved them in phosphate-buffered saline, extracted the retinoids from the crystals using hexane and analysed them by HPLC and UV–Vis spectroscopy. The extract showed only a single peak that co-eluted precisely with retinol, indicating that the crystals do in fact contain only retinol. It therefore appears that the retinoid derivative seen in these structures may arise from X-ray-induced damage during data collection (Supplementary Fig. S5). Consistent with this is the fact that an obvious orange colour is seen upon exposure of the crystal to the X-ray beam. It is interesting to note that for the rat CRBPII–retinol complex, the only retinol-bound CRBPII structure that was not plagued by the retinoid derivative, the crystal data were collected using a Cu Kα home X-ray source (Winter et al., 1993 ▶). All of the other data sets, the human, zebrafish and both of our hCRBPII complex structures, were collected at synchrotron sources using about 1 Å (11 KeV) wavelength radiation. Indeed, X-ray-induced rearrangements of retinoid species have been documented previously (Borshchevskiy et al., 2011 ▶; Matsui et al., 2002 ▶; Schreckenbach et al., 1977 ▶).
In an effort to produce a bona fide hCRBPII–retinol complex, we collected a variety of data sets at the synchrotron. Since most X-ray damage to protein crystals is thought to be solely related to dosage, we collected data at 11 keV but with a beam that was attenuated to less than 20% of the intensity of the beam used previously. However, electron density from the data collected was still consistent with only the rearranged retinol in the active site (data not shown). We then investigated the effect of X-ray energy by collecting data at 7, 8 and 9 keV, all using a beam attenuated to 20% of the original intensity, and an additional data set at 11 keV. Gratifyingly, the retinoid electron density in the 7 keV data set clearly shows an unrearranged all-trans-retinol (Figs. 4 ▶ a and 4 ▶ b). Together, these data show unequivocally that the rearranged product is a result of both dosage and wavelength-dependent X-ray damage, and that data collection at lower energy results in the first structure of the bona fide hCRBPII–retinol complex.
Figure 4.
Retinal versus retinol binding in hCRBPII. (a) The structure of the retinol-bound WT hCRBPII. (b) 2F o − F c electron-density map contoured at 1.0σ around the retinol for the 7 keV data (PDB entry 4qzt). (c) The structure of all-trans-retinal bound to WT hCRBPII (PDB entry 4qyp). (d) The overlaid structures of all-trans-retinal (green) and retinol (magenta). The position of an ordered water molecule (W) occupied by the hydroxyl group of retinol is indicated by a dashed circle.
3.4. The structure of retinol-bound hCRBPII contrasted with the retinal-bound structure
The difference in the binding of retinal versus retinol lies in the position of C15 and the hydroxyl group (Figs. 4 ▶ c and 4 ▶ d). In the hCRBPII–retinol complex the hydroxyl is positioned to make simultaneous hydrogen bonds to both the carbonyl O and N atoms of Gln108, an interaction that would be impossible for retinal, which cannot act as hydrogen-bond donor to the amide carbonyl O atom. Therefore, retinal shifts its orientation, making room for the water molecule between Gln108 and Lys40 and the creation of the hydrogen bond to this water molecule. The position of the retinol hydroxyl group in hCRBPII would in fact collide with this water molecule, rendering incompatible the conformation seen in the retinol-bound structure and the presence of this water molecule (Fig. 4 ▶ d). This difference in interaction may explain the difference in affinity for retinol versus retinal that was observed for hCRBPII. However, the physiological relevance of this difference in binding constant is unclear. The retinol-bound rat CRBPII structure is similar to that of hCRBPII, showing identical interactions between protein and ligand.
3.5. Ligand binding in CRBPI versus CRBPII
Rat CRBPI and CRBPII have similar binding affinities for all-trans-retinol (Li et al., 1991 ▶). An overlay of the crystal structures of retinal-bound and retinol-bound hCRBPII and the solution structure of hCRBPI (PDB entry 1kgl; Franzoni et al., 2002 ▶) and rat CRBPI (PDB entry 1crb; Cowan et al., 1993 ▶) illustrate that in hCRBPII the ligand is more than 1 Å deeper in the binding pocket. The identity of the amino acid at position 51 appears to be the deciding factor for ligand position (Fig. 5 ▶). Position 51 is isoleucine in CRBPI (both rat and human), while it is threonine in hCRBPII. Inspection of the structures of human and rat CRBPI (PDB entry 1crb; Cowan et al., 1993 ▶) shows that retinal cannot bind to hCRBPI in the same way that it does to hCRBPII. This is owing to a steric clash between the side chain of isoleucine and the retinal carbonyl group in hCRBPI. The combination of the addition of the larger isoleucine at position 51 and the altered position of the ligand leaves no room for a water molecule between Gln108 and Lys40.
Figure 5.
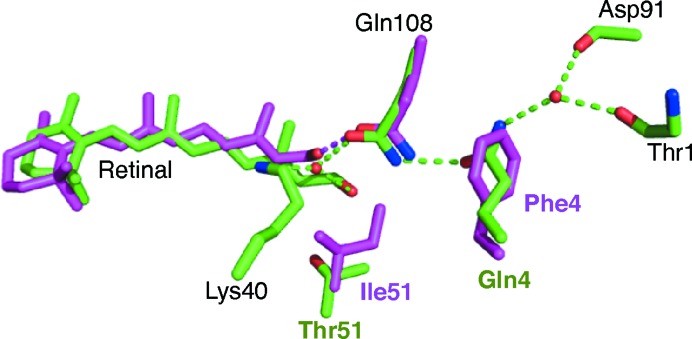
The structures of retinol-bound rat CRBPI (magenta C atoms and hydrogen bonds) and retinal-bound hCRBPII (green C atoms and hydrogen bonds) are overlaid. Amino-acid labels are coloured similarly when different in the two structures. Note the change in the water network introduced by Phe4.
In both the rat and human CRBPI structures a hydrogen bond is formed between Gln108 and the retinol hydroxyl group (Fig. 5 ▶ and Supplementary Fig. S2d). Although there is no structure available of a CRBPI bound to retinal, it is clear that a hydrogen bond to Gln108 can only be made if the amide N atom is pointing towards the retinal carbonyl, because it lacks a hydrogen donor. This is opposite to the orientation of Gln108 in hCRBPII because the orientation of Gln108 is determined by Gln4 and a water network as described above (Figs. 3 ▶ and 5 ▶). However, CRBPI proteins have a phenylalanine at position 4 (Phe3 in hCRBPI) instead of a glutamine, which may allow Gln108 to rotate and present the amide N atom to hydrogen bond to the carbonyl, which would allow CRBPI to bind retinal in a similar way to retinol, possibly explaining the similar affinity seen for the two ligands in CRBPI (Supplementary Fig. S2). In an attempt to better understand the role of the residue at position 51, the hCRBPII T51I mutant was produced and its binding affinities for retinol and retinal were measured. The dissociation constants of this mutant for both all-trans-retinol and all-trans-retinal were similar to those of the wild-type protein and therefore it still had much weaker binding affinity for all-trans-retinal than all-trans-retinol (Supplementary Fig. S1). Unfortunately, attempts to crystallize hCRBPII T51I bound to all-trans-retinal were not successful.
4. Conclusion
The crystal structure of holo wild-type hCRBPII with all-trans-retinal reveals that there are at least three distinct binding modes for retinoids in CRBPI and CRBPII. In the human and rat CRBPII retinol-bound structures, retinol is able to make hydrogen bonds to both the carbonyl and amide N atoms of the side chain of Gln108. Since this mode of interaction is precluded in retinal, owing to the lack of a hydrogen-bond donor, there is a conformational change of the carbonyl, leading to a water-mediated interaction between the retinal carbonyl and Lys40 and Gln108. This is consistent with the higher binding affinity for retinol relative to retinal in hCRBPII. In contrast, the presence of Ile51 causes a translation of the ligand in the CRBPI binding pocket, resulting in only a single hydrogen bond being made between Gln108 and the retinol hydroxyl group. This difference in the binding can explain the higher affinity of retinol versus retinal in hCRBPII, because retinol is able to make two hydrogen bonds to Gln108. We have also shown that wavelength-dependent X-ray damage during data collection is probably responsible for the retinoid derivatives seen in the structures of human and zebrafish CRBP.
Supplementary Material
PDB reference: hCRBPII, bound to retinol, 4qyn
PDB reference: 4qzt
PDB reference: 4qzu
PDB reference: bound to retinal, 4qyp
Supporting Information.. DOI: 10.1107/S1399004714023839/qh5018sup1.pdf
Acknowledgments
Generous support was provided by the NIH (GM101353) (to Borhan and Geiger). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). We would also like to thank Dr Spencer Anderson for his assistance in collecting the attenuated data at multiple wavelengths.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: QH5018).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Banaszak, L., Winter, N., Xu, Z., Bernlohr, D. A., Cowan, S. & Jones, T. A. (1994). Adv. Protein Chem. 45, 89–151. [DOI] [PubMed]
- Borshchevskiy, V. I., Round, E. S., Popov, A. N., Büldt, G. & Gordeliy, V. I. (2011). J. Mol. Biol. 409, 813–825. [DOI] [PubMed]
- Calderone, V., Folli, C., Marchesani, A., Berni, R. & Zanotti, G. (2002). J. Mol. Biol. 321, 527–535. [DOI] [PubMed]
- Chytil, F. (1984). Pharmacol. Rev. 36, 93S–100S. [PubMed]
- Cowan, S. W., Newcomer, M. E. & Jones, T. A. (1993). J. Mol. Biol. 230, 1225–1246. [DOI] [PubMed]
- Crow, J. A. & Ong, D. E. (1985). Proc. Natl Acad. Sci. USA, 82, 4707–4711. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Folli, C., Calderone, V., Ottonello, S., Bolchi, A., Zanotti, G., Stoppini, M. & Berni, R. (2001). Proc. Natl Acad. Sci. USA, 98, 3710–3715. [DOI] [PMC free article] [PubMed]
- Folli, C., Calderone, V., Ramazzina, I., Zanotti, G. & Berni, R. (2002). J. Biol. Chem. 277, 41970–41977. [DOI] [PubMed]
- Franzoni, L., Lücke, C., Pérez, C., Cavazzini, D., Rademacher, M., Ludwig, C., Spisni, A., Rossi, G. L. & Rüterjans, H. (2002). J. Biol. Chem. 277, 21983–21997. [DOI] [PubMed]
- Kane, M. A., Bright, F. V. & Napoli, J. L. (2011). Biochim. Biophys. 1810, 514–518. [DOI] [PMC free article] [PubMed]
- Li, E., Demmer, L. A., Sweetser, D. A., Ong, D. E. & Gordon, J. I. (1986). Clin. Res. 34, A685. [DOI] [PMC free article] [PubMed]
- Li, E. & Norris, A. W. (1996). Annu. Rev. Nutr. 16, 205–234. [DOI] [PubMed]
- Li, E., Qian, S. J., Winter, N. S., d’Avignon, A., Levin, M. S. & Gordon, J. I. (1991). J. Biol. Chem. 266, 3622–3629. [PubMed]
- MacDonald, P. N. & Ong, D. E. (1987). J. Biol. Chem. 262, 10550–10556. [PubMed]
- Malpeli, G., Stoppinf, M., Zapponi, M. C., Folli, C. & Berni, R. (1995). Eur. J. Biochem. 229, 486–493. [PubMed]
- Matsui, Y., Sakai, K., Murakami, M., Shiro, Y., Adachi, S., Okumura, H. & Kouyama, T. (2002). J. Mol. Biol. 324, 469–481. [DOI] [PubMed]
- McBee, J. K., Kuksa, V., Alvarez, R., de Lera, A. R., Prezhdo, O., Haeseleer, F., Sokal, I. & Palczewski, K. (2000). Biochemistry, 39, 11370–11380. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ong, D. E. (1984). J. Biol. Chem. 259, 1476–1482. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Schreckenbach, T., Walckhoff, B. & Oesterhelt, D. (1977). Eur. J. Biochem. 76, 499–511. [DOI] [PubMed]
- Sorensen, T. S. (1965). J. Am. Chem. Soc. 87, 5075–5084.
- Tarter, M., Capaldi, S., Carrizo, M. E., Ambrosi, E., Perduca, M. & Monaco, H. L. (2008). Proteins, 70, 1626–1630. [DOI] [PubMed]
- Vaezeslami, S., Mathes, E., Vasileiou, C., Borhan, B. & Geiger, J. H. (2006). J. Mol. Biol. 363, 687–701. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vasileiou, C., Vaezeslami, S., Crist, R. M., Rabago-Smith, M., Geiger, J. H. & Borhan, B. (2007). J. Am. Chem. Soc. 129, 6140–6148. [DOI] [PubMed]
- Veerkamp, J. H., van Kuppevelt, T. H. M. S. M., Maatman, R. & Prinsen, C. F. M. (1993). Prostaglandins Leukot. Essent. Fatty Acids, 49, 887–906. [DOI] [PubMed]
- Vogel, S., Mendelsohn, C. L., Mertz, J. R., Piantedosi, R., Waldburger, C., Gottesman, M. E. & Blaner, W. S. (2001). J. Biol. Chem. 276, 1353–1360. [DOI] [PubMed]
- Wang, W., Nossoni, Z., Berbasova, T., Watson, C. T., Yapici, I., Lee, K. S. S., Vasileiou, C., Geiger, J. H. & Borhan, B. (2012). Science, 338, 1340–1343. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winter, N. S., Bratt, J. M. & Banaszak, L. J. (1993). J. Mol. Biol. 230, 1247–1259. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: hCRBPII, bound to retinol, 4qyn
PDB reference: 4qzt
PDB reference: 4qzu
PDB reference: bound to retinal, 4qyp
Supporting Information.. DOI: 10.1107/S1399004714023839/qh5018sup1.pdf