Abstract
In malaria-endemic regions, many medical facilities have limited capacity to diagnose non-malarial etiologies of acute febrile illness (AFI). As a result, the etiology of AFI is seldom determined, although AFI remains a major cause of morbidity in developing countries. An outbreak of AFI was reported in the Afar region of Ethiopia in August of 2011. Retrospectively, 12,816 suspected AFI cases were identified by review of medical records. Symptoms were mild and self-limiting within 3 days after the date of onset; no fatalities were identified. All initial test results of AFI patient specimens were negative for selected pathogens using standard microbiological and molecular techniques. High-throughput sequencing of nucleic acid extracts of serum specimens from 29 AFI cases identified 17 (59%) of 29 samples as positive for Sandfly Fever Sicilian Virus (SFSV). These results were further confirmed by specific reverse transcription polymerase chain reaction. This is the first study implicating SFSV as an etiological agent for AFI in Ethiopia.
Introduction
Acute febrile illnesses (AFI) caused by a variety of pathogens pose a major public health challenge, in part because clinical examination cannot distinguish specific etiologies. Furthermore, practical and affordable diagnostic tests for the diagnosis of non-malarial etiologies of AFI are often not available in developing countries.1–3 Consequently, the incidence and relative importance of the etiologic agents responsible for AFI remain unknown in many parts of the world.2 This leads to potential misdiagnosis, inappropriate patient management, and an inability to effectively control or prevent additional cases. Sandfly fever, caused by infection with the Sandfly Fever Sicilian Virus (SFSV), is common in the Mediterranean region.4 SFSV is an arthropod-borne virus that was first identified in Sicily, Italy in 1943 during World War II as the etiology of sandfly fever that was a cause of AFI in Allied armed forces.4 The virus is spread during the summer season, which is the active period for Phlebotomus papatasi,5 the main vector of SFSV. Other sandflies, such as P. ariasi and sandflies of Larroussius group, also transmit SFSV. Sandfly fever is a self-limited mild illness including fever, headache, and muscle and joint pain; patients usually recover fully within a few days.5 Serologic evidence of sandfly fever indicated positive titers of SFSV from samples collected in Bangladesh, Djibouti, Ethiopia, Iraq, Morocco, Saudi Arabia, Somalia, Sudan, Tunisia, former republics of the Soviet Union, and Yugoslavia.6 This investigation identified SFSV as the etiologic agent in an AFI outbreak that was initially assumed to be caused by malaria and later thought to be caused by dengue. To our knowledge, this is the first report of sandfly fever in Ethiopia.
Methods and Materials
Samples were collected as a public health response under the direction of the Ethiopian Health and Nutrition Research Institute. As a public health response, this investigation was not categorized as research, and informed consent was not required.
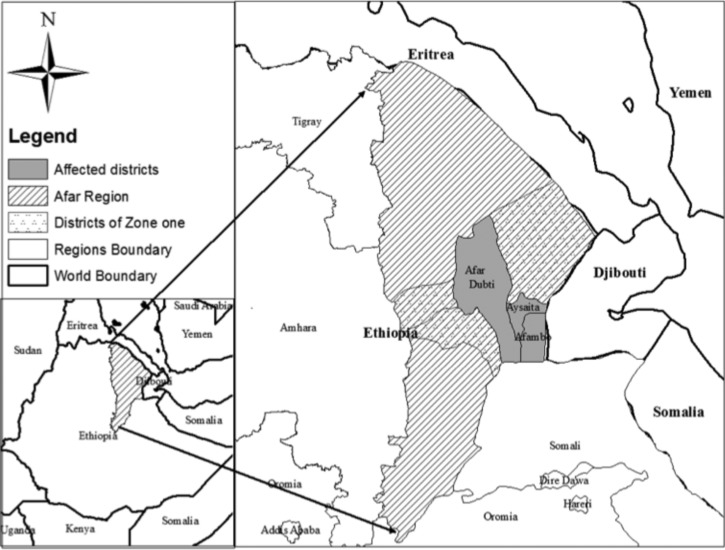
In August of 2011, an outbreak of AFI was reported in Asayta District, Afar Regional State, Ethiopia. The outbreak later spread to two neighboring districts of Dubti and Afambo (Figure 1). The illness was characterized by acute onset of fever, chills, headache, and myalgia.
Figure 1.
Acute febrile illness outbreak affected districts of Afar region, Ethiopia, 2011.
The regional health bureau (RHB) in the affected districts sought assistance from the Ethiopian Health and Nutrition Research Institute (EHNRI) when blood samples from patients with AFI tested negative for malaria. The national outbreak investigation team from the EHNRI initiated an investigation by examining medical records from five local health facilities. Using medical records, a suspect AFI case was defined as acute onset of fever, chills, headache, and myalagia from August 7 to September 12, 2011.
The national outbreak investigation team collected blood specimens from 29 acutely ill patients with AFI. These specimens were tested for malaria using a rapid diagnostic test (CareStart Malaria HRP2 [Pf] Test; Access Bio, Inc., Somerset, NJ) or microscopy on blood smears. In addition, samples were subcultured onto blood, chocolate, and MacConkey agar.
Sera, which had been stored at −80°C, from 29 acutely ill patients were transported to the Centers for Disease Control and Prevention (CDC) laboratories at the Kenya Medical Research Institute (KEMRI) in Kenya for real-time polymerase chain reaction (PCR) testing. Total nucleic acid extraction was performed using the MagMAX Viral RNA Isolation Kit (Life Technologies, Foster City, CA) as per the manufacturer's instructions. Amplification was done using the AgPath-ID One-Step RT-PCR Kit (Applied Biosystems, Carlsbad, CA) in individual PCR assays for dengue, Rift Valley fever (RVF), yellow fever (YF), filoviruses (Ebola zaire virus, Ebola bundibugyo virus, Ebola sudan virus, and Marburg virus), chikungunya, and Crimean-Congo Hemorrhagic Fever (CCHF) viruses. Primers and probes for the filoviruses were obtained from the CDC, Viral Special Pathogens Branch in Atlanta, Georgia. Primers and probes for YF, dengue, and RVF have previously been described, whereas those of chikungunya virus were obtained from the Division of Vector-Borne Diseases, CDC, in Fort Collins, Colorado.7 Thermal cycling conditions included reverse transcription at 45°C for 10 minutes, AmpliTaq Polymerase activation at 95°C for 10 minutes, and then, 45 cycles of denaturation at 95°C for 15 seconds and primer annealing/extension at 55°C for 1 minute.
Total nucleic acids from 29 of the serum specimens were transported to Washington University in St. Louis for next-generation sequencing analysis. Nucleic acids were randomly amplified by sequence-independent PCR using barcoded primers, pooled, and sequenced in one run on the Roche Titanium/FLX platform as described previously.8 Sequences were compared with publicly available sequence databases using a customized bioinformatics platform as previously described to identify microbial sequences present in the sample.9 Contig assembly was performed with the Newbler assembler.
For result confirmation, RNA was extracted from specimens positive by next-generation sequencing and subjected to a phlebovirus-specific consensus reverse transcription PCR (RT-PCR) assay. Resultant amplicons were sequenced using previously described methods at the CDC Division of Vector-Borne Diseases in Fort Collins, Colorado.10 All generated partial S-segment sequences were deemed identical in the amplified region. A phylogenetic analysis was conducted on a representative partial S-segment sequence (284 bp; GenBank accession no. KJ372529), which maps to nucleotides 139–422 of the GenBank reference SFSV-Turkey sequence (GenBank accession no. NC_015413.1) along with diverse SFSV sequences that are available in GenBank. Alignments were generated using the Clustal W function of MEGA, version 4 software.11,12 A neighbor-joining tree was generated and analyzed with 2,000 replicates for bootstrap testing.13
Results
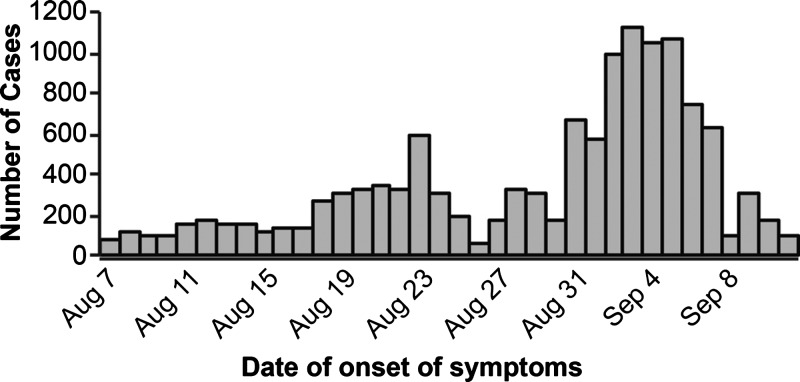
In total, 12,816 suspect AFI cases were identified between August 7 and September 12, 2011 through the medical records review from five health centers in Afambo, Asayta, and Dubti Districts of the Afar region (Figure 2); 9,107 (71%) cases were male. For 29 patients from whom sera was collected, the age distribution ranged from 2 to 55 years of age, with a mean age of 25 years old. Patient symptoms were mild and self-limiting, with patients recovering within 3–4 days after date of symptom onset.
Figure 2.
Suspect AFI cases by date of onset, Afar, August 7 to September 11, 2011.
All 29 blood samples tested negative for malaria (Plasmodium falciparum and P. vivax) by microscopy. In addition, bacterial culture failed to identify any potential etiologies. The 29 serum specimens tested negative for dengue, RVF, YF, filoviruses, chikungunya, and CCHF viruses by real-time PCR. Attempts to culture virus from selected samples on Vero cells were unsuccessful.
After next-generation sequencing, computational analysis detected one or more nucleic acid sequences with ≥ 97% nucleotide identity to SFSV in 17 (59%) of 29 specimens. In addition, one specimen was positive for hepatitis B and SFSV; two specimens were positive for hepatitis GBV-C sequences, one of which was positive for SFSV.
To further analyze the SFSV-like sequence, data from one sample, where more than 1,000 of approximately 30,000 total reads derived from SFSV, were assembled. Contigs of 6,355 nucleotides (GenBank accession no. KM042102) and 4,349 nucleotides (GenBank accession no. KM042103) that correspond to the nearly complete L and M segments of SFSV, respectively, were generated. In addition, two shorter contigs of 790 and 767 nucleotides (GenBank accession nos. KM042104 and KM042105) were generated that aligned to different regions of the S segment of SFSV (with a gap of approximately 100 nucleotides). The four contigs shared 97–99% nucleotide identity to the reference SFSV-Turkey strain in GenBank (accession nos. NC_015411.1, NC_015412.1, and NC_015413.1).
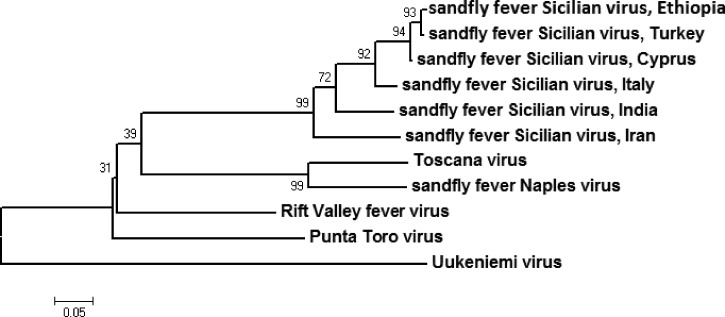
Of 17 specimens sent to the CDC in Ft. Collins, Colorado for confirmatory testing, 7 (41%) specimens were SFSV-positive by RT-PCR, and 2 (12%) specimens were indeterminate. All seven positive samples yielded identical amplicons. The resultant tree reveals a strongly supported association between the SFSV sequences detected in Ethiopia and strains that were isolated in recent years from Cyprus (2002) and Turkey (2008), with Uukeniemi virus as the outgroup (Figure 3).
Figure 3.
Phylogeny of the partial S segment of a geographic diversity of sandfly fever viruses. A neighbor-joining tree with bootstrap values determined by 2000 replicates is shown. The scale bar represents the number of nucleotide substitutions per site.
Discussion
This is the first report of SFSV as the etiology of AFI in Ethiopia. The symptoms, which were mild and self-limiting within 3–4 days, are identical to those in previous reports of SFSV infection, also known as 3-day fever.6,14
Human case reports and seroprevalence studies have suggested that SFSV or an SFSV-like virus circulates in the Mediterranean region.4 Recently, molecular evidence showed the presence of two distinct phleboviruses closely related to SFSV in Algeria and Tunisia.15,16 Nucleic acids detected in this study within positive samples shared approximately 99% nucleotide sequence identity with strains that were associated with human illness in Cyprus and Turkey in 2008, implicating a possible Eastern Mediterranean origin of the Ethiopian strain of SFSV.
This large outbreak of AFI was initially identified through an extensive medical records review, and although only 29 specimens were ultimately collected, 17 specimens contained nucleic acid sequences that were ≥ 97% identical to SFSV using a next-generation sequencing-based assay. These results were independently corroborated by RT-PCR in 7 of 17 specimens in a different laboratory (CDC, Fort Collins, CO). Sequences obtained in these seven specimens were identical to those obtained in the Washington University laboratory.
There were a number of limitations to this study. An attempt to isolate a pathogen on Vero cells was unsuccessful (data not shown). Because of resource limitations, no vector assessment was conducted. Case identification was a challenge because of the pastoral nature of the affected community. Limited specimens were collected relative to the great size of the affected area, in part because cases were on the wane by the time that the investigative team arrived.
Although the epidemiologic investigation provided limited information on risk factors for infection or clues to potential etiologic agents, detection of SFSV in these specimens by high-throughput sequencing underscores the power of unbiased metagenomic strategies for pathogen detection. It is unclear at this point whether detection of sandfly fever reflects a recent expansion in the geographic range of the virus beyond the Mediterranean region or alternately, previously unrecognized endemicity uncovered by the increased diagnostic breadth of the unbiased next-generation sequencing assay. Broader studies are needed to determine the prevalence of sandfly fever in this geographic area. Furthermore, disease surveillance should be implemented in endemic regions to monitor the trends and quickly identify an outbreak. Surveillance would also provide data to the national level on disease burden and etiological agents of focus in resource-limited settings.
This outbreak highlights the diagnostic challenges in identifying etiologic agents responsible for AFI outbreaks in developing countries. AFI cases usually go uncharacterized in resource-limited settings and are mainly treated as malaria or typhoid. In our study, the outbreak etiological identification was inefficient primarily because of the lack of advanced molecular techniques, which necessitated international collaboration. Although etiological identification was successful in this instance, this approach is not cost-effective considering the burden of AFI outbreaks in sub-Saharan Africa. There is a great need to develop laboratory capacity for both traditional microbiology and molecular techniques to improve identification of agents responsible for causing outbreaks of AFI in developing countries.
ACKNOWLEDGMENTS
The authors thank Mr. Ali Hussen, Head of Afar Regional Health Bureau (RHB), Mr. Mohammed Ahimed, Deputy Head of Afar RHB, and Mr. Mussa Ali, Head of Malaria Prevention and Control, for their assistance in field investigation. We also thank Ibrahim Adem, regional Public Health Emergency Management officer, Dr. Kebir Hussen, United Nations Children's Fund field officer, and Dr. Woisa Ahimed, World Health Organization surveillance officer, for their technical assistance and information sharing. Special thanks to Mr. Yasin Hebib, Head of Afar PHEM Center, for his close communication assigning technical expertise and accommodation arrangement.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Financial support: This work was supported, in part, by National Institutes of Health Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Grant U54 AI057160.
Authors' addresses: Abyot Bekele Woyessa, Workenesh Ayele, Abdi Ahmed, Negga Asamene Abera, and Daddi Jima, Ethiopian Health and Nutrition Research Institute, E-mails: sifanbashu@yahoo.com, wayele@gmail.com, abdiseid04@yahoo.com, negaasamene@gmail.com, and daddi_jima@yahoo.com. Victor Omballa, Lilian Waiboci, and Melvin Ochieng, Global Disease Detection Program, Centers for Disease Control and Prevention–Kenya, E-mails: VOmballa@kemricdc.org, waiboci@uonbi.ac.ke, and MOchieng@kemricdc.org. David Wang and Song Cao, Departments of Molecular Microbiology and Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, E-mails: davewang@borcim.wustl.edu and scao@wustl.edu. Amy Lambert, Division of Vector-Borne Disease, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mail: ahk7@cdc.gov. Joel M. Montgomery and Barry Fields, Global Disease Detection Program, Centers for Disease Control and Prevention–Kenya, and Global Disease Detection Branch, Division of Global Health Protection, Center for Global Health, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: ztq9@cdc.gov and bsf2@cdc.gov.
References
- 1.Archibald LK, Reller LB. Clinical microbiology in developing countries. Emerg Infect Dis. 2001;7:302–305. doi: 10.3201/eid0702.010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump JA, Youssef FG, Luby SP, Wasfy MO, Rangel JM, Taalat M, Oun SA, Mahoney FJ. Estimating the incidence of typhoid fever and other febrile illnesses in developing countries. Emerg Inf Dis. 2003;9:539–544. doi: 10.3201/eid0905.020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Animut A, Mekonnen Y, Shimelis D, Ephrahim E. Febrile illnesses of different etiology among outpatients in four health centers in northwestern Ethiopia. Jpn J Infect Dis. 2009;62:107–110. [PubMed] [Google Scholar]
- 4.Depaquit J, Grandadam M, Fouque F, Andry PE, Peyrefitte C. Arthropod- borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro Surveill. 2010;15:19507. [PubMed] [Google Scholar]
- 5.Dionisio D, Esperti F, Vivarelli A, Valassina M. Epidemiological, clinical and laboratory aspects of sandfly fever. Curr Opin Infect Dis. 2003;16:383–388. doi: 10.1097/00001432-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Tesh RB, Saidi S, Gajdamovic SJ, Rodhain F, Vesenjak-Hirjan J. Serological studies on the epidemiology of sandfly fever in the old world. Bull World Health Organ. 1976;54:663–674. [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C, Göttig S, Schilling S, Asper M, Panning M, Schmitz H, Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao G, Krishnamurthy S, Cai Z, Popov VL, Travassos da Rosa AP, Guzman H, Cao S, Virgin HW, Tesh RB, Wang D. Identification of novel viruses using VirusHunter–an automated data analysis pipeline. PLoS ONE. 2013;8:e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert AJ, Lanciotti RS. Consensus amplification and novel multiplex sequencing method for s segment species identification of 47 viruses of the Orthobunyavirus, Phlebovirus, and Nairovirus genera of the family Bunyaviridae. J Clin Microbiol. 2009;47:2398–2404. doi: 10.1128/JCM.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Guler S, Guler E, Caglayik DY, Kokoglu OF, Ucmak H, Bayrakdar F, Uyar Y. A sandfly fever virus outbreak in the east Mediterranean region of Turkey. Int J Infect Dis. 2012;16:244–246. doi: 10.1016/j.ijid.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Moureau G, Bichaud L, Salez N, Ninove L, Hamrioui B, Belazzoug S, de Lamballerie X, Izri A, Charrel RN. Molecular and serological evidence for the presence of novel phleboviruses in sandflies from northern Algeria. Open Virol J. 2010;4:15–21. doi: 10.2174/1874357901004010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhioua E, Moureau G, Chelbi I, Ninove L, Bichaud L, Derbali M, Champs M, Cherni S, Salez N, Cook S, de Lamballerie X, Charrel RN. Punique virus, a novel phlebovirus, related to sandfly fever Naples virus, isolated from sandflies collected in Tunisia. J Gen Virol. 2010;91:1275–1283. doi: 10.1099/vir.0.019240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]