Abstract
Ascamycin (ACM) and dealanylascamycin (DACM) are nucleoside antibiotics elaborated by Streptomyces sp. JCM9888. The later shows broad spectrum inhibition activity to various gram-positive and gram-negative bacteria, eukaryotic Trypanosoma and is also toxic to mice, while ascamycin is active against very limited microorganisms, such as Xanthomonas. Both compounds share an unusual 5′-O-sulfonamide moiety which is attached to an adenosine nucleoside. In this paper, we first report on the 30 kb gene cluster (23 genes, acmA to acmW) involved in the biosynthesis of these two antibiotics and a biosynthetic assembly line was proposed. Of them, six genes (AcmABGKIW) are hypothetical genes involved in 5′-O-sulfonamide formation. Two flavin adenine dinucleotide (FAD)-dependent chlorinase genes acmX and acmY were characterized which are significantly remote from acmA-W and postulated to be required for adenine C2-halogenation. Notably gene disruption of acmE resulted in a mutant which could only produce dealanylascamycin but was blocked in its ability to biosynthesize ascamycin, revealing its key role of conversion of dealanylascamycin to ascamycin.
Introduction
A number of nucleoside antibiotics have been discovered in the past half century and some of them were demonstrated to be particularly active antibiotics, pesticides and fungicides which have been developed into agrochemicals or pharmaceuticals [1]. Examples of nucleoside antibiotics include puromycin [2], nucleocidin [3], toyocamycin and sangivamycin [4], polyoxin [5], pacidamycin [6] [7], blasticidin S [8], nikkomycin [9] [10], gougerotin [11], A-500359s and A-503803s [12], caprazamycin [13], liposidomycin and A-90289 [14], muraymycin [15], mildiomycin [16], tunicamycin [15] [17] and amicetin [18]. Most of the biosynthetic pathways for the above nucleoside compounds have been reported in the past decade, which paved the way to the characterization of new enzymes and provided useful insights in devising novel antimicrobial agents [19].
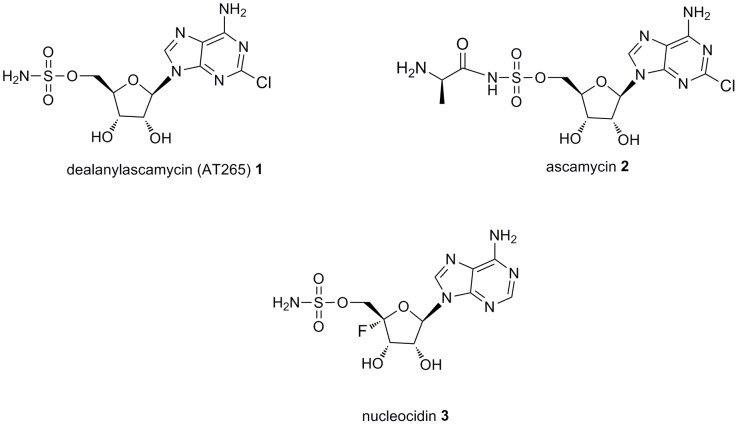
Dealanylascamycin (DACM) (1) and ascamycin (ACM) (2) [20] (also identified as AT-265 [21]) (Fig. 1) are two adenosine antibiotics produced by Streptomyces sp. JCM9888 whose chemical scaffold resemble that of the fluorine-containing antibiotic nucleocidin (3) [3] (Fig. 1). Ascamycin, dealanylascamycin and nucleocidin are closely related 5′-O-sulfonamide ribonucleosides. The ascamycins have C2-chloroadenine as the base on C-1′, whereas nucleocidin uses adenine, which lacks the chlorine. Nucleocidin also has a unique fluorine atom on its C-4′ position. Ascamycin and dealanylascamycin differ by N-alanylation of the 5′-O-sulfonamide moiety in the former but not the latter (Fig. 1) [3]. Although ascamycin and dealanylascamycin have similar chemical structures, they possess very different bactericidal activities. Ascamycin exhibited toxicity only to a few bacterium genus, such as Xanthomonas [22], while dealanylascamycin is broad-spectrum and active against both gram-positive, gram-negative bacteria and even some eukaryotic cells [22]. The later also showed unusual trypanocidal (such as Trypanosoma equiperdum) activity and is an anti-amoebae (such as Endamoeba histolytica) agent and thus it has been found valuable in treating animal diseases caused by these microorganisms [23]. Their differences in biological activity has been attributed to an aminopeptidase that cleaves the alanyl group from ascamycin. Since ascamycin is only active against a few microorganisms (Xanthomonas) in which this aminopeptidase was found, it was proposed that dealanylascamycin is the active form of this antibiotic, and most microorganisms (such as E. coli etc.) lack this aminopeptidase and are therefore not susceptible to ascamycin [22], [24]. Ascamycin and dealanylascamycin both possess a chlorine atom on the C2 position of adenine. It is often the case that incorporation of halogen into natural products plays an important role in increasing their biological scope and activity [25]. Some naturally occurring halogenated secondary metabolites, such as vancomycin, chlorotetracycline are of clinical importance. The mechanisms of biohalogenation, particularly for chlorination and bromination were widely discussed [25] [26] [27]. Enzymes involved in halogenation of aromatic groups have been extensively described, which require a two-component flavin-dependent halogenase/reductase as follows: A flavin reductase uses NADH (Nicotinamide adenine dinucleotide) to reduce FAD (flavin adenine dinucleotide) and produces FADH2 (reduced flavin adenine dinucleotide) and a halogenase utilize the FADH2 formed and O2 as cofactors to perform electrophilic halogenation via an intermediate halonium ion equivalent [25]. The mechanism of dealanylascamycin inhibition has been studied in the analogous antibiotic nucleocidin, which demonstrated that it inhibits amino acid incorporation into human liver cell protein in vivo and in vitro [28]. Due to their special biological activities, organic chemists have been attracted to synthesize ascamycin, dealanylascamycin, nucleocidin and related nucleoside derivatives [29] [30] [31].
Figure 1. Chemical structures of nucleoside antibiotics in this study: dealanylascamycin 1, ascamycin 2, nucleocidin 3.
In this paper we first report the entire gene cluster (acmA-acmW) responsible for the biosynthesis of two nucleoside antibiotics ascamycin/dealanylascamycin and a biosynthetic machinery was purposed from in silico analysis. Intriguingly, six genes (AcmABGKIW) are postulated to be involved in 5′-O-sulfonamide formation. Two FADH2-dependent halogenases (acmX and acmY) which are involved in the pathway were found not clustered with acmA-acmW and we postulate that they are required for chlorination on the C2-position of the adenine ring. Notably, inactivation of an esterase acmE resulted in a mutant which only produce dealanylascamycin but blocked in its ability to biosynthesize ascamycin which suggest its role of converting dealanylascamycin to ascamycin. These results provide biosynthetic profiles for the 5′-O-sulfonamide containing antibiotic ACM/DACM and lay a solid foundation for target improvement of their production via synthetic biology strategy..
Results
Identification and characterization of the ascamycin/dealanyl-ascamycin biosynthetic genes
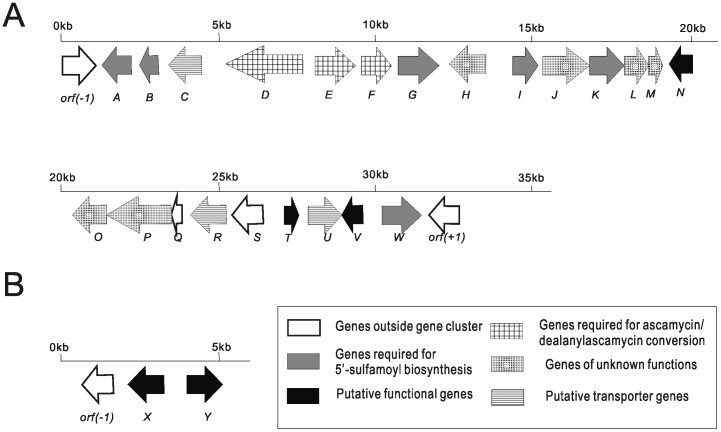
Streptomyces sp. JCM9888 is unusual in its ability to biosynthesize two nucleoside antibiotics: dealanylascamycin (1) and ascamycin (2) [20] (Fig. 1). As they are both featured with a 5′-O-sulfonamide moiety, we suspected sulfate metabolite related genes are required for the antibiotic production. Partial genome sequencing of Streptomyces genome revealed a 30,488 bp contiguous DNA sequence with an overall GC content of 66.7% (GenBank accession number KJ817374). Bioinformatics analysis of the sequence revealed 23 ORFs (acmA-W) and the deduced functions of individual ORFs are annotated and summarized in Table 1. Of them, 6 genes are proposed to be related to 5′-O-sulfonamide biosynthesis (acmA, acmB, acmG, acmI, acmK, acmW) (Fig. 2A). Notably, no halogenases were obvious around the gene cluster while BLASTp searches highlighted that two hypothetical flavin-dependent chlorinases (acmX and acmY) (GenBank accession number KJ817375) lying adjacent to each other on the chromosome in a distance of approx. 1 million basepair from acmA-W (Fig. 2B). AcmX and AcmY were shown to share high homology to all known FAD-dependent chlorinases, such as ChlB4 (accession number AAZ77674) from Streptomyces antibiotics which participate in chlorothricin biosynthesis (65% and 57% protein sequence identity respectively) [32]. Their analogy to proteins required for aromatic moiety chlorination suggested that they are potential candidates of the ascamycin/dealanylascamycin chlorinases as both two antibiotics possess a chlorine atom at C2-position of adenine.
Table 1. Deduced ORFs and their predicted functions in the ascamycin/dealanylascamycin gene cluster.
| Gene | Product size | Purposed function | Closest relative, origin, accession number | %identity |
| orf(-1) | 350 | transposase | Orf3, Streptomyces rishiriensis, AAR11882 | 69 |
| acmA | 320 | Sulfate adenylyltransferase subunit 2 | MmcV, Streptomyces lavendulae, Q9X5U0 | 80 |
| acmB | 179 | adenylylsulfate kinase | OrfQ, Streptomyces sp. AM-7161, BAC79014 | 67 |
| acmC | 414 | major facilitator superfamily protein | Micromonospora sp. CNB394, WP_018786560 | 29 |
| acmD | 871 | alanyl-tRNA synthetase | M901_2280, Bacteriovorax sp. DB6_IX, EQC50866 | 44 |
| acmE | 455 | esterase | Xanthomonas campestris, BAA11623 | 20 |
| acmF | 370 | alanyl-tRNA synthetase | CGL2_10933004, Leptospirillum sp. Group II '5-way CG', EDZ38785 | 27 |
| acmG | 480 | Sulfatase | Francci3_1756, Frankia sp. CcI3, YP_480861 | 47 |
| acmH | 453 | Radical SAM domain/B12 binding domain-containing protein | MCON_2786, Methanosaeta concilii, YP_004384990 | 24 |
| acmI | 302 | acylsulfatase | Streptomyces sp. CNB091, WP_018955485 | 69 |
| acmJ | 557 | Putative Fe-S oxidoreductase | Streptomyces clavuligerus, WP_003962555 | 79 |
| acmK | 356 | sulfotransferase | SGR_905, Streptomyces griseus, YP_001822417 | 59 |
| acmL | 250 | methyltransferase | UbiE, Leifsonia rubra CMS 76R, EPR77097 | 30 |
| acmM | 142 | hypothetical protein | BN159_7117, Streptomyces davawensis, YP_007525623 | 77 |
| acmN | 319 | amidinotransferase | Francci3_1759, Frankia sp. CcI3, YP_480864 | 57 |
| acmO | 444 | hypothetical protein | Francci3_1760, Frankia sp. CcI3, YP_480865 | 54 |
| acmP | 658 | DNA topoisomerase II | Francci3_1761, Frankia sp. CcI3, YP_480866 | 68 |
| acmQ | 155 | rubrerythrin | Francci3_1762, Frankia sp. CcI3, YP_480866 | 54 |
| acmR | 438 | major facilitator superfamily protein | Streptomyces sp. W007, WP_007453428 | 53 |
| acmS | 395 | hypothetical proteins | Streptomyces griseus, WP_003963977 | 31 |
| acmT | 220 | phosphatase | Streptomyces sulphureus, WP_019549800 | 52 |
| acmU | 418 | sodium/hydrogen antiporter | flH, Streptomyces cattleya, CAJ20009 | 28 |
| acmV | 194 | adenine phosphoribosyl-transferase | Streptomyces albulus, WP_016574607 | 58 |
| acmW | 451 | sulfate adenylyltransferase large subunit | Azi10, Streptomyces sahachiroi, ABY83149 | 78 |
| orf(+1) | 323 | oxidoreductase | Streptomyces scabrisporus, WP_020552419 | 85 |
| acmX | 449 | flavin-dependent halogenase | ChlB4, Streptomyces antibioticus, AAZ77674 | 65 |
| acmY | 438 | flavin-dependent halogenase | ChlB4, Streptomyces antibioticus, AAZ77674 | 57 |
Figure 2. Gene organization of ascamycin/dealanylascamycin biosynthesis pathway.
A) AcmA to AcmW. B) Chlorinases acmX and acmY.
5′-O-sulfonamide and 5′-(N-alanyl-O-sulfonamide) groups formation
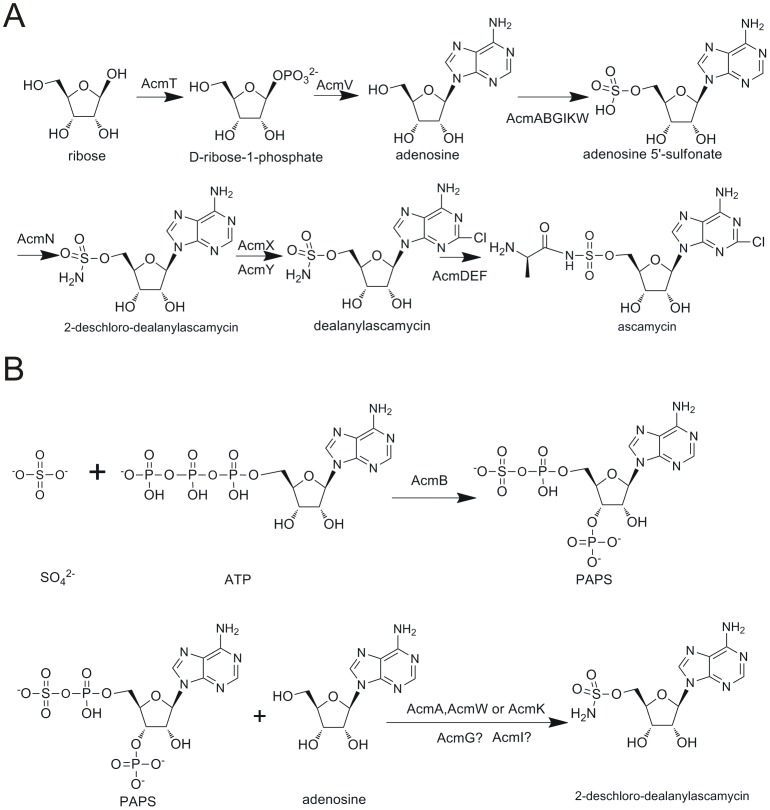
Ascamycin and dealanylascamycin are decorated with unusual sulfonamide groups at the 5′-hydroxyl of the ribose ring which are rare paradigms in secondary metabolites [33]. Six genes are postulated to be involved in 5′-O-sulfonamide moiety activation, loading and transfer in the ascamycin/dealanylascamycin gene cluster (acmA, B, G, I, K, W). Sulfate ions(SO4 2−) are not utilised directly but must first be activated by the formation of adenosyl phosphosulfate (APS) or phosphoadenosine phosphosulfate (PAPS). These are activated forms of sulfate generated by coupling inorganic sulfate onto ATP (adenosine triphosphate) (Fig. 3B). Sulfurylation of ATP is probably catalysed by adenylsulfate kinase (AcmB): SO4 2− + ATP → PAPS + PPi. Next, AcmK (sulfotransferase) [34] or AcmA and AcmW (sulfate adenyltransferase) [35] act upon the 5′ position of adenosine, utilizing PAPS as an activated sulfate donor to generate adenosine 5′-sulfonate. The manner in which AcmG (sulfatase) and AcmI (acylsulfatase) participate in this complex sulfate metabolite pathway is still elusive although bioinformatics analyses suggest that they have relevant functions (Fig. 3B). An aminotransferase AcmN was postulated to generate the 5′-O-sulfonamide moiety by replacing a hydroxyl group of the sulfonate with an amine group (Fig. 3B).
Figure 3. Proposed model for Acm/Dacm and 5′-O-sulfonamide group biosynthesis.
A) Biosynthetic pathway of ascamycin/dealanylascamycin. B) Biosynthetic pathway for 5′-O-sulfonamide group formation.
Verification of the ascamycin/dealanylascamycin biosynthetic genes
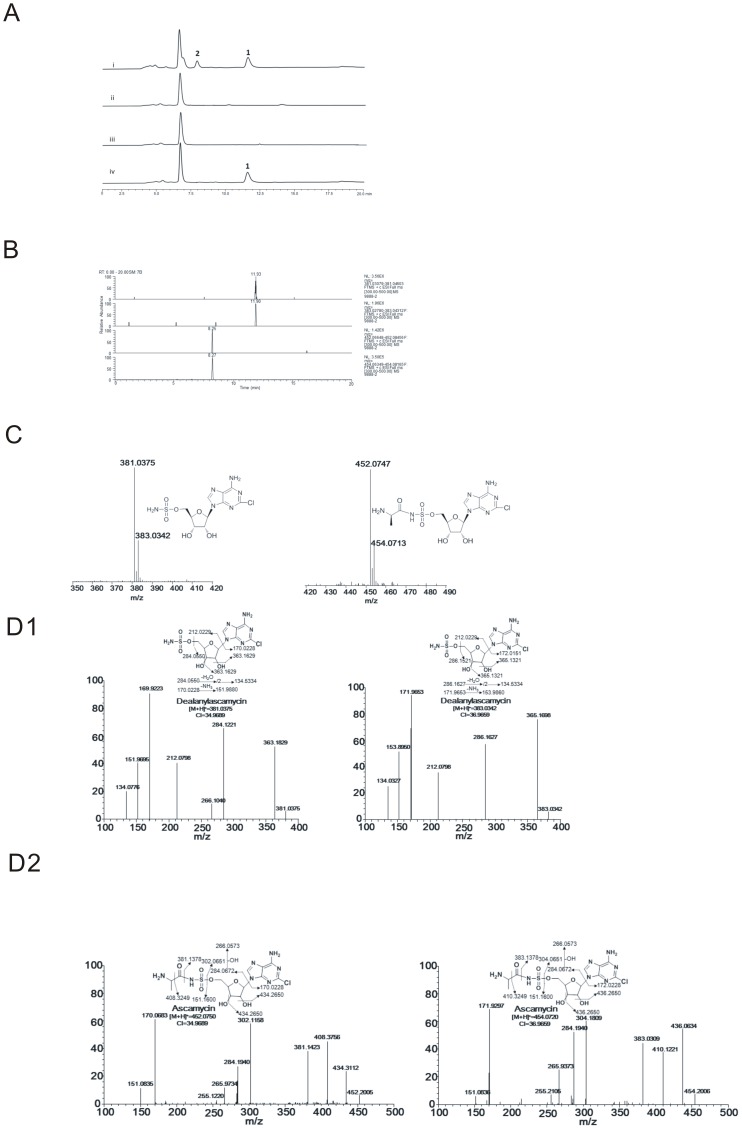
To verify our hypothesis that this sulfate related pathway is responsible for ascamycin/dealanylascamycin biosynthesis, a genomic cosmid library of JCM9888 was constructed on shuttle vector SuperCos1. Using acmG gene as a DNA probe, four positive clones were identified from some 4,000 packaged cosmids. Gene inactivation was carried out to disrupt acmG (sulfatase) or acmK (sulfotransferase) respectively (See material and methods) and the resulted mutants were validated by PCR analysis and subsequent sequencing using primers listed in Table S2 (See results in Fig. S1). When the Streptomyces mutants were fermented on petri dishes, as anticipated they lose their ability to elaborate both ascamycin and dealanylascamycin, as showed in HPLC-UV analysis (Fig. 4A). The molecular formula of compounds 1 and 2 were determined to be C10H13ClN6O6S (m/z = 381.0384, 383.0354 [M+H]+) and C13H18ClN7O7S (m/z = 452.0755, 454.0725 [M+H]+) on the basis of ESI-HRMS, the isotopic pattern indicated ascamycin and dealanylascamycin each contained one Cl atom (Fig. 4BC). Detailed MS/MS pattern of ascamycin and dealanylascamycin are shown (Fig. 4D). The detected data is closely consistent with anticipated fragmentation ions and every mass of debris have been elucidated (Fig. 4D1: dealanylascamycin; Fig. 4D2: ascamycin).
Figure 4. Analysis of metabolite profiles of Streptomyces JCM9888 and related mutants.
A) HPLC-UV analysis of metabolite profiles of Streptomyces: i) Wild type JCM9888, ii) ΔacmG mutant, iii) ΔacmK mutant, iv) ΔacmE mutant. The products DACM (1), ACM (2) are indicated. B) HPLC-ESI-MS analysis of metabolite profile of JCM9888 under the same program of HPLC-UV. C) The molecular formula of compounds DACM (1), ACM (2) were determined to be C10H13ClN6O6S (m/z = 381.0384, 383.0354 [M+H]+) and C13H18ClN7O7S (m/z = 452.0755, 454.0725 [M+H]+) on the basis of ESI-HRMS. D) MS/MS pattern of ascamycin and dealanylascamycin are shown and the detected data is closely consistent with anticipated fragmentation ions. D1: dealanylascamycin; D2: ascamycin.
AcmE is an esterase which is responsible for the conversion of dealanylascamycin to ascamycin
Analysing ascamycin/dealanylascamycin gene cluster also revealed two alanyl-tRNA synthetases (acmD and acmF). Intriguingly, ascamycin has an alanyl-cap while dealanylascamycin lacks it. Gene acmE located between acmD and acmF whose translated product weakly resemble an aminopeptidase from Xanthomonas campestris (accession number BAA11623, 20% identity) which have been demonstrated in vitro to catalyse a hydrolysis reaction of ascamycin to dealanylascamycin causing the host to become susceptible to ascamycin [36]. Most bacteria lack this enzyme and therefore are resistant to ascamycin and this explain the very different bacterial activity spectrum of ascamycin and dealanylascamycin [37]. The location of acmCDE in the biosynthetic pathway suggests their role in converting dealanylascamycin to ascamycin via a tRNA-dependent manner, in which acmE may catalyse a condensation reaction between alanyl-tRNA and dealanylascamycin to generate ascamycin [38].
To verify above hypothesis, gene inactivation was carried out on acmE and the Streptomyces mutant was validated by PCR on appropriate primers (Fig. S1-A). Intriguingly, acmE-null mutant could only produce dealanylascamycin but was blocked in its ability to biosynthesize ascamycin, as illustrated in HPLC-UV analysis (Fig. 4A(iv)), confirming the role of acmE to catalyse the condensation reaction of dealanylascamycin and L-alanine to generate ascamycin. (Fig. 3A)
A proposed model for ACM/DACM biosynthesis
Thus we propose a model for ascamycin/dealanylascamycin biosynthesis as illustrated in Fig. 3A. Firstly, a phosphatase (AcmT) mediates phosphorylation at C1 of D-ribose to generate D-ribose-1-phosphate [39]. The next enzyme, a phosphoribosyltransferase (AcmV) catalyze the displacement of the phosphate group by adenine to form adenosine [40]. 5′-O-sulfonamide biosynthesis related enzymes (AcmABGIKW) activate inorganic sulfate anions and install a sulfonate group onto the 5′ position of adenosine. Then AcmN, an aminotransferase transfers an amine group onto the sulfonate moiety to generate the corresponding O-sulfonamide, which is dealanylascamycin. Finally, AcmE catalyse an amide formation between the carboxyl end of L-alanine and the NH2 on the O-sulfonamide group of dealanylascamycin in a tRNA-dependent manner to furnish ascamycin [38].
Discussion
Nucleoside antibiotics are a large family of compounds with significant activities and some of them have been developed into antibacterial, antifungal and anti-parasite agents. The pace of discovering their biosynthetic pathways were increasing in the past decade, revealing new enzymes and paving the way to new “unnatural” natural products by synthetic biology. Ascamycin/dealanylascamycin are unusual nucleoside antibiotics which are elaborated by Streptomyces sp. They share a special 5′-O-sulfonamide moiety on adenosine which is not present in other antibiotics to the best of our knowledge. In this study we identified the putative genes which are involved in their biosynthetic pathway, including those genes required for sulfonamide formation, chlorination and N-alanylation. The details of how dealanylascamycin is converted into ascamycin is an intriguing question that has yet to be answered. Here we present in vivo evidence to prove our hypothesis that an esterase AcmE are responsible for that. Bioinformatics analysis showed that an esterase AcmE was present which also had some similarity to the only aminopeptidase identified to-date that cleaves ascamycin to generate dealanylascamycin. The host only becomes (Xanthomonas sp.) susceptibile to ascamycin [24] when they have this aminopeptidase activity. We suspect acmE is involved in a reversible N-alanylation enzymatic reaction in the ascamycin/dealanylascamycin pathway.
Ascamycin and dealanylascamycin are rare secondary metabolites decorated with sulfonamide groups at the 5′-hydroxyl of the ribose ring. Six organosulfur metabolism genes (AcmABGKIW) and aminotransferase AcmN are candidate genes required for 5′-O-sulfonamide formation. These genes showed little homology to other genes of sulfur-groups metabolism [eg. The caprazamycin metabolite pathway which involves a two-step sulfation:A triketide pyrones that are sulfated by an PAPS-dependent sulfotransferase (Cpz8) to generate phenolic sulfate esters which play a role as sulfate donors for a PAPS-independent arylsulfate sulfotransferase (Cpz4) to produce sulfated licaprazamycin(s) [41]]. Another intriguing question is halogenation of ACM/DACM, which probably involved AcmX and/or AcmY. Though they are 1Mb remote from AcmA-AcmW gene cluster, they can be regarded as candidate for adenine C-2 halogenase because they showed considerable homology to all FAD-type halogenase which are believed to be required for aromatic adenine ring chlorination. When JCM9888 genome was aligned with non-heme iron halogenases, SAM-dependent halogenases or vanadium and heme-dependent halogenses, no counterpart genes or homologues were identified [25] [26] [27]. We also checked the mycelium and spore formation of Streptomyces mutant ΔacmE, ΔacmG, ΔacmK but did not observed any difference between them and wild type JCM9888, suggesting that these genes or secondary metabolites ACM/DACM are not absolutely required for morphological differentiation.
It is an important aim of our project to provide more information to investigate the enigmatic fluorinated natural compound nucleocidin, which was discovered in 1969 [3], while all efforts to identify this famous fluorinated natural products from Streptomyces deposited in public collections failed in the past twenty years (Prof. David O′Hagan, University of St Andrews, UK, personal communication). Nucleocidin possess a very similar chemical structure with dealanylascamycin except that nucleocidin has a fluorine atom at C4′ of the ribose while the later is chlorinated at C2 position of the adenine ring. We suspect that these two metabolic pathways use a set of very similar genes and proteins. Given access to the recently published genome of the putative nucleocidin producer Streptomyces calvus DSM40010 [42], we aligned the ascamycin/dealanylascamycin gene cluster against the S. calvus genome database, but no homologous counterpart was identified, suggesting that commercially obtained cultures of S. calvus DSM40010 (or ATCC13382) may not be the correct nucleocidin-producing strain. We suspect that a true nucleocidin-producing Streptomyces would also carry the related genes of ACM/DACM biosynthesis. Thus we envisage that with certain effort, it would be possible to re-isolate a nucleocidin-producing Streptomyces strain from such colonies by soil screening approach. Knowledge of the genes of related ACM/DACM biosynthesis would be necessary in order to devise PCR screening primers. This target-narrowing using gene PCR provides obvious advantages over verification by fermentation, extraction and metabolite analyses of candidate Streptomyces colonies individually.
Methods
Bacterial strains, plasmids and reagents
Bacterial strains and plasmids used and constructed in this study are listed in Table S1. Primers were chemically synthesized by commercial resources and listed in Table S2. strains JCM9888 were grown at 28°C on ISP4 medium for growth and sporulation [43]. MS medium (soy bean flour 20 g, mannitol 20 g, agar 20 g, 2.5 mM MgSO4, 2.5 mM KCl in 1 L of tap water, pH 8.0) is used for petri dish fermentation of antibiotics ascamycin/dealanylascamycin [20], [21]. ISP4 medium supplemented with 0.2% yeast extract is used for E. coli-Streptomyces bi-parental conjugation.
DNA extraction, manipulation and sequencing
Total DNA isolations, plasmid DNA preparations, restriction endonuclease digestions, ligations, and other DNA manipulations were performed according to standard procedures for E. coli and Streptomyces [43] [44]. The sequencing service at the Chinese National Genome Centre (Shanghai) was used for DNA sequencing. Computer-assisted database searching and sequence analysis were carried out online using the frame-plot 4.0 sequence analysis software (http://nocardia.nih.go.jp/fp4/) and the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Gene Inactivation Experiments
The lambda-RED-mediated gene replacements were performed as standard procedures [45]. In all cases, the aac(3)IV gene from pIJ773 was inserted within the coding region of the gene to be disrupted and in the same direction of transcription. The introduction of DNA into JCM9888 was achieved through intergeneric conjugation from E. coli ET12567/pUZ8002. After the introduction of the constructs into Streptomyces by intergeneric conjugation, exconjugants, in which a double crossover occurred, were identified by their resistance to apramycin and susceptibility to kanamycin.
To generate a mutant containing a deletion or an insertion mutation, the following primers were designed: (i) Mutagenesis of AcmE (esterase): AcmE-FP: CCC GGC CAA CCG CGC TTC ATC TGC TCG ACG TTG ACC GTC att ccg ggg atc cgt cga cc, AcmE-RP: CCG CAA CGA CCC GCG TGC GCC GCC CCG ATG GTC AAG CGG tgt agg ctg gag ctg ctt c. (ii) Mutagenesis of AcmG (Sulfatase): AcmG-FP: GAG GGG AGA CGC TCT TCA CCT GGG CGC GCC GAC GCG GCT att ccg ggg atc cgt cga cc, AcmG-RP: CGA GGA TGA TCG GGA AGT CGC TCT TGA AGT ACG TCC GCA tgt agg ctg gag ctg ctt c. (iii) Mutagenesis of AcmK (sulfotransferase): AcmK-FP: GTC CGA GAC CGC ACG TCG CGC CGC GAC CCG GGA CGC CCT att ccg ggg atc cgt cga cc, AcmK-RP: TCA CAG CCC CAC GGG TGG ATG TGC GTG CGC AGG AGG GCC tgt agg ctg gag ctg ctt c.
The apramycin resistance cassette pIJ773 was used as a template throughout the experiments. The PCR products were transferred into competent cells of BW25113 (pIJ790, cosmid 15H6) to isolate apramycin-resistant and amplicillin-resistant E. coli colonies. These mutated cosmids were introduced into ET12567 (pUZ8002) and then transferred by conjugation to JCM9888 to screen for apramycin resistant and kanamycin-sensitive colonies. This resulted in ΔacmE, ΔacmG and ΔacmK mutants respectively, which were validated by PCR and subsequent sequencing. (Fig. S1)
Production, purification and analysis of ascamycin/dealanylascamycin
Ascamycin/dealanylascamycin isolation were performed according to the literature [20] [46]. Fermentation agar from petri dishes (weighing approx. 200 g) was extracted with 60% aqueous acetone. After removal of acetone by rotatory evaporation, the residual extract was passed through a Dowex 80WX8(H) resin column (20 ml), which was then eluted by 0.5 M ammonia solution.
The HPLC-MS analysis of the antibiotics obtained from above was performed on a C-18 reversed-phase column (150 by 4.6 mm; particle size, 5 mm [Phenomenex]) with UV detection at 258 nm. The solvent system consisted of solvent A (0.1% formic acid in water), and solvent B (CH3CN). The HPLC gradient was as follows: 10 % to 55 % solvent B (0 to 15 min), 55 % solvent B (15 to 18 min), 55 % to 10 % solvent B (18 to 20 min), and 10 % solvent B (20 to 25 min) at a flow rate of 0.5 ml/min.
Nucleotide sequence accession number
The complete sequence of the ascamycin/dealanylascamycin biosynthetic gene cluster and annotated proteins have been deposited in GenBank under accession number (GenBank accession number KJ817374 and KJ817375).
Supporting Information
PCR validation of Streptomyces mutants.
(TIF)
Strains, plasmids and cosmids used in this study.
(DOCX)
PCR primers used in this study.
(DOCX)
Supplementary data associated with this article.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The complete sequence of the ascamycin/dealanylascamycin biosynthetic gene cluster and annotated proteins have been deposited in GenBank under accession number (GenBank accession number KJ817374 and KJ817375).
Funding Statement
The authors gratefully acknowledge grants from the 973 Program (2009CB118901, 2012CB721002), Ministry of Science and Technology, China, and the National Natural Science Foundation of China. CZ is a recipient of China postdoctoral training project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Winn M, Goss RJ, Kimura K, Bugg TD (2010) Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep 27:279–304. [DOI] [PubMed] [Google Scholar]
- 2. Lacalle RA, Tercero JA, Jimenez A (1992) Cloning of the complete biosynthetic gene cluster for an aminonucleoside antibiotic, puromycin, and its regulated expression in heterologous hosts. EMBO J 11:785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morton GO, Lancaster JE, Van Lear GE, Fulmor W, Meyer WE (1969) The structure of nucleocidin. 3. (A new structure). J Am Chem Soc 91:1535–1537. [DOI] [PubMed] [Google Scholar]
- 4. McCarty RM, Bandarian V (2008) Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol 15:790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen W, Huang T, He X, Meng Q, You D, et al. (2009) Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J Biol Chem 284:10627–10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ (2010) Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. Chembiochem 11:1700–1709. [DOI] [PubMed] [Google Scholar]
- 7. Zhang W, Ostash B, Walsh CT (2010) Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc Natl Acad Sci U S A 107:16828–16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM (2003) The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4:821–828. [DOI] [PubMed] [Google Scholar]
- 9. Bruntner C, Lauer B, Schwarz W, Mohrle V, Bormann C (1999) Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet 262:102–114. [DOI] [PubMed] [Google Scholar]
- 10. Li J, Li L, Tian Y, Niu G, Tan H (2011) Hybrid antibiotics with the nikkomycin nucleoside and polyoxin peptidyl moieties. Metab Eng 13:336–344. [DOI] [PubMed] [Google Scholar]
- 11. Niu G, Li L, Wei J, Tan H (2013) Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem Biol 20:34–44. [DOI] [PubMed] [Google Scholar]
- 12. Funabashi M, Nonaka K, Yada C, Hosobuchi M, Masuda N, et al. (2009) Identification of the biosynthetic gene cluster of A-500359s in Streptomyces griseus SANK60196. J Antibiot (Tokyo) 62:325–332. [DOI] [PubMed] [Google Scholar]
- 13. Kaysser L, Lutsch L, Siebenberg S, Wemakor E, Kammerer B, et al. (2009) Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem 284:14987–14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Funabashi M, Baba S, Nonaka K, Hosobuchi M, Fujita Y, et al. (2010) The biosynthesis of liposidomycin-like A-90289 antibiotics featuring a new type of sulfotransferase. Chembiochem 11:184–190. [DOI] [PubMed] [Google Scholar]
- 15. Cheng L, Chen W, Zhai L, Xu D, Huang T, et al. (2011) Identification of the gene cluster involved in muraymycin biosynthesis from Streptomyces sp. NRRL 30471. Mol Biosyst 7:920–927. [DOI] [PubMed] [Google Scholar]
- 16. Wu J, Li L, Deng Z, Zabriskie TM, He X (2012) Analysis of the mildiomycin biosynthesis gene cluster in Streptoverticillum remofaciens ZJU5119 and characterization of MilC, a hydroxymethyl cytosyl-glucuronic acid synthase. Chembiochem 13:1613–1621. [DOI] [PubMed] [Google Scholar]
- 17. Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, et al. (2012) Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat Chem 4:539–546. [DOI] [PubMed] [Google Scholar]
- 18. Zhang G, Zhang H, Li S, Xiao J, Zhang G, et al. (2012) Characterization of the amicetin biosynthesis gene cluster from Streptomyces vinaceusdrappus NRRL 2363 implicates two alternative strategies for amide bond formation. Appl Environ Microbiol 78:2393–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walsh CT, Zhang W (2011) Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS Chem Biol 6:1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isono K, Uramoto M, Kusakabe H, Miyata N, Koyama T, et al. (1984) Ascamycin and dealanylascamycin, nucleoside antibiotics from Streptomyces sp. J Antibiot (Tokyo) 37:670–672. [DOI] [PubMed] [Google Scholar]
- 21. Takahashi E, Beppu T (1982) A new nucleosidic antibiotic AT-265. J Antibiot (Tokyo) 35:939–947. [DOI] [PubMed] [Google Scholar]
- 22. Osada H, Isono K (1986) Purification and characterization of ascamycin-hydrolysing aminopeptidase from Xanthomonas citri. Biochem J 233:459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaffe JJ, McCormack JJ, Meymerian E (1970) Trypanocidal properties of 5'-O-Sulfamoyladenosine, a close structural analog of nucleocidin. Exp Parasitol 28:535–543. [DOI] [PubMed] [Google Scholar]
- 24. Osada H, Isono K (1986) Occurrence of an ascamycin dealanylating enzyme, Xc-aminopeptidase, in mammalian cell membranes and susceptibility to ascamycin. J Antibiot (Tokyo) 39:286–293. [DOI] [PubMed] [Google Scholar]
- 25. Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT (2006) Nature's inventory of halogenation catalysts: oxidative strategies predominate. Chem Rev 106:3364–3378. [DOI] [PubMed] [Google Scholar]
- 26. Butler A, Sandy M (2009) Mechanistic considerations of halogenating enzymes. Nature 460:848–854. [DOI] [PubMed] [Google Scholar]
- 27. Wagner C, El Omari M, Konig GM (2009) Biohalogenation: nature's way to synthesize halogenated metabolites. J Nat Prod 72:540–553. [DOI] [PubMed] [Google Scholar]
- 28. Florini JR, Bird HH, Bell PH (1966) Inhibition of protein synthesis in vitro and in vivo by nucleocidin, an antitrypanosomal antibiotic. J Biol Chem 241:1091–1098. [PubMed] [Google Scholar]
- 29.Ubukata M, Osada H, Isono K (1985) Synthesis and biological activity of nucleoside antibiotics, ascamycin and its amino acid analogs. Nucleic Acids Symp Ser: 81–83. [PubMed]
- 30. Shuman DA, Robins MJ, Robins RK (1970) The synthesis of nucleoside sulfamates related to nucleocidin. J Am Chem Soc 92:3434–3440. [DOI] [PubMed] [Google Scholar]
- 31. Shuman D, Robbins RK, Robins MJ (1969) The synthesis of adenine 5'-O-sulfamoyl nucleosides related to nucleocidin. J Am Chem Soc 91:3391–3392. [DOI] [PubMed] [Google Scholar]
- 32. Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, et al. (2006) Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem Biol 13:575–585. [DOI] [PubMed] [Google Scholar]
- 33. Chapman E, Best MD, Hanson SR, Wong CH (2004) Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl 43:3526–3548. [DOI] [PubMed] [Google Scholar]
- 34. Kaysser L, Eitel K, Tanino T, Siebenberg S, Matsuda A, et al. (2010) A new arylsulfate sulfotransferase involved in liponucleoside antibiotic biosynthesis in streptomycetes. J Biol Chem 285:12684–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobashi K, Fukaya Y, Kim DH, Akao T, Takebe S (1986) A novel type of aryl sulfotransferase obtained from an anaerobic bacterium of human intestine. Arch Biochem Biophys 245:537–539. [DOI] [PubMed] [Google Scholar]
- 36. Sudo T, Shinohara K, Dohmae N, Takio K, Usami R, et al. (1996) Isolation and characterization of the gene encoding an aminopeptidase involved in the selective toxicity of ascamycin toward Xanthomonas campestris pv. citri. Biochem J 319 (Pt 1):99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Osada H, Isono K (1985) Mechanism of action and selective toxicity of ascamycin, a nucleoside antibiotic. Antimicrob Agents Chemother 27:230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang W, Ntai I, Kelleher NL, Walsh CT (2011) tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci U S A 108:12249–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng X, Gao F, May WS (2009) Protein phosphatase 2A inactivates Bcl2's antiapoptotic function by dephosphorylation and up-regulation of Bcl2-p53 binding. Blood 113:422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatzios SK, Iavarone AT, Bertozzi CR (2008) Rv2131c from Mycobacterium tuberculosis is a CysQ 3'-phosphoadenosine-5'-phosphatase. Biochemistry 47:5823–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang X, Eitel K, Kaysser L, Kulik A, Grond S, et al. (2013) A two-step sulfation in antibiotic biosynthesis requires a type III polyketide synthase. Nat Chem Biol 9:610–615. [DOI] [PubMed] [Google Scholar]
- 42. Kalan L, Gessner A, Thaker MN, Waglechner N, Zhu X, et al. (2013) A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem Biol 20:1214–1224. [DOI] [PubMed] [Google Scholar]
- 43.Kieser T, Bibb M, Butter M, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich, United Kingdom.
- 44.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45. Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kirst HA, Dorman DE, Occolowitz JL, Jones ND, Paschal JW, et al. (1985) The structure of A201A, a novel nucleoside antibiotic. J Antibiot (Tokyo) 38:575–586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR validation of Streptomyces mutants.
(TIF)
Strains, plasmids and cosmids used in this study.
(DOCX)
PCR primers used in this study.
(DOCX)
Supplementary data associated with this article.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The complete sequence of the ascamycin/dealanylascamycin biosynthetic gene cluster and annotated proteins have been deposited in GenBank under accession number (GenBank accession number KJ817374 and KJ817375).