Abstract
Sensitization to environmental allergens remains one of the strongest risk factors for asthma, and there is likely a genetic basis. We sought to identify genetic determinants for the development of allergic sensitization to environmental allergens, particularly cockroach allergen, in early childhood. A total of 631 children with the information about genotypic data on 895 single nucleotide polymorphisms (SNPs) in 179 candidate genes were selected from an existing dataset (Boston Birth Cohort). Genetic analysis was performed for allergic sensitizations among all subjects and sub-population, Black/African, respectively. Eight SNPs in seven genes showed significant association with allergic sensitization with P < 0.05, including two top SNPs, rs7851969 in JAK2 (P = 0.003) and rs11739089 in CNOT6 (P = 0.008). When analyses were specifically performed for cockroach sensitization, 16 SNPs in 13 genes showed P < 0.05, including five genes with SNPs at P < 0.01 (JAK1, JAK3, IL5RA, FCER1A, and ADAM33). Particularly, haplotype analyses demonstrated that multiple-haplotypes in FCER1A were significantly associated with cockroach sensitization with the strongest association for a 2-marker haplotype (rs6665683T-rs12136904T, P = 0.001). Furthermore, SNP rs6665683 was marginally associated with the levels of cockroach allergen specific IgE. When a similar analysis was performed for house dust mite, four SNPs in three genes (JAK2, MAML1, and NOD1) had P < 0.01. Of these, JAK2 appeared to be an only gene showing association across the sensitizations we analyzed. Some of findings were further validated when analysis was limited to black population. Our study identified several loci that may confer the susceptibility to allergic sensitization, and suggested that sensitization to allergens may depend on their unique loci.
Keywords: Asthma, allergic sensitization, cockroach sensitization, house dust mites, single nucleotide polymorphisms
Introduction
Asthma is the leading serious chronic illness of children in the United States [1]. It is largely accepted that gene–environment interactions are responsible for the development of asthma; cockroach sensitization has been demonstrated to be one of the major risk factors for the development of asthma [2–4]. In particular, it has been suggested that cockroach sensitized exposure may be closely associated with increased asthma morbidity among inner-city residents [5–7]. While there appears to be a rather clear relationship between allergen exposure and allergen sensitization, a dose–response relationship is mostly relevant for “susceptible” individuals [8]. Similarly, although no difference was observed in cockroach sensitization between those with and without asthma, environmental allergen exposure was suggested to play a critical role in the development of asthma among genetically susceptible individuals [9]. Furthermore, although many individuals were exposed to very high concentrations of allergen, they never become sensitized [10]. These findings suggest that cockroach sensitization is not a function of cockroach allergen exposure alone, and genetic susceptibility may be important. Indeed, Hopp et al. [11] reported that skin test reactivity is determined by genetic and environmental factors with a greater genetic influence as reflected in the higher correlation in monozygotic twins. Recent studies have demonstrated a significant familial aggregation of allergic sensitization to cockroach allergen in the Chinese population [12] and suggested a role of genetic factors in allergic sensitization. Previously Togias et al. [13] demonstrated that individuals sensitized to cockroach allergens, especially African Americans, have more severe symptoms than the general asthma population, exhibiting higher IgE levels, a high degree of steroid dependency and greater chronicity of disease. Collectively, these studies suggest that there is a genetic basis for environmental allergen sensitization, which contributes to the risk of asthma and/or the severity of asthma.
Although it is clear that there is a genetic basis for allergen induced sensitization, few studies have been performed to identify genetic factors specifically for cockroach sensitization. In a study on HLA-D associations and cockroach sensitization, Donfack et al. [14] observed associations with alleles of the HLA-DR molecule, DRB1*0101 in Hutterites and DRB1*0102 in African Americans, and hypothesized that the DRB1*0102 allele may have a higher affinity for cockroach allergens and therefore may elicit a stronger response to bind antigens than DRB1*0101 allele. In a genome-wide linkage study of asthma-related phenotypes on 2551 individuals from 533 families, Xu et al. [15] provided suggestive evidence of linkage at a possible QTL, D4S1647 for skin reactivity to cockroach defined by skin prick tests (SPT) (pointwise P = 0.0003). Several similar epidemiological studies concerning the association between genetic and SPT reactivity have found several SPT-related gene [16,17]. Furthermore, Hunninghake et al. [18] reported that there was significant evidence of linkage to IgE to cockroach on chromosome 5q23 (peak LOD, 4.14 at 127 cM) in female subjects. Within this genomic region, there is a compelling candidate gene with experimental evidence of female-specific effects on lung disease, thymic stromal lymphopoietin (TSLP). In a sex-stratified analysis, the single-nucleotide polymorphism (SNP) rs2289276, in the 5′untranslated region of TSLP was associated with reductions in specific IgE to cockroach in Costa Rican girls. In addition, Leung et al. [19] observed that polymorphisms in the Mannose-binding lectin (MBL) gene, a member of the innate immune system, may protect against cockroach sensitization in Chinese children, and Pistiner et al. [20] demonstrated that polymorphisms in IL-12A were associated with cockroach sensitization among children with asthma in both Costa Rica and Caucasian children with asthma in the Childhood Asthma Management Program (CAMP). In addition, new information is emerging from hypothesis-free approaches such as genome-wide association studies. Recent study by Bønnelykke et al. [21] have identified ten loci influencing allergic sensitization by meta-analysis of genome-wide association studies, including SNPs in or near TLR6, C11orf30, STAT6, SLC25A46, HLA-DQB1, IL1RL1, LPP, MYC, IL2, and HLA-B on adult population. Interestingly, some of those identified genes have been shown to be critical in the pathogenesis of allergic diseases.
In this study, we examined the genetic determinants for the development of sensitization to specific environmental allergens in early childhood in a total of 631 Children consisting of allergen sensitized and non-sensitized individuals, all of whom were identified from Boston Birth Cohort (BBC) and have the genotyping data of 895 SNPs in 179 highly selected genes. Our initial analysis was performed for allergic sensitization to common environmental allergens, followed by an analysis specific to cockroach allergen and house dust mite. Findings from these initial analyses were further validated when analyses were limited to black population only. This study provided target genes for future replication in diverse ethnic populations and further functional investigation.
Materials and Methods
Study population
A total of 1219 children from the BBC were initially genotyped for 1188 SNPs. Detailed information on the recruitment of BBC has been described previously [22]. In this study, we specifically focused on subjects who have both genotypic data and data on allergic sensitization. As shown in Figure 1, there are a total of 756 children from BBC with 1188 SNPs genotyped by the Illumina HumanOmni-Quad Beadchip. Of these, 125 subjects were further removed because of failure in quality control (QC) measures. In addition, we removed 293 SNPs because of one or more of the followings: (1) minor allele frequency (MAF) ≤5% (n = 52), (2) Hardy–Weinberg equilibrium (HWE) P-value <0.001 (n = 148), (3) SNPs in strong linkage disequilibrium (LD—r2 ≥ 0.8; n = 118), and (4) SNPs with 2 or more than 2 mentioned criteria above (n = 25) [23]. Thus, our final analysis was performed in 631 subjects consisting of allergen sensitized (AS, n = 207) and non-sensitized to environmental allergens (n = 424; Table1) with the information about genotypic data on 895 single nucleotide polymorphisms (SNPs) in 179 candidate genes (Table S1). Allergic sensitization was defined as at least 1 positive allergen-specific IgE measurement. These allergic individuals incldued those who are sensitized to: (1) cockroach allergen (n = 75), (2) dermatophagoides Pteronyssinus (n = 99), Dermatophagoides Farinae (N = 33), (3) alternaria alternata (n = 89), (4) danger to dog (n = 79), and (6) cat (n = 60) (Fig. 1). For the black population, there were 132 allergic sensitized and 259 were non-sensitized subjects. Among these subjects with allergic sensitization, a total of 48 subjects were sensitized to cockroach, 59 were sensitized to house dust mite, and 160 were sensitized to others (Table1, Table S2). Among those subjects, the most common races are Black (62.1%), Hispanic (21.7%), and White (5.1%), Cap Verdean (5.1%), respectively. The rest are less than 5% (Asian/Pacific Islander, Caribbean, and unknown, Table2). The study protocols were approved by the Institutional Review Boards (IRBs) of Children's Memorial Hospital in Chicago and Boston University Medical Center. A data repository protocol of the BBC was approved by the Johns Hopkins Bloomberg School of Public Health IRB. All participating families provided written informed consent.
Figure 1.
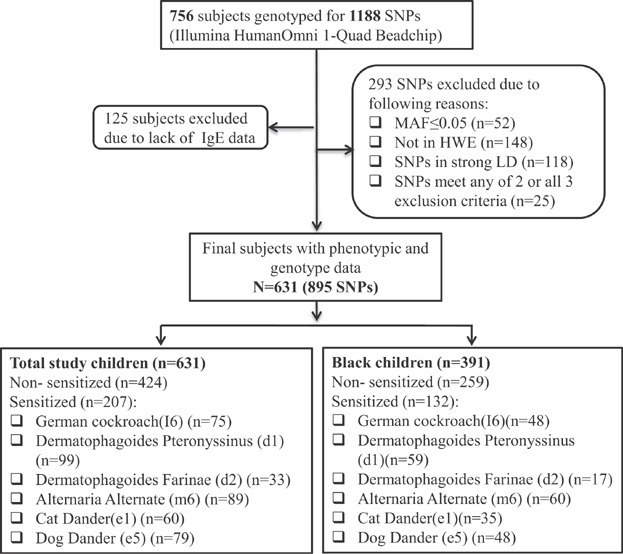
Flow chart on study sample selection from the Boston Birth Cohort.
Table 1.
Characteristics of study children (N = 631)
| Subjects | Total | Total study children |
P-value | Total | Black children |
P-value | ||
|---|---|---|---|---|---|---|---|---|
| Non-sensitized (424) | Sensitized (207) | Non-sensitized (259) | Allergen-sensitized (132) | |||||
| Age (M ± SD) | 2.5 ± 2.07 | 2.17 ± 1.88 | 3.21 ± 2.27 | <0.001 | 2.6 ± 2.08 | 2.29 ± 1.91 | 3.26 ± 2.25 | 0.006 |
| Gender (F) N (%) | 313 (49.6) | 205 (48.3) | 108 (52.2) | 0.397 | 197 (50.4) | 130 (50.2) | 67 (50.8) | 1.000 |
| Maternal continuous smoking N (%) | 55 (8.7) | 42 (9.9) | 13 (6.3) | 0.136 | 35 (9.0) | 27 (10.4) | 8 (6.1) | 0.191 |
| Maternal atopy N (%) | 232 (36.8) | 155 (36.6) | 77 (37.2) | 0.930 | 142 (36.3) | 96 (37.1) | 46 (34.8) | 0.775 |
| Postnatal IgE (median) | 1.37 | 1.17 | 1.87 | <0.001 | 1.43 | 1.17 | 1.90 | <0.001 |
Table 2.
Sensitization to all allergens, cockroach, and house dust mite in different races
| Total | Non-sensitized | Sensitized |
|||
|---|---|---|---|---|---|
| All allergens | Cockroach | House dust mite | |||
| Black/African American | 391 | 259 (66.2%) | 132 (33.8%) | 48 (36.4%) | 59 (44.7%) |
| White | 32 | 24 (75.0%) | 8 (25.0%) | 1 (12.5%) | 4 (50.0%) |
| Hispanic | 137 | 88 (64.2%) | 49 (35.8%) | 23 (46.9%) | 32 (65.3%) |
| Asian/Pacific | 9 | 9 (100.0%) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Cap Verdean | 32 | 22 (68.8%) | 10 (31.2%) | 1 (10.0%) | 3 (30.0%) |
| Mixed | 2 | 1 (50.0%) | 1 (50.0%) | 0 (0.00) | 0 (0.00) |
| Caribbean | 5 | 5 (100.0%) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Unknown | 23 | 16 (72.7%) | 7 (27.3%) | 2 (28.6%) | 1 (14.3%) |
| Total | 631 | 424 (67.3%) | 207 (32.7%) | 75 (36.2%) | 99 (47.8%) |
Measurement of specific IgE
Venous blood samples were collected and stored at −80°C for IgE measurement. Tests for serum allergen specific IgE were performed by using ImmnunoCAP Total Low Range Assay (Phadia AB, Uppsala, Sweden) according to the manufacturer's prescribed protocol for the following six common indoor extract allergens characterized by company: German cockroach (I6); dermatophagoides Pteronyssinus (d1); dermatophagoides Farinae (d2); alternaria alternata (m6); cat dander (e1); and dog dander (e5). The calibration range for tIgE and sIgE was 2.0–5000 kU/L and 0.1–100 kUA/L, respectively. These assays were measured by the Clinical Immunology Laboratory at Children's Memorial Hospital, Chicago, Illinois.
Rationale of SNP selection
We have provided a list of selected 895 SNPs with the information about allele, chromosome, chromosome position, and possible function predicted by Regulome DB score (http://regulome.stanford.edu/) in supplementary materials (Table S1). In addition to tagging SNPs with LD cut-off as r2 ≥ 0.8 [23], we selected potentially functional SNPs with a minor allele frequency of 0.05 or greater that met the following criteria: (1) coding SNPs, (2) SNPs creating/disrupting a splicing site, or (3) SNPs predicted to be functional variants based on bioinformatics tools (e.g., PupaSuite [http://pupasuite.bioinfo.cipf.es/], FuncPred [http://snpinfo.niehs.nih.gov/snpfunc.htm], and F-SNP [http://compbio.cs.queensu.ca/F-SNP/]; for example, this included SNPs in transcription factor binding sites, in exonic splicing enhancers or silencers, in microRNAs and their target sequences, and/or in a DNA triplex [triplex]); and (4) SNPs within the expression quantitative trait locus. Additional clinical and genetic data are reported with details in our previous published articles [23–28]. Furthermore, for candidate gene selection, we selected those that may be associated with asthma, eczema and other allergic diseases, and genes that are located in functionally important pathways such as antigen presentation, IgE production, innate immunity, Th1 and Th2 skewing, and airway remodeling.
SNP genotyping
SNPs were genotyped in the genotyping center at Washington University at St Louis by using the Illumina GoldenGate custom panel (Illumina, Inc., San Diego, CA). For quality control and quality assurance, four duplicate DNA control samples were included in each 96-well plate and were genotyped. The concordance rate of these duplicate samples was greater than 99.5%.
Statistical analysis
To control for potential confounding caused by population stratification, 150 ancestry informative markers, with averaged δ (allele frequency difference between two ancestral populations) ≥0.5, were randomly selected from a recently reported genome-wide admixture map [29]. Of those, 144 ancestry informative markers (with a call rate ≥98.0%) were included in the estimation of ancestral proportion for three ancestral populations (Asian, European, and African) using the STRUCTURE program (version 2.3.1; http://pritch.bsd.uchicago.edu/structure.html). These estimated ancestral proportion was then included as a covariate in subsequent analyses. Detailed information can be found previously published paper [23].
For each SNP genotyped in this study, a χ2 test was applied to test HWE in the total population. Pairwise LD of SNPs in each gene was also calculated. Association between each individual SNP and allergic sensitization was performed using logistic regression analysis under an additive model, with adjustments for age, gender and race. The levels of total and specific IgE were log-transformed from a non-normal distribution into a normal distribution. The association with IgE was assessed using a linear regression analysis adjusted for age, gender and race. Similar analyses were performed for the black population adjusted for age and gender. Expectation–maximization algorithm was used to analyze haplotype frequencies and P values. To represent final results omnibus P values were recorded. Omnibus test is a general name refers to an overall or a global test and in commonly omnibus test is called in other expressions such as: F-test or Chi-squared test. Omnibus test as a statistical test is implemented on an overall hypothesis that tends to find general significance between parameters' variance. All data analyses were conducted by using the PLINK program (http://pngu.mgh.harvard.edu/∼purcell/plink/). A 2-tailed P value of less than 0.05 was considered statistically significant.
Results
Characteristics of study population
Total study population consisted of approximately equal proportion of male subjects between allergens sensitized (52.2%) and non-sensitized (48.3%; P = 0.397; Table1) and equal proportion of maternal atopy between allergen sensitized (37.2%) and non- sensitized (36.6%; P = 0.93), and equal proportion of maternal continuous smoking between allergen sensitized (6.3%) and non- sensitized (9.9%; P = 0.136). However, there were significant differences between two groups for age (allergens sensitized vs non-sensitized to environmental allergens, 3.21 ± 2.27 vs. 2.17 ± 1.88 (years), P < 0.001). Total serum IgE levels (KU/L) were significantly higher among those who are sensitized to one of the tested allergens as compared to those who are not sensitized to any of these allergens (median of log-transformed IgE, 1.87 vs. 1.17, P < 0.001). When analysis was limited to Black children, the significant difference was also observed for age (allergens sensitized vs. non-sensitized to environmental allergens, 3.26 ± 2.25 vs. 2.29 ± 1.91 (years), P = 0.006), and postnatal IgE (median of log-transformed IgE, KU/L) allergens (sensitized vs. non-sensitized to environmental allergens, 1.90 vs. 1.17, P < 0.001). When all these allergic subjects were divided by race, there was no significant difference among these most common races (n ≥ 10) [Black (33.8%), Hispanic (35.8%), and Cap Verdean (31.2%)]. The same is also true for those who are specifically allergic to cockroach [Black (36.4%) and Hispanic (46.9%)]. However, the difference was observed for those who are allergic to house dust mite in Hispanic and Black/African (65.3% vs. 44.7%, P = 0.0027; Table2).
Association of SNPs with allergic sensitization
We examined the association between those genotyped SNPs and allergic sensitization (Table3, Fig. 2A). A total of 8 SNPs in or near seven genes showed P < 0.05. Of these, three SNPs had P < 0.01, including rs7851969 in JAK2 [Odds ratio (OR) (95% CI), 2.08 (1.29–3.37), P = 0.003], rs11739089 in CNOT6 [OR (95% CI), 1.54 (1.17–2.11), P = 0.008], and rs6627 in MAML1 [OR (95% CI), 0.70 (0.54–0.92), P = 0.009]. Furthermore, several candidate genes also showed significant association with allergic sensitization, including IL4R (rs3024633, rs3024576), JAK1 (rs7524842), IL12B (rs3212227), and IL5RA (rs163550). When the same analyses was limited to black population, we found that, except for SNPs in gene MAML1 and IL5RA, the associations we observed for others in total population still remained. Of these, SNP rs11739089 in CNOT6 showed even stronger association in the black population (P = 0.001).
Table 3.
Association between genotyped single gene polymorphisms and sensitization to environmental allergens
| rs ID | Gene | Chromosome | SNP | Minor allele frequency total |
OR (95% CI) | P-value | Black Non-sensitized (259) vs. sensitized (132) |
||
|---|---|---|---|---|---|---|---|---|---|
| Non-sensitized (N = 424) | Sensitized (N = 207) | OR (95% CI) | P-value | ||||||
| rs7851969 | JAK2 | 9 | C/G | 0.05 | 0.09 | 2.08 (1.29–3.37) | 0.003 | 2.02 (1.17–3.49) | 0.011 |
| rs11739089 | CNOT6 | 5 | T/C | 0.13 | 0.18 | 1.54 (11.70–2.11) | 0.008 | 1.89 (1.28–2.78) | 0.001 |
| rs6627 | MAML1 | 5 | G/A | 0.38 | 0.31 | 0.70 (0.54–0.92) | 0.009 | 0.84 (0.61–1.14) | 0.835 |
| rs3024633 | IL4R | 16 | A/G | 0.08 | 0.04 | 0.53 (0.30–0.91) | 0.022 | 0.47 (0.24–0.92) | 0.027 |
| rs7524842 | JAK1 | 1 | T/C | 0.05 | 0.08 | 1.87 (1.12–3.13) | 0.017 | 1.81 (1.03–3.17) | 0.040 |
| rs3212227 | IL12B | 5 | A/C | 0.33 | 0.41 | 1.36 (1.06–1.75) | 0.017 | 1.38 (1.01–1.88) | 0.044 |
| rs3024576 | IL4R | 16 | G/A | 0.10 | 0.07 | 0.57 (0.35–0.91) | 0.020 | 0.45 (0.25–0.83) | 0.010 |
| rs163550 | IL5RA | 3 | G/C | 0.16 | 0.22 | 1.39 (1.02–1.90) | 0.035 | 1.22 (0.80–1.85) | 0.357 |
OR, odds ratio; CI, confidence interval; all P values were analyzed under additive model adjusted for age, gender and race.
Figure 2.
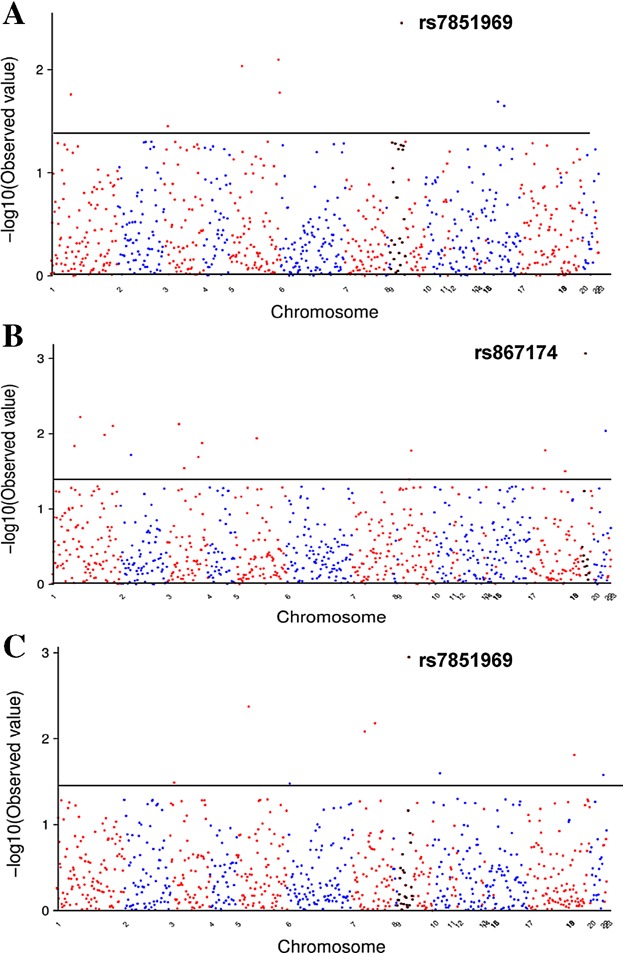
The −log10 (P value) for associations of genotyped SNPs with (A) allergic sensitization, (B) cockroach allergen, and (C) house dust mite. The x-axis represents individual SNP. The gray line represents P = 0.05.
Association of SNPs with sensitization to cockroach allergen
When analysis was performed specifically for cockroach sensitization as defined by positive specific IgE measurement of German cockroach (I6), 17 SNPs in 11 genes showed P < 0.05 (Table4, Fig. 2B). The strongest association was observed for SNP rs867174 in JAK3 [OR (95% CI), 0.28 (0.13–0.61), P = 0.001]. Additionally, five SNPs in four genes had P < 0.01, including, SNP rs10889502 in JAK1 (P = 0.006), rs334807 in IL5RA (P = 0.007), both rs6665683 and rs12135788 in FCER1A (P = 0.008 and 0.010, respectively), and rs2787094 in ADAM33 (P = 0.009). Furthermore, several well-recognized candidate genes also showed association with allergic sensitization, including IL13 (rs1800925, P = 0.016, rs2069743, P = 0.04), JAK2 (rs10815160, P = 0.017), DPP10 (rs4516432, P = 0.019), and CD86 (rs1129055, P = 0.013; rs11717893, P = 0.020). Similarly, when analysis was restricted to black population, we found similar associations for several top SNPs with P < 0.01, including SNP in JAK3 (rs867174, P = 0.002), JAK1 (rs10889502, P = 0.011), FCER1A (rs6665683, P = 0.013; rs12135788, P = 0.005), and ADAM33 (rs2787094, P = 0.036), and for few others, IL13 (rs1800925, P = 0.016) and CD86 (rs1129055, P = 0.011; rs11717893, P = 0.007).
Table 4.
Association between genotyped single gene polymorphisms and cockroach sensitization
| rs ID | Gene | Chromosome | SNP | Minor allele frequency total |
OR (95% CI) | P-value | Black non-sensitized (259) vs. sensitized (48) |
||
|---|---|---|---|---|---|---|---|---|---|
| Non-sensitized (N = 424) | Sensitized (N = 75) | OR (95% CI) | P-value | ||||||
| rs867174 | JAK3 | 19 | A/G | 0.16 | 0.05 | 0.28 (0.13–0.61) | 0.001 | 0.19 (0.07–0.55) | 0.002 |
| rs10889502 | JAK1 | 1 | C/G | 0.42 | 0.54 | 1.71 (1.70–2.50) | 0.006 | 1.88 (1.16–3.07) | 0.011 |
| rs334807 | IL5RA | 3 | G/T | 0.46 | 0.57 | 1.67 (1.15–2.41) | 0.007 | 1.45 (0.92–2.28) | 0.106 |
| rs6665683 | FCER1A | 1 | C/T | 0.15 | 0.25 | 1.81 (1.17–2.82) | 0.008 | 1.87 (1.14–3.05) | 0.013 |
| rs2787094 | ADAM33 | 20 | C/G | 0.38 | 0.27 | 0.59 (0.39–0.87) | 0.009 | 0.59 (0.37–0.97) | 0.036 |
| rs12135788 | FCER1A | 1 | G/T | 0.24 | 0.35 | 1.63 (1.12–2.37) | 0.010 | 1.94 (1.22–3.09) | 0.005 |
| rs1800925 | IL13 | 5 | T/C | 0.31 | 0.43 | 1.61 (1.12–2.33) | 0.011 | 1.71 (1.10–2.64) | 0.016 |
| rs1129055 | CD86 | 3 | A/G | 0.21 | 0.12 | 0.51 (0.30–0.87) | 0.013 | 0.39 (0.19–0.81) | 0.011 |
| rs17127114 | JAK1 | 1 | G/A | 0.16 | 0.08 | 0.46 (0.25–0.86) | 0.015 | 0.48 (0.22–1.07) | 0.072 |
| rs10815160 | JAK2 | 9 | G/T | 0.38 | 0.27 | 0.61 (0.40–0.92) | 0.017 | 0.61 (0.36–1.05) | 0.074 |
| rs1635278 | CCL16 | 17 | A/G | 0.42 | 0.31 | 0.62 (0.42–0.92) | 0.017 | 0.66 (0.42–1.03) | 0.070 |
| rs4516432 | DPP10 | 2 | T/C | 0.17 | 0.24 | 1.75 (1.09–2.79) | 0.019 | 1.77 (0.95–3.29) | 0.073 |
| rs11717893 | CD86 | 3 | C/T | 0.23 | 0.32 | 1.59 (1.08–2.35) | 0.020 | 1.92 (1.19–3.09) | 0.007 |
| rs11079339 | EPX | 17 | A/G | 0.13 | 0.2 | 1.71 (1.07–2.73) | 0.024 | 2.01 (1.12–3.62) | 0.020 |
| rs4054760 | IL5RA | 3 | T/A | 0.22 | 0.32 | 1.57 (1.05–2.23) | 0.029 | 1.48 (0.88–2.48) | 0.138 |
| rs7851969 | JAK2 | 9 | C/G | 0.05 | 0.09 | 1.99 (1.03–3.87) | 0.040 | 1.99 (1.03–3.87) | 0.082 |
OR, odds ratio; CI, confidence interval; all P values were analyzed under additive model adjusted for age, gender and race.
FCER1A haplotypes contribute to cockroach sensitization
Haplotype analyses were performed for several genes with at least two SNPs showing association with cockroach sensitization, including JAK1, IL5RA, FCER1A, IL13, and CD86. Significance was illustrated by different colors from green (P > 0.1) to red (P < 0.005). Of these, multiple haplotypes of 2–5 SNPs in FCER1A were significantly associated with cockroach sensitization (P = 0.042–0.001, Fig. 3A), with the strongest association seen with a 2-SNP haplotype (T-T, P = 0.001) comprised of markers in the intron region for FCER1A (rs6665683 and rs1213904) among all allergen sensitized subjects. The same pattern was seen when analysis was limited to black population. No association for FCER1A haplotypes was observed for sensitization to house dust mite among all or black population (Data not shown). The results suggest that FCER1A SNPs may be specific to the susceptibility to cockroach sensitization.
Figure 3.
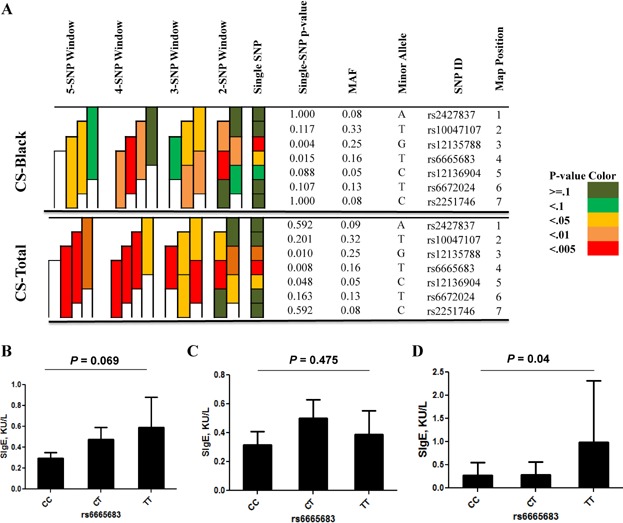
Association between FCER1A SNPs and cockroach sensitization. (A) Haplotype analysis for FCER1A SNPs and cockroach sensitization. Shading represents the magnitude and significance of SNP and 2–4 haplotype with a red to white gradient reflecting higher to lower P values. CS, cockroach sensitization. (B and C) Association of FCER1A SNP rs6665683 with sIgE was performed among all (B) or black (C) or non-black (D) subjects with cockroach sensitization.
FCER1A SNPs contribute to differential levels of cockroach allergen specific IgE
To explore the biological link of FCER1A SNPs to disease, we examined the association between all FCER1A SNPs and the levels of cockroach allergen specific IgE (sIgE). The analysis was performed by a linear regression analysis adjusting for age, sex, and race. The SNP rs6665683 that was associated with cockroach sensitization showed marginal association (P = 0.069, Fig. 3B). Those carrying CC genotype had much lower levels of sIgE (0.100 KU/L ± 0.096, n = 44) as compared to those carrying CT (0.476 KU/L ± 0.571, n = 25) and TT (0.589 KU/L ± 0.714, n = 6). While similar pattern was seen for black population, it was not statistically significant (P = 0.475, Fig. 3C). We also specifically analyzed the correlation between the genotype of FCER1A SNP rs6665683 and cockroach allergen specific IgE plasma levels in non-black subpopulation and observed that those carrying TT genotype have significantly higher levels of cockroach allergen specific IgE as compared with those with CC and CT (P < 0.04, Fig. 3D). No significant correlation was observed for other FCER1A SNPs in these populations analyzed (data not shown).
Association of SNPs with sensitization to house dust mite
For comparaison, association analyses were also performed for allergic sensitization to house dust mites, as determined by positive specific IgE for dermatophygoides (Table5, Fig. 2C). Nine SNPs in or near eight genes showed significant association with allergic sensitization to house dust mite with P < 0.05. Interestingly, rs7851969 in JAK2, a top associated SNP with allergic sensitization, also showed the strongest association with the sensitization to house dust mites [OR (95% CI), 2.87 (1.56–5.26), P = 0.001]. In addition, three SNPs in 2 genes showed P < 0.01, including SNP rs6627 in MAML1 [OR (95% CI), 0.57 (0.39–0.83), P = 0.004] and two SNPs in NOD1 [rs5743334, OR (95% CI), 0.37 (0.18–0.75), P = 0.006; rs5743356, 0.36 (0.17–0.77), P = 0.008, respectively]. When the analysis was performed for black population; the associations with SNPs rs7851969 in gene JAK2 [OR (95% CI), 3.08 (1.55–6.14), P = 0.001] and both SNPs in NOD1 [rs5743334, OR (95% CI), 0.42 (0.19–0.90), P = 0.026; rs5743356, OR (95% CI), 0.33 (0.15–0.77), P = 0.010] were further confirmed.
Table 5.
Association between genotyped single gene polymorphisms and house dust mite sensitization
| rs ID | Gene | Chromosome | SNP | Minor allele frequency total |
OR (95% CI) | P-value | Black non-sensitized (259) vs. sensitized (59) |
||
|---|---|---|---|---|---|---|---|---|---|
| Non-sensitized (N = 424) | Sensitized (N = 99) | OR (95% CI) | P-value | ||||||
| rs7851969 | JAK2 | 9 | C/G | 0.05 | 0.11 | 2.87 (1.56–5.26) | 0.001 | 3.08 (1.55–6.14) | 0.001 |
| rs6627 | MAML1 | 5 | G/A | 0.38 | 0.27 | 0.57 (0.39–0.83) | 0.004 | 0.74 (0.48–1.14) | 0.741 |
| rs5743334 | NOD1 | 7 | G/C | 0.11 | 0.05 | 0.37 (0.18–0.75) | 0.006 | 0.42 (0.19–0.90) | 0.026 |
| rs5743356 | NOD1 | 7 | T/G | 0.09 | 0.05 | 0.36 (0.17–0.77) | 0.008 | 0.33 (0.15–0.77) | 0.010 |
| rs12982518 | FCER2 | 19 | C/G | 0.48 | 0.39 | 0.66 (0.48–0.93) | 0.015 | 0.71 (0.47–1.07) | 0.104 |
| rs1058240 | GATA3 | 10 | A/G | 0.22 | 0.13 | 0.58 (0.37–0.93) | 0.025 | 0.58 (0.33–1.01) | 0.053 |
| rs6131034 | CD40 | 20 | C/G | 0.05 | 0.11 | 2.05 (1.09–3.85) | 0.026 | 1.41 (0.56–3.56) | 0.470 |
| rs163550 | IL5RA | 3 | G/C | 0.16 | 0.24 | 1.56 (1.04–2.31) | 0.032 | 1.28 (0.73–2.24) | 0.384 |
| rs3749985 | HLA-DPB1 | 6 | G/C | 0.07 | 0.02 | 0.31 (0.11–0.91) | 0.033 | 0.38 (0.11–1.31) | 0.125 |
OR, odds ratio; CI, confidence interval; all P values were analyzed under additive model adjusted for age, gender and race.
Discussion
Previous studies have demonstrated that sensitization to cockroach allergen is one of the strongest risk factors for asthma. Although it is widely recognized that there is a genetic basis for allergen-induced sensitization, few studies have been performed to identify gene factors specifically for cockroach sensitization [8,19,30,31]. In this study, we have examined the association between allergic sensitization and SNPs in well-recognized candidate genes in the pathogenesis of allergic diseases. Our study populations were drawn from a prospective birth cohort consisting of allergens sensitized and non-sensitized individuals to aeroallergens. Thought it was birth cohort study, however, all selected individuals were greater than 5 months in age with average age (y) at 2.5 ± 2.07. Allergic sensitization was defined using a cut-point of 0.1 KU/L in the analyses, rather than a commonly used cutoff (0.35 KU/L), because we have previously shown no change for the direction and significance of the effects observed in analyses when either 0.1 or 0.35 KU/L was used as cutoffs [25]. Furthermore, genotyping data of a total of 895 SNPs in 179 genes are also available for those selected individuals. Those selected SNPs were either tagging SNPs or SNPs within the functional regions of gene or SNPs that were significantly associated with allergic diseases like asthma.
For this study, we selected German cockroach, Dermatophagoides Pteronyssinus, Dermatophagoides Farinae, Alternaria Alternata, Cat Dander, and Dog Dander, these most common indoor allergens for the tests for allergic sensitization. Our initial analysis was performed for allergic sensitization, which has an estimated heritability of 0.40–0.85 [32,33]. While many candidate genes have been previously reported for allergic sensitization, few have been firmly established. In our analysis, SNPs in seven selected candidate genes were associated with allergic sensitization, including several top ranking genes JAK2, CNOT6, and MAML1. Among these, SNP rs7851969 in JAK2 showed consistent association with allergic sensitization, or specific sensitization to house dust mite or cockroach allergens. While these genes are functionally important in diverse diseases, none of the studied SNPs in these genes have been previously shown to be associated with allergic sensitization or other allergic diseases. For instance, JAK2 are a family of tyrosine kinases that associated with cytokine receptors (e.g., IL-3, IFN-gamma). Upon receptor activation JAKs phosphorylate the transcription factors and initiate the JAK-STAT signaling pathway that is critical in the pathogenesis of allergic diseases like asthma [34]. CNOT6 (CCR4-NOT transcription complex) belongs to a family of chemoattractant molecules involved in the directed migration of immune cells. MAML1 (mastermind-like 1 (Drosophila)), as a protein-coding gene, is a novel modulator for NF-κB signaling and regulates cellular survival [35]. In addition, SNPs in or near IL-4R, JAK1, and IL12B showed modest association with allergic sensitization. These genes are well-known to be critical in mediating Th1/Th2 mediated immune responses, and SNPs in these genes have been associated with allergic diseases like asthma [36–39]. More importantly, very recent meta-analysis of genome wide association studies have identified ten loci that are associated with allergic sensitization [40], including SNPs in or near TLR6, C11orf30, STAT6, SLC25A46, HLA-DGB1, IL1RL1, LPP, MYC, IL2 and HLA-B. While SNPs in TLR6, C11orf30, STAT6, IL1RL1, and IL2 were also included in our dataset for analysis, none of SNPs in these genes showed significant association with allergic sensitization. In contrast, our study suggests several novel genes for allergic sensitization, such as JAK2 and CNOT6.
We next performed an analysis on sub-groups, specifically those who are sensitized to cockroach allergen, one of our major focuses. It is well-known that sensitization to cockroach allergens is one of the strongest identified risks for greater asthma morbidity in low-income urban population [41,42]. Several genes (e.g., HLA-DR, TSLP, MBL2, and IL-12A) have been reported to be associated with different cockroach sensitization-related phenotypes [19,43,44]. In this analysis, 16 SNPs showed association with cockroach allergen sensitization, including top-ranking SNPs in several genes (JAK1, JAK3, IL5RA, FCER1A, and ADAM33) and genes with at least two associated SNPs (JAK1, IL5RA, FCERA1, IL13, and CD86). Of these, associations with SNPs in or near JAK1, JAK3, FCER1A, ADAM33, IL13, and CD86 were further validated when analysis was limited to black population. When haplotype analysis was performed for several genes with multiple-associated SNPs, multiple haplotypes in FCER1A centered on SNP rs6665683 showed significant association with cockroach sensitization in total and black population only. It is known that FCER1A plays an important role in the development of allergy diseases [45]. Previous studies have suggested the linkage of two FCER1A functional polymorphisms −66T>C (rs2251746) and −315C>T (rs2427827) to allergic disorder [46], and total serum IgE levels [47,48]. SNP rs2427827, next to the promoter SNP rs2251746 in FCER1A, affects FCER1A transcriptional activity/FceRIa expression in mast cells and/or basophils [48,49]. In contrast, few haplotypes showed association with all allergen sensitization or particularly with sensitization to house dust mites features (data not shown), suggesting that cockroach sensitization may have unique features.
Interestingly, we found a correlation between the associated SNP rs6665683 and the levels of cockroach allergen specific IgE, and the effect on sIgE levels is allele dose dependent among all sensitized subjects with TT genotype having higher sIgE levels. The extremely higher levels of sIgE were also observed in the TT genotype of the non-black subpopulation, but not in the black population. The difference in two sub-populations may be due to (1) the source of SIgE and (2) population stratification. For the source of sIgE, it is suggested that some of those may be from maternofetal transfusion of IgE for these younger infants. However, maternal IgE does not cross the placenta [50] and the fetus is capable of producing IgE during the 11th week of gestation [51], most IgE detected in cord blood is likely to be produced by the fetus itself. Although a small amount of maternal blood might enter the fetal circulation during pregnancy and delivery [52], these will be completely replaced by the blood and allergen specific IgE produced by infants themselves. Further evidence by Kamemura et al. [53] demonstrated that IgE antibodies in CB are of fetal origin by using a novel allergen microarray of diamond-like-carbon-coated chip. Furthermore, in an ex vivo placental perfusion model, transfer of IgE-allergen complexes has been described for food allergen, but inhalant allergens were not transferred [54]. To control for potential confounding caused by population stratification, we genotyped ancestry informative markers for the estimation of ancestral proportion, and then used the estimated ancestral proportion as a covariate for the race variable in regression analysis.
For comparison, we also investigated the association of SNPs with allergic sensitization to house dust mites, one of the most common allergens. Among all those associated SNPs with house dust mites, several of them showed association with allergic sensitization, one SNP rs7851969 in JAK2 showed association with both house dust mite and cockroach sensitization, suggesting that sensitization to house dust mite and cockroach allergens may share some of genetic features, which may be due to that (1) some of study subjects who are allergic multiple allergens, and (2) cross reactivity between allergens. Indeed, as shown in Table S3, among all these sensitized subjects, 48% of them are allergic to one allergen, 22.7% allergic to two allergens, 12.1% are allergic to three allergens, and <10% of subjects are allergic to more than 4, or 5 or 6 allergens.
In this study, there was a significant difference for the age between sensitized and non-sensitized subjects. Age and environmental factors may affect the allergen sensitization. To control for the possible confounding by age, our analysis was performed by using age as a covariate in logistic regression analysis. In the future, a longitudinal study may be essential to confirm whether the associations hold true over long periods of time in an age-matched population.
We recognize that, in this study, none of SNPs reached statistical significance after Bonferroni correction for multiple testing. It is widely recognized that the Bonferroni correction offers the most conservative approach to control for false positive and may conceal important functional variants. The overall modest P values seen in this study are more likely to be due to the modest sample size rather than a type-I error I (false positive). In addition, we have included multiple populations including Black/African, White, Hispanic, Cap Verdean, Asian/Pacific Islander, Caribbean, and unknown. Among these, the most common races are Black (62.1%), Hispanic (21.7%), and White (5.1%), and Cap Verdean (5.1%). Although it may be preferable to remove some races with too low-sample sizes from the analyses, we still included these in order to increase the total sample size to strengthen the power for data analysis.
In summary, we performed association analysis aiming at identifying genetic determinants for allergic sensitization, specifically cockroach sensitization. SNPs in or near JAK2, CNOT6, MAML1, CD40, IL-4R, and IL-5RA genes were associated with allergic sensitization and SNPs in or near JAK1, JAK3, IL5RA, FCER1A, and ADAM33 genes were associated with cockroach sensitization. Specifically, SNP rs6665683 in FCER1A was associated with cockroach sensitization and levels of sIgE to cockroach allergen. Furthermore, SNP rs7851969 in JAK2 showed consistent association with allergic sensitization, sensitization to HDM and cockroach allergen. Future studies will target on these genes to replicate the findings in larger samples of diverse ethnic populations, and subsequently investigate their biological relevance to disease.
Acknowledgments
We wish to thank all of the study participants. We are also grateful for the dedication and hard work of the field team at the Department of Pediatrics, Boston University School of Medicine. This study was supported by National Institutes of Health (NIH) grant RO1ES021739-01A1 and R21AI109062 to P. Gao and the Boston Birth Cohort (the parent study) was supported in part by the March of Dimes PERI grants (20-FY02-56, #21-FY07-605), the Food Allergy Initiative, and the National Institutes of Health grants R21 ES011666, R01HD041702, R21HD066471, R21AI088609, U01AI090727 to X. Wang. P. Tripathi & X. Hong: data analysis; D. Caruso: collect and store the data; and X. Wang & P. Gao: study design; and P. Tripathi and P. Gao: paper writing.
Conflict of Interest
None declared.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Table S1. List of selected genes, SNPs, location and predicted function.
Table S2. Clinical information.
Table S3. Children with one or multiple (2–6) allergens.
References
- 1.Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, Scalia MR. Akinbami LJ. National surveillance for asthma-United States, 1980–2004. MMWR Surveill. Summ. 2007;56(8):1–54. and. [PubMed] [Google Scholar]
- 2.Amr S, Bollinger ME, Myers M, Hamilton RG, Weiss SR, Rossman M, Osborne L, Timmins S, Kimes DS, Levine ER. Blaisdell CJ. Environmental allergens and asthma in urban elementary schools. Ann. Allergy Asthma Immunol. 2003;90(1):34–40. doi: 10.1016/S1081-1206(10)63611-3. and. [DOI] [PubMed] [Google Scholar]
- 3.Arruda LK, Vailes LD, Ferriani VP, Santos AB, Pomés A. Chapman MD. Cockroach allergens and asthma. J. Allergy Clin. Immunol. 2001;107(3):419–428. doi: 10.1067/mai.2001.112854. and. [DOI] [PubMed] [Google Scholar]
- 4.Crain EF, Walter M, O'Connor GT, Mitchell H, Gruchalla RS, Kattan M, Malindzak GS, Enright P, Evans R, Morgan W. Stout JW. Home and allergic characteristics of children with asthma in seven U.S. urban communities and design of an environmental intervention: the Inner-City Asthma Study. Environ. Health Perspect. 2002;110(9):939–945. doi: 10.1289/ehp.02110939. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang B. Study on cockroach antigen as a probable causative agent in bronchial asthma. J. Allergy Clin. Immunol. 1976;58(3):357–365. doi: 10.1016/0091-6749(76)90115-9. [DOI] [PubMed] [Google Scholar]
- 6.Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, Mitchell H, McNiff-Mortimer K, Lynn H, Ownby D. Malveaux F. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. N. Engl. J. Med. 1997;336(19):1356–1363. doi: 10.1056/NEJM199705083361904. and. [DOI] [PubMed] [Google Scholar]
- 7.Matsui EC, Wood RA, Rand C, Kanchanaraksa S, Swartz L, Curtin-Brosnan J. Eggleston PA. Cockroach allergen exposure and sensitization in suburban middle-class children with asthma. J. Allergy Clin. Immunol. 2003;112(1):87–92. doi: 10.1067/mai.2003.1588. and. [DOI] [PubMed] [Google Scholar]
- 8.Huss K, Adkinson NF, Jr, Eggleston PA, Dawson C, Van Natta ML. Hamilton RG. House dust mite and cockroach exposure are strong risk factors for positive allergy skin test responses in the Childhood Asthma Management Program. J. Allergy Clin. Immunol. 2001;107(1):48–54. doi: 10.1067/mai.2001.111146. and. [DOI] [PubMed] [Google Scholar]
- 9.Diette GB, Hansel NN, Buckley TJ, Curtin-Brosnan J, Eggleston PA, Matsui EC, McCormack MC, Williams DL. Breysse PN. Home indoor pollutant exposures among inner-city children with and without asthma. Environ. Health Perspect. 2007;115(11):1665–1669. doi: 10.1289/ehp.10088. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehr J, Frischer T, Meinert R, Barth R, Forster J, Schraub S, Urbanek R. Karmaus W. Mite allergen exposure is a risk for the incidence of specific sensitization. J. Allergy Clin. Immunol. 1994;94(1):44–52. doi: 10.1016/0091-6749(94)90070-1. and. [DOI] [PubMed] [Google Scholar]
- 11.Hopp RJ, Bewtra AK, Watt GD, Nair NM. Townley RG. Genetic analysis of allergic disease in twins. J. Allergy Clin. Immunol. 1984;73(2):265–270. doi: 10.1016/s0091-6749(84)80018-4. and. [DOI] [PubMed] [Google Scholar]
- 12.Jin Y, Wang W, Xu Y, Zhao J, Liu H. Xue S. Familial aggregation of skin sensitization to aeroallergens in a rural area in China. Int. Arch. Allergy Immunol. 2009;148(1):81–86. doi: 10.1159/000151509. and. [DOI] [PubMed] [Google Scholar]
- 13.Togias A, Horowitz E, Joyner D, Guydon L. Malveaux F. Evaluating the factors that relate to asthma severity in adolescents. Int. Arch. Allergy Immunol. 1997;113(1–3):87–95. doi: 10.1159/000237515. and. [DOI] [PubMed] [Google Scholar]
- 14.Donfack J, Tsalenko A, Hoki DM, Parry R, Solway J, Lester LA. Ober C. HLA-DRB1*01 alleles are associated with sensitization to cockroach allergens. J. Allergy Clin. Immunol. 2000;105(5):960–966. doi: 10.1067/mai.2000.106926. and. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Fang Z, Wang B, Chen C, Guang W, Jin Y, Yang J, Lewitzky S, Aelony A, Parker A, Meyer J, Weiss ST. Xu X. A genomewide search for quantitative-trait loci underlying asthma. Am. J. Hum. Genet. 2001;69(6):1271–1277. doi: 10.1086/324650. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouzigon E, Chaudru V, Carpentier AS, Dizier MH, Oryszczyn MP, Maccario J, Kauffmann F. Demenais F. Familial correlations and inter-relationships of four asthma-associated quantitative phenotypes in 320 French EGEA families ascertained through asthmatic probands. Eur. J. Hum. Genet. 2004;12(11):955–963. doi: 10.1038/sj.ejhg.5201241. and. [DOI] [PubMed] [Google Scholar]
- 17.Bouzigon E, Dizier MH, Krähenbühl C, Lemainque A, Annesi-Maesano I, Betard C, Bousquet J, Charpin D, Gormand F, Guilloud-Bataille M. Clustering patterns of LOD scores for asthma-related phenotypes revealed by a genome-wide screen in 295 French EGEA families. Hum. Mol. Genet. 2004;13(24):3103–3113. doi: 10.1093/hmg/ddh340. et al. [DOI] [PubMed] [Google Scholar]
- 18.Hunninghake GM, Lasky-Su J, Soto-Quirós ME, Avila L, Liang C, Lake SL, Hudson TJ, Spesny M, Fournier E. Sylvia JS. Sex-stratified linkage analysis identifies a female-specific locus for IgE to cockroach in Costa Ricans. Am. J. Respir. Crit. Care Med. 2008;177(8):830–836. doi: 10.1164/rccm.200711-1697OC. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung TF, Tang NL, Sung YM, Li CY, Ma SL, Lam CW. Wong GW. Genetic association study between mbl2 and asthma phenotypes in Chinese children. Pediatr. Allergy Immunol. 2006;17(7):501–507. doi: 10.1111/j.1399-3038.2006.00446.x. and. [DOI] [PubMed] [Google Scholar]
- 20.Pistiner M, Hunninghake GM, Soto-Quiros ME, Avila L, Murphy A, Lasky-Su J, Schuemann B, Klanderman BJ, Raby BA. Celedón JC. Polymorphisms in IL12A and cockroach allergy in children with asthma. Clin. Mol. Allergy. 2008;6:6. doi: 10.1186/1476-7961-6-6. and. doi: 10.1186/1476-7961-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bønnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, Linneberg A, Curtin JA, Warrington NM, Standl M. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat. Genet. 2013;45(8):902–906. doi: 10.1038/ng.2694. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Zuckerman B, Pearson C, Kaufman G, Chen C, Wang G, Niu T, Wise PH, Bauchner H. Xu X. Maternal cigarette smoking, metabolic gene polymorphism, infant birth weight. JAMA. 2002;287(2):195–202. doi: 10.1001/jama.287.2.195. and. [DOI] [PubMed] [Google Scholar]
- 23.Hong X, Tsai HJ, Liu X, Arguelles L, Kumar R, Wang G, Kuptsova-Clarkson N, Pearson C, Ortiz K. Bonzagni A. Does genetic regulation of IgE begin in utero? Evidence from T(H)1/T(H)2 gene polymorphisms and cord blood total IgE. J. Allergy Clin. Immunol. 2010;126(5):1059–1067. doi: 10.1016/j.jaci.2010.08.029. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R, Story RE, Pongracic JA, Hong X, Arguelles L, Wang G, Kuptsova-Clarkson N, Pearson C, Ortiz K, Bonzagni A. Maternal pre-pregnancy obesity and recurrent wheezing in early childhood. Pediatr. Allergy Immunol. Pulmonol. 2010;23(3):183–190. doi: 10.1089/ped.2010.0032. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar R, Ouyang F, Story RE, Pongracic JA, Hong X, Wang G, Pearson C, Ortiz K, Bauchner H. Wang X. Gestational diabetes, atopic dermatitis, and allergen sensitization in early childhood. J. Allergy Clin. Immunol. 2009;124(5) doi: 10.1016/j.jaci.2009.06.052. and ): 1031-8.e1–1034.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Arguelles L, Zhou Y, Wang G, Chen Q, Tsai HJ, Hong X, Liu R, Price HE, Pearson C. Longitudinal trajectory of vitamin D status from birth to early childhood in the development of food sensitization. Pediatr. Res. 2013;74(3):321–326. doi: 10.1038/pr.2013.110. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong X, Wang G, Liu X, Kumar R, Tsai HJ, Arguelles L, Hao K, Pearson C, Ortiz K, Bonzagni A. Gene polymorphisms, breast-feeding, and development of food sensitization in early childhood. J. Allergy Clin. Immunol. 2011;128(2):374–381. doi: 10.1016/j.jaci.2011.05.007. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, Divall S, Radovick S, Paige D, Ning Y, Chen Z, Ji Y, Hong X, Walker SO, Caruso D. Preterm birth, random plasma insulin levels at birth, in early childhood. JAMA. 2014;311(6):587–596. doi: 10.1001/jama.2014.1. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G. A genome wide admixture map for Latino populations. Am. J. Hum. Genet. 2007;80(6):1024–1036. doi: 10.1086/518313. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo ML, Mahdi F, Kolte D. Shariat-Madar Z. Upregulation of prolylcarboxypeptidase (PRCP) in lipopolysaccharide (LPS) treated endothelium promotes inflammation. J. Inflamm. (Lond) 2009;6:3. doi: 10.1186/1476-9255-6-3. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdez Y, Diehl GE, Vallance BA, Grassl GA, Guttman JA, Brown NF, Rosenberger CM, Littman DR, Gros P. Finlay BB. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cell Microbiol. 2008;10(8):1646–1661. doi: 10.1111/j.1462-5822.2008.01155.x. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Los H, Postmus PE. Boomsma DI. Asthma genetics and intermediate phenotypes: a review from twin studies. Twin Res. 2001;4(2):81–93. doi: 10.1375/1369052012191. and. [DOI] [PubMed] [Google Scholar]
- 33.Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA. Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin. Exp. Allergy. 2006;36(11):1382–1390. doi: 10.1111/j.1365-2222.2006.02512.x. and. [DOI] [PubMed] [Google Scholar]
- 34.Busse WW. Sedgwick JB. Eosinophils in asthma. Ann. Allergy. 1992;68(3):286–290. and. [PubMed] [Google Scholar]
- 35.Jin B, Shen H, Lin S, Li JL, Chen Z, Griffin JD. Wu L. The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J. Biol. Chem. 2010;285(19):14356–14365. doi: 10.1074/jbc.M109.078865. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benito Pescador D, Isidoro-García M, García-Solaesa V, Pascual de Pedro M, Sanz C, Hernández-Hernández L, Sánchez-López J, Lorente SF, Picado C, Valero A. Dávila I. Genetic association study in nasal polyposis. J. Investig. Allergol. Clin. Immunol. 2012;22(5):331–340. and. [PubMed] [Google Scholar]
- 37.Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat. Genet. 1998;19(2):119–120. doi: 10.1038/472. et al. [DOI] [PubMed] [Google Scholar]
- 38.Leung DY, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, Taylor PA, Boguniewicz M, Canniff J, Armstrong B. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-γ response. J. Allergy Clin. Immunol. 2011;127(4):965–973. doi: 10.1016/j.jaci.2011.02.010. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fagerås Böttcher M, Hmani-Aifa M, Lindström A, Jenmalm MC, Mai XM, Nilsson L, Zdolsek HA, Björkstén B, Söderkvist P. Vaarala O. A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J. Allergy Clin. Immunol. 2004;114(3):561–567. doi: 10.1016/j.jaci.2004.04.050. and. [DOI] [PubMed] [Google Scholar]
- 40.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 2013;45(8):907–911. doi: 10.1038/ng.2686. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Togias A, Fenton MJ, Gergen PJ, Rotrosen D. Fauci AS. Asthma in the inner city: the perspective of the National Institute of Allergy and Infectious Diseases. J. Allergy Clin. Immunol. 2010;125(3):540–544. doi: 10.1016/j.jaci.2010.01.040. and. [DOI] [PubMed] [Google Scholar]
- 42.Perzanowski MS, Chew GL, Divjan A, Jung KH, Ridder R, Tang D, Diaz D, Goldstein IF, Kinney PL, Rundle AG. Early-life cockroach allergen and polycyclic aromatic hydrocarbon exposures predict cockroach sensitization among inner-city children. J. Allergy Clin. Immunol. 2013;131(3):886–893. doi: 10.1016/j.jaci.2012.12.666. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C. A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin. Immunol. 2010;125(2):336–346. doi: 10.1016/j.jaci.2009.08.031. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieuwenhuizen N, Herbert DR, Brombacher F. Lopata AL. Differential requirements for interleukin (IL)-4 and IL-13 in protein contact dermatitis induced by Anisakis. Allergy. 2009;64(9):1309–1318. doi: 10.1111/j.1398-9995.2009.02002.x. and. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Cheng X, Xiang MX, Alanne-Kinnunen M, Wang JA, Chen H, He A, Sun X, Lin Y, Tang TT. IgE stimulates human and mouse arterial cell apoptosis and cytokine expression and promotes atherogenesis in Apoe-/- mice. J. Clin. Invest. 2011;121(9):3564–3577. doi: 10.1172/JCI46028. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Duan S, Lin X, Zhang W, Meng N, Zhao L, Zhao Y, Han D. Zhang L. SNPs in the FCER1A gene region show no association with allergic rhinitis in a Han Chinese population. PLoS ONE. 2010;5(12):e15792. doi: 10.1371/journal.pone.0015792. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potaczek DP, Okumura K. Nishiyama C. FCER1A genetic variability and serum IgE levels. Allergy. 2009;64(9):1383. doi: 10.1111/j.1398-9995.2009.02090.x. and. doi: 10.1111/j.1398-9995.2009.02090.x. Epub 2009 Jul 17. [DOI] [PubMed] [Google Scholar]
- 48.Bae JS, Kim SH, Ye YM, Yoon HJ, Suh CH, Nahm DH. Park HS. Significant association of FcepsilonRIalpha promoter polymorphisms with aspirin-intolerant chronic urticaria. J. Allergy Clin. Immunol. 2007;119(2):449–456. doi: 10.1016/j.jaci.2006.10.006. and. [DOI] [PubMed] [Google Scholar]
- 49.Kanada S, Nakano N, Potaczek DP, Maeda K, Shimokawa N, Niwa Y, Fukai T, Sanak M, Szczeklik A, Yagita H. Two different transcription factors discriminate the -315C > T polymorphism of the Fc epsilon RI alpha gene: binding of Sp1 to -315C and of a high mobility group-related molecule to -315T. J. Immunol. 2008;180(12):8204–8210. doi: 10.4049/jimmunol.180.12.8204. et al. [DOI] [PubMed] [Google Scholar]
- 50.Avrech OM, Samra Z, Lazarovich Z, Caspi E, Jacobovich A. Sompolinsky D. Efficacy of the placental barrier for immunoglobulins: correlations between maternal, paternal and fetal immunoglobulin levels. Int. Arch. Allergy Immunol. 1994;103(2):160–165. doi: 10.1159/000236622. and. [DOI] [PubMed] [Google Scholar]
- 51.Miller DL. Hiravonen T. Gitlin D synthesis of IgE by the human conceptus. J. Allergy Clin. Immunol. 1973;52(3):182–188. doi: 10.1016/0091-6749(73)90035-3. and. [DOI] [PubMed] [Google Scholar]
- 52.Depner M, Ege MJ, Genuneit J, Pekkanen J, Roponen M, Hirvonen MR, Dalphin JC, Kaulek V, Krauss-Etschmann S, Riedler J. Atopic sensitization in the first year of life. J. Allergy Clin. Immunol. 2013;131(3):781–788. doi: 10.1016/j.jaci.2012.11.048. et al. [DOI] [PubMed] [Google Scholar]
- 53.Kamemura N, Tada H, Shimojo N, Morita Y, Kohno Y, Ichioka T, Suzuki K, Kubota K, Hiyoshi M. Kido H. Intrauterine sensitization of allergen-specific IgE analyzed by a highly sensitive new allergen microarray. J. Allergy Clin. Immunol. 2012;130(1):113–121.e2. doi: 10.1016/j.jaci.2012.02.023. and. [DOI] [PubMed] [Google Scholar]
- 54.Szépfalusi Z, Loibichler C, Hänel-Dekan S, Dehlink E, Gerstmayr M, Pichler J, Eiwegger T, Horvat R. Urbanek R. Most of diaplacentally transferred allergen is retained in the placenta. Clin. Exp. Allergy. 2006;36(9):1130–1137. doi: 10.1111/j.1365-2222.2006.02559.x. and. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of selected genes, SNPs, location and predicted function.
Table S2. Clinical information.
Table S3. Children with one or multiple (2–6) allergens.


