Abstract
Reusable, publicly available data is a pillar of open science. The Cancer Imaging Archive (TCIA) is an open image archive service supporting cancer research. TCIA collects, de-identifies, curates and manages rich collections of oncology image data. Image data sets have been contributed by 28 institutions and additional image collections are underway. Since June of 2011, more than 2,000 users have registered to search and access data from this freely available resource. TCIA encourages and supports cancer-related open science communities by hosting and managing the image archive, providing project wiki space and searchable metadata repositories. The success of TCIA is measured by the number of active research projects it enables (>40) and the number of scientific publications and presentations that are produced using data from TCIA collections (39).
I. INTRODUCTION
The volume of scientific data doubles each year with single experiments now generating petabytes of data annually [1]. Data-driven research and decision-making, though broadly recognized as critical, suffer a gap between potential and realization due, in part, to the challenge of effectively managing the exploding volume of data [2, 3].
NIH research funding for genomics and medical imaging, two Big Data disciplines, has shifted to a paradigm supporting large public databases and encouraging funded researchers to publicly share their data in hopes of using open-data to stimulate open-science collaboration. Genomics has spawned numerous knowledge-sharing databases (model organisms, nucleotide, protein, structure, taxonomy) [4, 5]. Imaging projects such as the Bioinformatics Research Network (BIRN) [6] and recently the Human Connectome Project [7] are accumulating vast amounts of image data in order to accelerate our understanding of brain structure and function and have firmly established medical imaging in the realm of Big Data based science. In cancer imaging, the National Cancer Institute (NCI) has funded The Cancer Imaging Archive (TCIA), described here, as a public repository of cancer images and related clinical data for the express purpose of enabling open science research [8].
II. OPEN SCIENCE AND OPEN DATA
The concept of open science is perhaps most generally assumed to mean the free sharing of tools, data and results among scientists; a process that began with the Renaissance. In more recent literature the term open science has become somewhat nebulous and has been used to encompass a wide variety of concepts [9, 10] including:
Using Open Source software in scientific research;
Making data and tools available to the public to enhance basic science education;
Making scientific results available in Open Access journals;
Finding innovative solutions to scientific problems via crowd sourcing;
Using Open Source software to capture and manage Open Data to encourage and support research and education;
Creating Research Communities around an Open Data resource.
TCIA utilizes open source software to create and support research communities around an open access information resource. TCIA data was originally collected for clinical diagnosis or a specific research project but is now being offered to the research community to enable new lines of research.
III. THE CANCER IMAGING ARCHIVE (TCIA)
A. TCIA’s Multi-component Architecture
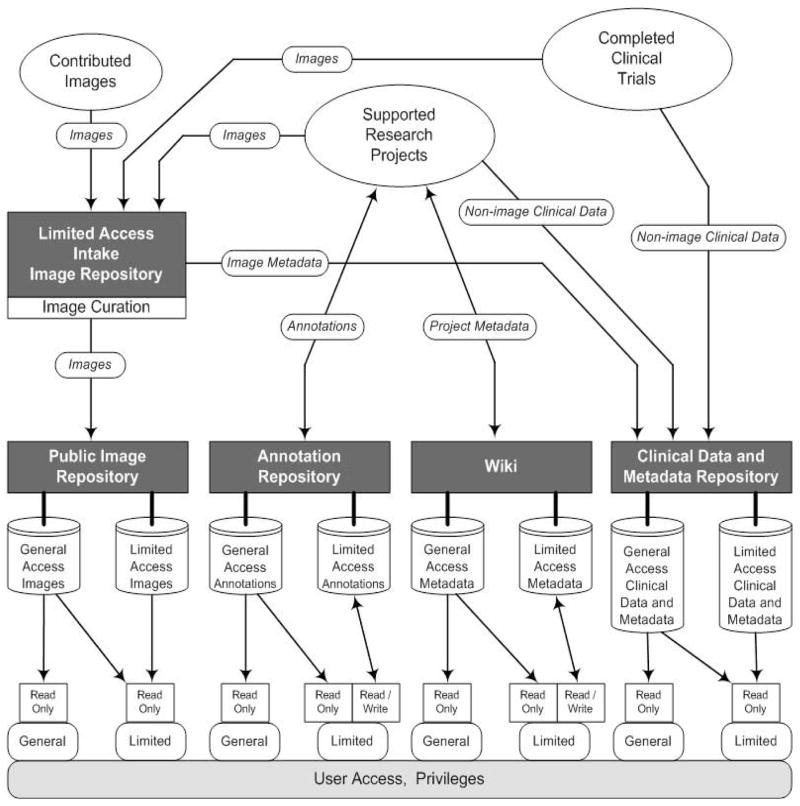
Figure 1 illustrates the various ways by which images and non-image data are added to TCIA, stored in TCIA, and harvested from TCIA. Images may be provided as completed collections from ongoing supported research projects or from completed clinical trials. Inbound images, de-identified at their contributing source, are deposited with an intake server until they have been curated, after which they are placed with TCIA’s public server, either among general-access (fully public) collections or among limited-access collections, with placement determined by NCI. While most collections are publicly available, about 5% are limited-access for groups of investigators needing to share images but not quite ready to release their images to the public. Image metadata may be extracted from the images and deposited with clinical-trial non-image data, in the TCIA Clinical Data and Metadata Repository. Some image collections arrive with annotation and markup objects either in DICOM format [11] or study specific format [12]. Ongoing research projects may add annotations, created by Annotation and Image Markup (AIM) compliant applications [13], to the TCIA Annotation Repository [14] or project metadata to the TCIA wiki. All users have read-access to the Public Image Repository, the Annotation Repository, wiki, and the Clinical Data and Metadata Repository. Users with project specific privileges, including those connected with supported research projects, may harvest images and data from the limited-access portions of the repositories and wiki and contribute (write privileges) to the Annotation Repository and the wiki.
Figure 1.
TCIA collects multiple types of de-identified data documenting supported research projects and completed clinical trials, and makes these data available to enable ongoing research.
B. Contributed Images
TCIA is a managed archive of contributed radiology images of cancer in DICOM format. TCIA supports the de-identification, submission and curation of image data so that they can be made publicly available in a HIPAA compliant form while maximizing their scientific value. Image data are de-identified with open-source software [15] configured and provided to the contributor for the transmission of images to TCIA’s intake server. Arriving images are visually inspected for image corruption and visible protected health information (PHI), while image headers are automatically scanned for potential PHI. Preparation of de-identification scripts tailored to individual image collections and in-coming image quality control require significant effort and attention to detail. These efforts are essential, however, to high-quality curation [3], the activity of organizing biological information such that they are easily digestible by both humans and their computers. Upon such effort rests the efficient proof or disproof of hypotheses put forward by image-consuming researchers in hopes of biological discovery.
TCIA groups images into collections. A collection typically includes studies (groups of images and associated study data) from several human subjects. In some collections, there may be only one study per subject. In other collections, subjects may have been followed over time, in which case there will be multiple studies per subject. The subjects typically have in common a particular disease and/or particular anatomical site (e.g., lung, brain). Collections are labeled so that a TCIA user can easily identify the related research project and cancer type (e.g., TCGA-GBM) or imaging modality and anatomy imaged (e.g., Prostate-MRI).
C. Image Retrieval
Images passing quality control are posted to a public server from which anyone with a TCIA account (free) may view and download images. The primary image management application is the open-source National Biomedical Imaging Archive (NBIA) [16]. NBIA presents the user with over ninety DICOM tags upon which to refine queries on the image data. Once an investigator has selected desired images, the images may be downloaded immediately or the investigator may save links to the images as a shared list; a list of image series stored in the NBIA database. The investigator may recall a shared list at any future time and download the associated images. The investigator may also inform collaborators who could then log into NBIA and access the specified shared list in order to download the same image set, thus enabling the collaboration with a simple mechanism for sharing images.
How does a researcher know what data are relevant to his research and how does one search for these data? Typically, one would be directed to the TCIA home page to find “For Researchers,” specific links for: gaining access to the images, image collections, related publications, and research projects. The how of searching is well described in the TCIA User Guide, available from the main system menu.
A public TCIA wiki space provides detailed information for most collections. Multi-site collections include links to the project in which the providers are participating. As users enquire about certain kinds of images, the answers are captured on a public-faced wiki page. The wiki gives data contributors a platform to describe the scope and intent of their image collection and to provide metadata and/or ways for users to contact them. The wiki supports research groups by summarizing the work of participants and posting conference abstracts and publications. The public space also provides access to user guides.
The TCIA Support Center services users via email and direct links from the TCIA web site. All user issues are documented and tracked using an open-source trouble-ticket program for problems in these areas: (1) normal user questions concerning account creation and credentialing, (2) use of the NBIA application, (3) direction to documentation on the collections.
IV. TCIA DATA COLLECTIONS
An NCI Cancer Imaging Program advisory group prioritizes new TCIA image collection candidates based on the extent to which the data comply with the following objectives:
NCI grant/contract award data sharing requirements;
Analysis of imaging features to be used as biomarkers;
Creation of correlative signatures for multi-platform biomarkers;
Creation of algorithms for detection of cancer;
Testing and validating quantitative analysis techniques;
Unique characteristics for clinical training.
TCIA image collections represent cancers affecting a variety of organs (brain, breast, head/neck, lung, colon, prostate, kidney) from a variety of imaging modalities (computed tomography, magnetic resonance, mammography, X-ray, positron-emission tomography, radiation treatment planning). There are also a few phantom collections available for algorithm and measurement process verification. Image collections are typically from completed studies, as TCIA does not manage ongoing clinical trials. Table 1 summarizes the number of images (e.g. single CT axial slice) in the TCIA image collections by anatomy and imaging modality. It includes over 20 million chest CT images belonging to the limited-access National Lung Screening Trial (NLST) [17, 18] collection.
Table 1.
TCIA image collections by anatomic region (number of images for each imaging modality and the types of cancer imaged).
| Anatomic Region | DX | CT | MR | PT | Cancer Type(s) |
|---|---|---|---|---|---|
| Brain | 6.482 | 959,401 | Glioma, Glioblastoma Multiforme | ||
| Breast | 6.980 | 257,062 | 5,492 | Breast Invasive Carcinoma | |
| Lung/Chest | 569 | 21,424,099 | 123,744 | Adenocarcinoma, Squamous cell carcinoma, Bronchioloalveolar carcinoma, Large-cell carcinoma, non-small-cell carcinoma, small cell carcinoma, carcinoid | |
| Colon | 941.771 | Adenocarcenoma | |||
| Head/Neck | 83,915 | 118.133 | Squamous cell carcinoma | ||
| Kidney | 50.852 | 25.630 | Renal Clear cell carcinoma | ||
| Prostate | 13,534 | 81.132 | 27,870 | Adenocarcinoma |
Imaging Modalities: DX -digital x-ray, CT - Computed Tomography, MR -Magnetic Resonance Imaging, PT - Positron Emission Tomography
Most collections have associated clinical and/or image metadata, which can be accessed via TCIA wiki pages. The NLST collection utilizes a Query Tool that allows an investigator to pose user-created queries against the non-image data collected during the trial and trial results (e.g., demographics, image-screening results, smoking history, medical history, work history, cancer diagnosis and tracking) and/or the imaging data extracted from the DICOM header (e.g., study year, kVp, mAs, pitch, series description, series instance UID). Once satisfied with the results of a query, the results can be saved to a text file, and/or a shared list, or the images can be downloaded from TCIA. In addition, the queries may be saved for later recall or for finer tuning.
While the Query Tool was developed with NLST data, it is now being deployed for use with other research groups with TCIA images and associated non-image data, thus allowing researchers to query non-image data and, among other things, choose images for downloading by invoking the TCIA image-download function from the Query Tool.
V. TCIA ENABLED RESEARCH
As an open-access archive linked to extensive meta-data, cross-disciplinary researchers can use TCIA to test biomedical hypotheses and develop analytic techniques. TCIA provides the international research community with free access to imaging data sets that have in the past been prohibitively costly or impossible to generate. Cancer researchers can use these data to test new hypotheses and develop new analysis techniques to advance the scientific understanding of cancer. Engineers and software developers can build new analysis tools and techniques using this data as test material for developing and validating algorithms. Educators can use it as a teaching tool for introducing students to medical imaging technology and cancer phenotypes. In addition, a number of active research communities have developed around specific TCIA collections. Table 2 lists the currently active communities and the associated TCIA collections.
Table 2.
Collaborative research groups that are enabled by the TCIA resource.
| Community | Collaborative Projects | Active Researchers | TCIA Collections Utilized |
|---|---|---|---|
| TCGA Glioma Phenotype Group | 11 | >20 | TCGA-GBM TCGA-LGG |
| TCGA Breast Phenotype Group | 4 | >12 | TCGA-BRCA |
| TCGA Renal Phenotype Group | 1 | >13 | TCGA-KIRC |
| Quantitative Imaging Network | >16 | >190 | QIN Breast QIN Phantom QIN Lung QIN Prostate |
| National Lung Screening Trial Related Groups | 8 | 25 | NLST |
TCIA is actively developing collections of image data from cases where genomic, clinical and histopathology data are available on The Cancer Genome Atlas [5] website, providing a unique resource for researchers in the relatively new field of imaging phenotype to genotype analysis. TCGA researchers are collecting tissue samples (brain, breast, gastrointestinal, head and neck, hematologic, skin, thoracic, and urologic) and are mapping the genetic changes in 20 cancers. The TCGA Data Portal provides a platform for researchers to search, download, and analyze data sets generated by TCGA while associated radiology images are available through TCIA. To date 16 active research projects are on going based on the data available from the TCGA Data Portal and TCIA. TCIA enabled researchers are advancing the use of image and genomics data in the fight against breast, brain, lung and renal cancers [19–21].
The Quantitative Imaging Network (QIN)[16] has contributed brain, breast, head-neck, and prostate cancer images. More than 16 active QIN research projects utilize TCIA data and many of these projects maintain limited access collections on TCIA to support the development and validation of quantitative imaging-derived biomarkers.
The National Lung Screening Trial was a decade-long multi-center trial to determine whether screening for lung cancer with low-dose helical computed tomography (CT) reduces mortality from lung cancer in high-risk individuals relative to screening with chest radiography. Approximately 54,000 participants were enrolled between August 2002 and April 2004. The primary outcome of the trial was the finding that lung cancer mortality was reduced by 20% in the CT arm of the trial [18]. This extensive data set is now available as a limited access collection with access permission granted by NCI [22]. Eight research groups are currently utilizing this resource.
A key metric of the value of TCIA is the dissemination of scientific research results that rely on the TCIA resource. Since the Cancer Imaging Archive went on-line in 2011, TCIA enabled research initiatives have produced 6 peer reviewed publications (with more in review) and 33 scientific presentations [23] with more in preparation as the work is ongoing and new projects and collections are continually being added.
VI. CONCLUSIONS
The Cancer Imaging Archive is an investment in Open Science by the National Cancer Institute and allows Open Access to cancer images, trial data, and mechanisms for collaborative research. TCIA is not primarily technology focused but rather a service, designed to give access to image collections to the broadest possible research community. Open Science initiatives such as TCIA are producing substantial scientific impact. Open science communities have formed around TCIA data collections and are gaining traction as evidenced by a steadily increasing output of abstracts, presentations and publications.
Footnotes
Research supported by the National Cancer Institute under Contract NO. HHSN261200800001E, and Washington University subcontract 10XS220.
Contributor Information
Fred W. Prior, Email: priorf@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA (phone: 314-747-0331; fax: 314-362-6971;).
Ken Clark, Email: clarkk@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
Paul Commean, Email: commeanp@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
John Freymann, Email: freymannj@mail.nih.gov, SAIC-Frederick, Inc., Frederick, MD 21702 USA.
Carl Jaffe, Email: carljaffe@gmail.com, Department of Radiology, Boston University School of Medicine, Boston, MA USA.
Justin Kirby, Email: kirbyju@mail.nih.gov, SAIC-Frederick, Inc., Frederick, MD 21702 USA.
Stephen Moore, Email: moores@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
Kirk Smith, Email: smithki@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
Lawrence Tarbox, Email: tarboxl@mir.wustl.edu, Lawrence Tarbox is with the Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
Bruce Vendt, Email: vendtb@mir.wustl.edu, Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO 63110 USA.
Guillermo Marquez, Email: marquezg@mail.nih.gov, National Cancer Institute, Bethesda, MD 20892 USA.
References
- 1.Szalay A, Gray J. 2020 Computing: Science in an exponential world. Nature. 2006;440:413–414. doi: 10.1038/440413a. [DOI] [PubMed] [Google Scholar]
- 2.Lynch C. Big data: How do your data grow? Nature. 2008;455:28–29. doi: 10.1038/455028a. [DOI] [PubMed] [Google Scholar]
- 3.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birney E, Bateman A, Clamp ME, Hubbard TJ. Mining the draft human genome. Nature. 2001;409:827–828. doi: 10.1038/35057004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hampton T. Cancer Genome Atlas. JAMA: The Journal of the American Medical Association. 2006;296:1958–1958. [Google Scholar]
- 6.Grethe JS, Baru C, Gupta A, James M, Ludaescher B, Martone ME, et al. Biomedical informatics research network: building a national collaboratory to hasten the derivation of new understanding and treatment of disease. Studies in health technology and informatics. 2005;112:100–110. [PubMed] [Google Scholar]
- 7.Van Essen D, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, et al. The human connectome project: a data acquisition perspective. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffe CC. Imaging and Genomics: Is There a Synergy? Radiology. 2012 Aug;264:329–331. doi: 10.1148/radiol.12120871. [DOI] [PubMed] [Google Scholar]
- 9.Woelfle M, Olliaro P, Todd MH. Open science is a research accelerator. Nat Chem. 2011;3:745–748. doi: 10.1038/nchem.1149. [DOI] [PubMed] [Google Scholar]
- 10.Molloy JC. The open knowledge foundation: open data means better science. PLoS Biology. 2011;9:e1001195. doi: 10.1371/journal.pbio.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clunie DA. DICOM structured reporting and cancer clinical trials results. Cancer informatics. 2007;4:33. doi: 10.4137/cin.s37032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNitt-Gray MF, Armato SG, III, Meyer CR, Reeves AP, McLennan G, Pais RC, et al. The Lung Image Database Consortium (LIDC) data collection process for nodule detection and annotation. Academic Radiology. 2007;14:1464. doi: 10.1016/j.acra.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL. The cabig™ annotation and image markup project. Journal of Digital Imaging. 2010;23:217–225. doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Pan T, Sharma A, Saltz J. Managing and querying image annotation and markup in XML,” in. Proceedings of SPIE. 2010:762805. doi: 10.1117/12.843606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freymann J, Kirby J, Perry J, Clunie D, Jaffe C. Image Data Sharing for Biomedical Research - Meeting HIPAA Requirements for De-identification. Journal of Digital Imaging. 2011:1–11. doi: 10.1007/s10278-011-9422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke LP, Croft BS, Nordstrom R, Zhang H, Kelloff G, Tatum J. Quantitative imaging for evaluation of response to cancer therapy. Translational Oncology. 2009;2:195. doi: 10.1593/tlo.09217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark K, Gierada D, Marquez G, Moore S, Maffitt D, Moulton J, et al. Collecting 48,00 CT Exams for the Lung Screening Study of the National Lung Screening Trial. Journal of Digital Imaging. 2009 Dec;22:667–680. doi: 10.1007/s10278-008-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aberle D, Adams A, Berg C, Black W, Clapp J, Fagerstrom R, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365:395. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R, Poisson L, Narang J, Gutman D, Scarpace L, Hwang SN, et al. Genomic Mapping and Survival Prediction in Glioblastoma: Molecular Subclassification Strengthened by Hemodynamic Imaging Biomarkers. Radiology. 2012 doi: 10.1148/radiol.12120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinn PO, Sathyan P, Mahajan B, Bruyere J, Hegi M, Majumder S, et al. A Novel Volume-Age-KPS (VAK) Glioblastoma Classification Identifies a Prognostic Cognate microRNA-Gene Signature. PloS one. 2012;7:e41522. doi: 10.1371/journal.pone.0041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinn PO, Majadan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PloS one. 2011;6:e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NCI. CDAS Cancer Data Access System. 2013 Feb 3; Available: https://biometry.nci.nih.gov/cdas/
- 23.TCIA. For Researchers; Related Publications. 2013 Jan 18; Available: http://cancerimagingarchive.net/publications.html.