Oncogenic mutations of the H3F3A gene, which encodes histone variant H3.3, are present in the majority of pediatric brainstem gliomas1,2. The most common H3F3A mutation causes a substitution of lysine 27 with methionine (i.e., K27M), abolishing a critical site of regulatory post-translational methylation. We, and others, have demonstrated that these oncogenic mutations have a dominant effect, sequestering polycomb repressive complex 2 (PRC2) and reducing cellular H3K27 methylation3–6. As such, tumor-derived histone gene mutations are thought to drive tumorigenesis by causing reduced histone K27 methylation and thereby altering gene expression in cells of the developing pons. We hypothesized that pharmacologic reversal of brainstem glioma H3K27 demethylation could serve as a therapeutic strategy for this uniformly lethal malignancy7.
To test this hypothesis, we studied two K27M mutant brainstem glioma cell lines (SF77618 and SF86289), four glioma cell lines with wild-type H3.3 (SF9012, SF9402, SF9427, and GBM43), a glioma cell line expressing G34V-mutant H3.3 (KNS42), and isogenic human astrocytes (HAs) with and without transgene expression of K27M H3F3A3 (all lines derived from human; see Supplementary Table 1 for relevant clinical information). Immunoblot and immunocytochemistry analysis showed that K27M glioma cells and K27M HAs (hereafter “K27M-expressing cells”) have less di- and trimethylated H3K27 (K27me2 and K27me3) than glioma cells with wild type and G34V H3.3 (Fig. 1a and Supplementary Figs. 1 and 2).
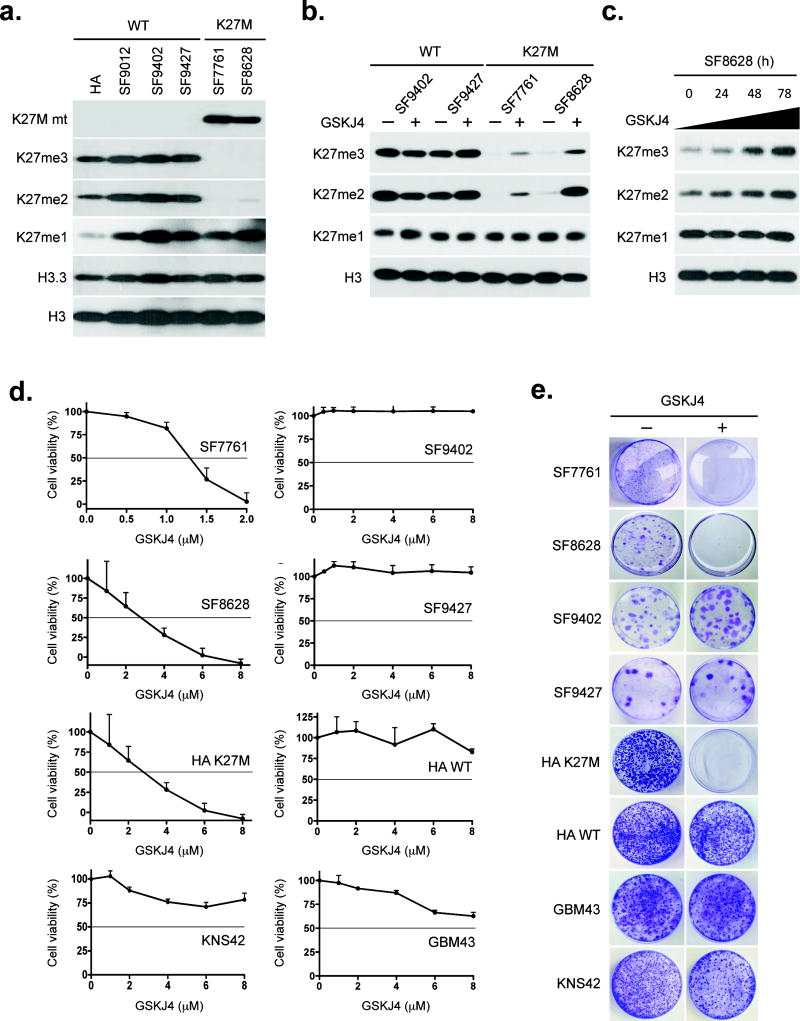
Figure 1. GSKJ4 increases K27 methylation, inhibits K27M glioma cell growth and prevents K27M colony formation.
(a) Immunoblot results showing K27M mutant (K27M mt), K27me3, K27me2, K27me1, histone H3.3 and total histone H3 expression in K27M brainstem glioma cell lines SF7761 and SF8628, wild-type (WT) H3.3 glioma cell lines SF9012, SF9402 and SF9427, and human astrocytes (HA). (b) Immunoblot results for K27me3, K27me2, K27me1, and total histone H3 in glioma cells either untreated or treated with 6 μM GSKJ4 for 48 h. (c) Temporal effect of GSKJ4 on H3K27 methylation in K27M SF8628 cells. (d) Proliferation response of glioma cells to increasing concentrations of GSKJ4. Values shown are the average (mean ± SEM) from quadruplicate samples for each incubation condition. (e) GSKJ4 colony-forming effect on cells with or without K27M.
H3K27 is methylated by histone-lysine N-methyltransferase EZH2, a component of PRC2, and is demethylated by the KDM6-subfamily K27 demethylases JMJD3 and UTX10–12. Treatment of all cell sources with GSKJ413, an ethyl ester derivative of the H3K27 demethylase inhibitor GSKJ1, led to higher K27me2 and K27me3 in K27M-expressing cells (Fig. 1b and Supplementary Figs. 2 and 3), with increasing length of treatment leading to increasing K27me2 and K27me3 (Fig. 1c).
We analyzed the effects of GSKJ4 treatment in vitro on tumor cell proliferation, apoptosis and colony formation. GSKJ4 treatment of K27M-expressing cells revealed a dose-dependent inhibition of cellular viability, with 50% growth inhibition reached at concentrations of 1.3–3.0 μM (Fig.1d). In contrast, GSKJ4 concentrations as high as 8 μM did not achieve 50% growth inhibition for any of tumor cells expressing wild-type and G34V H3.3. GSKJ4 treatment of HAs revealed that HAs expressing K27M were more sensitive to GSKJ4 than HAs without the mutation (Fig. 1d). Analysis of cell cycle distributions revealed S-phase as being most diminished cell cycle stage in tumor cells expressing K27M (Supplementary Fig. 4). GSKJ4 also led to more apoptosis of K27M cells, as indicated by annexin V staining (Supplementary Fig. 5) and by cell counts of GSKJ4-treated samples (Supplementary Fig. 6). Finally, GSKJ4 completely inhibited the clonal growth of all K27M-expressing cells but had no affect on the clonal growth of the cells expressing wild-type and G34V H3.3 (Fig. 1e). Analysis of tumor cell proliferation after inhibition of EZH2 by GSK12614 revealed little effect of the methyltransferase inhibitor on any cell line tested (Supplementary Fig. 7a) despite evidence of less K27me3 and K27me2 in treated cells (Supplementary Fig. 7b).
To confirm that the effect of GSKJ4 on tumor cell growth was due to activity against JMJD3, we studied the effects of depleting JMJD3 and UTX on tumor cell growth and the response of these depleted cells to GSKJ4. siRNA-mediated depletion of JMJD3 inhibited the growth of K27M cells. GSKJ4 had no significant effect on proliferation of JMJD3 depleted K27M cells (Supplementary Fig. 8). siRNA depletion of UTX had little effect on the growth of K27M cells, and pre-treatment of K27M cells with UTX (KDM6A) siRNA did not prevent GSKJ4 from significantly inhibiting K27M cell growth (P < 0.03 for K27M cells versus wild-type cells, two-tailed unpaired t-test; Supplementary Fig. 9). These results support the anti-tumor activity of GSKJ4 as involving inhibition of JMJD3, but do not exclude the possibility of other histone demethylase activities being affected (see http://www.thesgc.org/chemical-probes/GSKJ1).
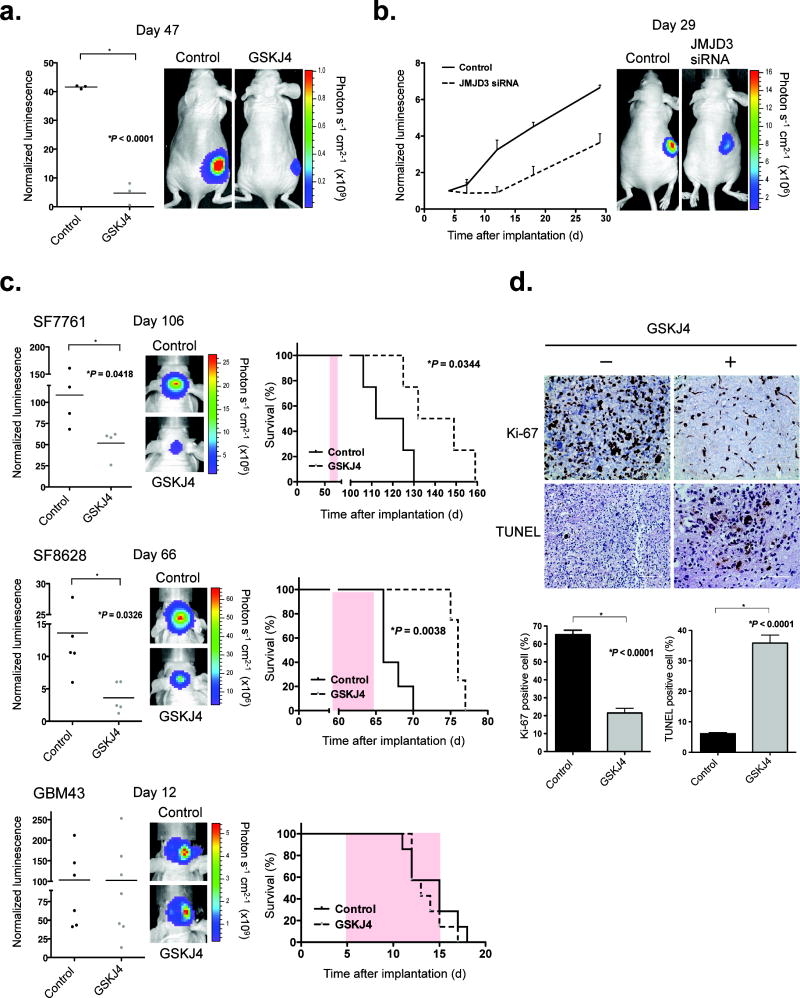
To determine whether GSKJ4 is active against K27M brainstem gliomas in vivo, we administered GSKJ4 by intraperitoneal injection, at 100 mg per kg per day for 10 consecutive days, to athymic mice harboring subcutaneous SF8628 K27M xenografts. The drug showed significant growth-inhibitory activity against SF8628 subcutaneous tumors (Fig. 2a). The same cells, when transfected with JMJD3 (KDM6B) siRNA and then injected subcutaneously, showed substantial growth delay relative to the growth of SF8628 cells transfected with scrambled siRNA (Fig. 2b).
Figure 2. GSKJ4 treatment shows K27M antitumor activity in vivo.
(a) Mice with SF8628 subcutaneous tumor were treated with either vehicle (DMSO) or GSKJ4 (100 mg per kg per day) daily for 10 d. Left, tumor bioluminescence in mice at day 47 d after tumor cell injection. Horizontal bars indicate the mean from 3 mice for each treatment group. A two-tailed unpaired t-test was used to compare vehicle and GSKJ4 treatment. Right, tumor bioluminescence overlay images showing relative bioluminescence intensities from representative vehicle- vs. GSKJ4-treated mice. (b) Left, growth plots for subcutaneous tumors, established by injection of SF8628 cells transfected with scrambled (Control) or JMJD3 (KDM6B) siRNA. Tumor bioluminescence values were normalized against bioluminescence values obtained 3 d after tumor cell injection. Values are mean ± s.e.m. from 3 mice at each time after implantation. Right, corresponding tumor bioluminescence intensity overlay images for representative scrambled and JMJD3 siRNA–treated mice 29 d after tumor cell implantation. (c) Left, bioluminescence distributions of SF7761 (top; 106 d after tumor cell implantation), SF8628 (66 d after tumor cell implantation) and GBM43 (12 d after tumor cell implantation) brainstem xenografts, following 10 d (SF7761 and SF8628) or 7 d (GBM43) of treatment with GSKJ4 or vehicle. Horizontal bars indicate the mean from 4 mice for each treatment group in SF7761, 5 for each group in SF8628, and 6 for Control and 7 for GSKJ4 treatment in GBM43. A two-tailed unpaired t-test was used to compare vehicle and GSKJ4 treatment. Center, corresponding tumor bioluminescence intensity overlay images for representative vehicle- and GSKJ4-treated mice. Right, survival plots for each experiment. Statistical analysis was performed using a log-rank test. Pink shaded areas indicate the duration of treatment. (d) Representative Ki-67 and TUNEL staining for vehicle- vs. GSKJ4-treated SF8628 tumors, from mice euthanized at the end of treatment. Bottom, average percentage of positive cells in four high-powered fields for each tumor. Error bars, s.e.m. Two mice were analyzed in each treatment. A two-tailed unpaired t-test was used to compare vehicle and GSKJ4 treatment.
We next tested for GSKJ4 activity against orthotopic K27M brainstem glioma xenografts (SF7761 and SF8628)8,15. Administration of GSKJ4 for 10 consecutive days significantly reduced the growth of K27M tumors engrafted in mouse brainstem and significantly extended animal survival (Fig. 2c). Analysis of K27M tumors obtained from mice at the end of therapy showed significantly reduced Ki-67 staining—a measure of cell proliferation—as well as significantly increased TUNEL positivity—a measure of apoptotic activity—after GSKJ4 treatment (Fig. 2d). In addition, GSKJ4 treatment significantly increased tumor cell K27me3 positivity (P = 0.0016, two-tailed unpaired t-test; Supplementary Fig. 10). In contrast to K27M tumors, xenografts established from GBM43 cells expressing wild-type H3.3 showed no response to GSKJ4 treatment (Fig. 2c).
To address GSKJ4 brain access, we administered GSKJ4 to three non-tumor-bearing mice that were euthanized 3 h after the third GSKJ4 treatment. Their brains were immediately resected, and each brainstem was dissected from the surrounding brain. HPLC analysis of tissue extracts revealed detectable GSKJ1, the hydrolysis product and activated derivative of GSKJ4, in all three brainstem samples (Supplementary Table 2). This finding indicated that the drug entered the brain: specifically, the site of brainstem tumor development.
Drawing on known associations between K27 methylation and gene regulation, we applied expression array and K27me3 chromatin immunoprecipitation–sequencing (ChIP-seq) analysis to SF8628 K27M cells. Results for sequences showing the most significant inverse correlation between GSKJ4-associated gene expression changes and K27me3 sequence association are indicated in Supplementary Fig. 11 and Supplementary Table 3 (see also Gene Expression Omnibus data set GSE8497).
The recent discovery of histone K27M mutation in pediatric gliomas and the identification of the altered epigenetic program resulting from these mutations has prompted the investigation of small-molecule inhibitors targeting H3 demethylases as new cancer therapies. Here we have demonstrated that pharmacologic modulation of histone K27 methylation, by inhibiting H3 demethylation, is an approach warranting further investigation for treating K27M pediatric gliomas.
Methods
Cell sources and propagation
Primary pediatric human glioma cells, designated with “SF”, were obtained from surgical biopsy of tumor from patients admitted to UCSF medical center, and in accord with an institutionally approved protocol by the UCSF Committee for Human Research (IRB# 10-01318). Subjects, or their legal guardians if the subject was a minor, provided informed consent. Human astrocytes (HAs) were obtained from Z. Zhang (Mayo Clinic). These cells were modified to express wild-type (WT) H3F3A or K27M H3F3A transgene as previously described3. Establishment of glioma cell cultures, from surgical specimens, and tumor cell modification for expression of firefly luciferase, for in vivo bioluminescence imaging, has been described8,9,15. GBM43 is maintained as a serially-passaged subcutaneous xenograft16, with xenograft tissue used to establish explant cultures for in vitro experiments that are described below. Cell line KNS42, with H3F3A G34V mutation (substitution of glycine 34 with valine)17, was obtained from the Japanese Collection of Bioresources and was established from a 16 year old male.
Nuclear protein extraction and immunoblotting
Nuclear proteins were extracted from proliferating cells in acid extraction buffer (0.2N HCL), following the extraction of cytoplasmic proteins with TNE buffer: 150 mM NaCl, 10 mM Tris pH 7.4, 1% Triton X-100, 5 mM EDTA, 1% NP40, 1 μM DTT, and proteinase (Roche) plus phosphatase (Sigma) inhibitor cocktails. Protein extracts were resolved by SDS-PAGE and transferred to poly(vinylidene difluoride) (PVDF) membranes. After probing with primary antibodies, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody, and signals visualized by ECL (GE Healthcare). Antibodies specific for total histone H3 (96C10, 1:1,000), H3K27me3 (C36B11, 1:1,000), H3K27me2 (D18C8, 1:1,000), H3K27me1 (#7693 1:1,000), and EZH2 (D2C9, 1:1,000), were obtained from Cell Signaling Technologies. Histone H3.3 antibody (ab97968, 1:1,000) was obtained from Abcam, and histone K27M mutant antibody (ABE419, 1:1,000) was from EMD Millipore.
Immunocytochemistry
Tumor cells were fixed on coverslips in 4% paraformaldehyde (PFA), rinsed in PBS, and blocked in PBS containing 0.3% Triton X-100 and 5% FBS for 1 h at room temperature. Coverslips were incubated with H3K27me3 antibody (C36B11, Cell Signaling, 1:800) or JMJD3 antibody (ab154985, Abcam, 1:800) in PBS containing 0.3% Triton X-100 and 5% FBS overnight at 4 °C. Overnight incubations were followed by incubations using goat-anti-rabbit Alexa568 secondary antibody (Invitrogen, 1:800) in PBS containing 5% FBS, for 50 min at room temperature and in the absence of light. Coverslips were rinsed in PBS four times and the nuclei were stained with DAPI in PBS at room temperature. Coverslips were then subjected to successive rinses in PBS and sterile water and then mounted on glass slides using Vectashield (Vector Laboratories) and analyzed with a Zeiss LSM 510 NLO Meta microscope at 1,000 × magnification.
Cell viability, cell proliferation, and clonogenic assays
For determination of cell viability effects of GSKJ4, tumor cells were seeded in 96-well plates, at 5,000 cells per well, and cultured in the presence of 0–8 μM GSKJ4 (R&D Systems) or GSK126 (Xcess Biosciences) for 72 h, with quadruplicate samples for each incubation condition. Relative numbers of viable cells were determined using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega). IC50 values were calculated by nonlinear least-squares curve-fitting. Inhibitor proliferation effects were determined by cell counting from samples obtained at 0, 1, 2, 3, and 6 d in the presence of vehicle (DMSO) or with IC50 concentrations of GSKJ4 (SF7761, SF8628, HA KM) or with 3 μM GSKJ4 (SF9402, SF9427, HA WT, GBM43, KNS42). Clonogenic assays were performed by plating at 300–1,000 cells/ml in 60-mm plates, with cells treated with vehicle, with IC50 concentrations of GSKJ4 (SF7761, SF8628, HA KM), or with 3 μM GSKJ4 (SF9402, SF9427, HA WT, GBM43, KNS42) for 72 h, beginning 3 d after plating. Cells were incubated for 3 weeks at 37 °C and then stained with 0.05% crystal violet.
Xenograft studies
Six-week-old female athymic mice (nu/nu genotype, BALB/c background) were purchased from Simonsen Laboratories and housed under aseptic conditions. All protocols, described below, were approved by the UCSF Institutional Animal Care and Use Committee.
Tumor cells were implanted into the pontine tegmentum of athymic mice as previously described8,15. Briefly, mice were anesthetized by intraperitoneal (ip) injection of a mixture containing ketamine (100 mg per kg) and xylazine (10 mg per kg) in 0.9% saline. A 1-cm sagittal incision was made along the scalp, and the skull suture lines were exposed. A small hole was created by puncture with a 25-gauge needle at 1.5 mm to the right of the bregma and posterior to the lambdoid suture. With use of a sterile Hamilton syringe (Stoelting), 1 × 105 cells/μl in Hank’s balanced salt solution, without Ca2+ and Mg2+ were slowly injected into the pontine tegmentum at 5-mm deep from the inner base of the skull. Mice were monitored daily and euthanized at indication of progressive neurologic deficit or found in a moribund condition. For subcutaneous tumor cell implantation, 4 × 106 cells, in 0.4 ml of cell culture media with matrigel (BD Bioscience) were injected in the right flank of mice. For testing the effect of JMJD3 siRNA in vivo, 2 × 105 tumor cells transfected with JMJD3 siRNA (Ambion oligo ID: s23110) or scrambled control siRNAs for 72 h were injected into the right flank of mice. Tumor growth and response to therapy were determined twice weekly by bioluminescence imaging. For imaging, mice were anesthetized as described above and examined for tumor bioluminescence 10 min following ip injection of D-luciferin (potassium salt, 150 mg per kg, Gold Biotechnology). Signal intensities were quantified within regions of interest, as defined by the Living Image software. Bioluminescence measurements for each animal at each time point were normalized against corresponding readings obtained at the beginning of therapy.
For the in vivo therapy-response analysis, ten mice were randomly assigned to vehicle (DMSO, n = 5) and GSKJ4 (n = 5) treatment groups, with GSKJ4 administration by ip injection at 100 mg per kg, with once-daily administration for 10 consecutive days. An additional two mice, not included in the survival analysis, were sacrificed 6 h after administration of final treatment, with the brains of these mice resected and placed in 4% PFA.
Immunohistochemical analysis
PFA-fixed mouse brains were paraffin-embedded and sectioned (10 μm) for immunohistochemical analysis of effect of therapy on cell proliferation, as indicated by positivity for Ki-67 staining (2 μg/ml, Ventana Inc.), and on K27 methylation, as indicated by positivity for H3K27me3 staining (C36B11, Cell Signaling, 1:500). To assay for apoptotic response to treatment, TUNEL staining was performed using the ApopTag® Peroxidase In Situ Apoptosis Detection Kit (S7100, Millipore) according to the manufacturer’s protocol.
Statistical analyses
The Kaplan–Meier estimator and Prism software were used to generate and analyze survival plots. Differences between survival plots were calculated using a log-rank test. Two-way ANOVA with Bonferroni post-test was used for comparison of annexin V staining. For all other comparisons, a two-tailed unpaired t-test was used (GraphPad Software). No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. The experiments were not randomized. In the in vivo therapy-response experiments, intracranial tumor bioluminescence signals were ranked from highest to lowest on the day of initiating therapy, and animals were assigned to treatment groups on an alternating basis, as determined by rank order, such that the mean tumor bioluminescence signals for each treatment group were approximately equal at the time of therapy initiation.
Online Methods
Cell cycle analysis and apoptosis assay
GSKJ4 cell cycle effects were determined by treating cells with 3 μM GSKJ4 for 72 h, pulsing cells with 10 μmol/l 5-ethynyl-2′-deoxyuridine (EdU) for 30 min, and then collecting and staining cells with Alexa Fluor 647-azide using a Click-iT assay kit (Life Technologies). DNA was stained using 1 mg/ml DAPI. Cells were then subjected to flow cytometric analysis, using a BD FACSCalibur™ instrument, with data analyzed using FlowJo 8.1 software. Cell apoptosis was assessed with the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis kit (Life Technologies).
siRNA oligonucleotide treatments
JMJD3 (KDM6B), UTX (KDM6A), and scrambled control siRNAs (Ambion oligo IDs JMJD3 siRNA A: s23109, B: s23110, and UTX siRNA A: s14737, B: s14736) were used to transfect tumor cells at a concentration of 30 nM using Oligofectamine reagent (Invitrogen) according to the manufacturer’s instructions. Cells were incubated with siRNAs for 96 h, after which relative number of viable cells was determined, as described above.
Quantitative PCR for determination of JMJD3 (KDM6B) and UTX (KDM6A) gene expression
Total RNA was extracted from cell lines following treatment of JMJD3 and UTX siRNA for 72 h, and cDNA was synthesized by reverse transcription (RT), using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed in a 15-μl reaction mixture using Maxima SYBR master mix (#K0221, Thermo Scientific). Fluorescence data were collected at annealing stages, and real-time analysis performed with SDS v2.3 software (Applied Biosystems). Ct values were determined using automatically set baseline and fluorescence thresholds. Fold changes were calculated using the ΔΔCt method and normalized against GAPDH and ACTB expression. Forward and reverse primers, respectively, used for PCR were 5′–CCTCGAAATCCCATCACAGT–3′ and 5′– CAGGGTCTTGGTGGAGAAGA–3′ for KDM6B (81 bp fragment), and 5′–ATTCATAGCAGCGAACAGCC–3′ and 5′–CTGGACAGCCGCCTCTT–3′ for KDM6A (91 bp fragment).
Immunoblotting for JMJD3 and UTX proteins
Cell lysates were collected from asynchronously proliferating cells in buffer (Cell Signaling) supplemented with proteinase (Roche) and phosphatase (Sigma) inhibitor cocktails. Lysates were resolved by SDS-PAGE, and detected protein expression as described above. Antibodies specific for JMJD3 (AP1022a, 1:1,000) was obtained from Abgent, and UTX antibody (A302-374A, 1:1,000) was from Bethyl.
Analysis of brain-tissue drug concentration
Athymic mice were administered GSKJ4 at 100 mg per kg per day for 3 d, with brains resected following mouse euthanasia 3 h after the third administration. Brainstem was dissected from surrounding brain, with tissues snap frozen and stored at −80 °C. GSKJ4 was extracted from homogenized tissues using a Bullet Blender (Next Advance, Inc.). Homogenates were extracted with organic solvent and further processed before transfer to an autosampler for high-performance liquid chromatography (HPLC) analysis (Shimadzu VP Series 10 System), and determination of GSKJ4 and GSKJ1 content (Integrated Analytical Systems).
Microarray analyses
RNAs from SF8628 K27M DIPG cells, either untreated or treated with GSKJ4 for 24 and 72 h, were extracted then labeled (Cy3 for treated and Cy5 for untreated). 100-ng quantities of corresponding treated–untreated pairs were hybridized to SurePrint G3 Human Gene Expression 8 × 60K Microarrays (Agilent Technologies), in duplicate, for each length of treatment, for 17 h at 65 °C. Subsequent to post-hybridization rinsing, arrays were scanned using an Agilent Microarray Scanner (model G2505C), and signal intensities were extracted using Agilent’s Feature Extraction 10.5.1 software. Agilent’s two-color technology option was used for thresholding of signal values to five, ratio computing (Cy3/Cy5), and log transformation. No baseline transformation was performed. Comparison of log2 intensity distributions among all eight samples indicated concordance. The distribution of Cy3/Cy5 intensity ratios plotted against the average intensity indicated the absence of bias, i.e. no normalization was necessary.
ChIP-seq
The ChIP-seq experiments were performed as described3. Briefly, 1 × 106 cells, treated with vehicle or 6 μM GSKJ4 for 24 and 72 h, were harvested, washed with PBS and fixed with 1% formaldehyde for 5 min at room temperature. Cells were then quenched with 125 mM glycine for 5 min. After washing, cell extracts were prepared using lysis buffer (50 mM HEPES/KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton-X100, 0.1% Na-deoxycholate, 1 mM PMSF, 1 mM Pefoblock, 1 mM benzamidine, 1 mg/ml bacitracin) on ice for 10 min. Chromatin was sheared by sonication (Bioruptor, High power, 15 × 2 cycles), to average lengths of 500 bp and then immunoprecipitated using an antibody against H3K27me3 (#9733, Cell Signaling, 1:50). After repeated washings, the DNA was recovered from the beads by incubating the beads in elution buffer (10 mM Tris at pH 8.0, 10 mM EDTA at pH 8.0, 1% SDS, 150 mM NaCl, 5 mM DTT) at 65 °C. Both input DNA and eluted DNA were subsequently purified using Qiagen Mini Elute PCR purification kit. ChIP DNA libraries were prepared with the Ovation Ultralow DR Multiplex system (NuGEN). The DNA libraries were sequenced using the paired-end method by an Illumina Hi-seq 2000. Reads were aligned to the human genome (hg19) using Bowtie218 software and preset parameters. Only uniquely mapping reads were used for further analysis.
Informative tags were selected from the aligned ChIP-seq data of each sequenced sample based on the cross-correlation profile of positive- and negative-strand tag densities. Background anomalies resulting from extremely high-density peaks at single chromosome positions were removed. After these filtering steps, the remaining tag list from each K27me3-immunoprecipitated sample was normalized against tag lists from sample-matched input DNA (treated only with DMSO). Broad regions of binding enrichment (i.e. ChIP signal over input) for each immunoprecipitated sample were obtained by comparing scaled ChIP and input tag counts to see whether their ratio exceeds that expected from a Poisson process. The R library SPP19 was used to perform all aforementioned steps. Distance to the nearest transcription start site (TSS) was obtained for each enriched binding region using Bioconductor package ChIPpeakAnno20 (function annotatePeakInBatch, AnnotationData: TSS.human.GRCh37).
Combined ChIP-seq and gene expression analysis
To identify both, differentially expressed and differentially immunoprecipitated genes, we derived P values and fold changes separately for differential expression and K27me3 immunoprecipitation as described below.
To identify differential gene expression associated with GSKJ4 treatment, we performed a t-test to verify the null hypothesis that the Cy3/Cy5 intensity log ratios derived from GSKJ4-treated cells (24 and 72 h; two replicates per time point) are distinct from 0. The R package DBChIP was used to identify sequences for which differential K27me3-immunoprecipitated is associated with GSKJ4 treatment. Enrichment sites estimated from vehicle-treated SF8628 cells and SF8628 cells incubated with 6 μM GSKJ4 (24 and 72 h) were merged into consensus binding sites (function site.merge), and fold changes of non-differential binding at each consensus site were calculated (function test.diff.binding). P values for each gene were obtained by performing a two-sided Wilcoxon signed-rank test of the hypothesis that fold changes associated with a gene come from a distribution whose median is 0. The resulting P values reflect both differential length of K27me3 immunoprecipitated regions between GSKJ4-treated and untreated samples, and differential sequencing depth within those regions. The 34 differentially expressed and differentially immunoprecipitated genes (P ≤ 0.1 and absolute fold change ≥ 1.5), and for which there was an inverse correlation between GSKJ4-associated gene expression change and corresponding change in K27me3 sequence association, are shown in Supplementary Fig. 11 and Supplementary Table 3.
Supplementary Material
Acknowledgments
Supported by US National Institutes of Health (NIH) Brain Tumor SPORE Grant CA97257 (R.H., C.D.J.), the Pediatric Brain Tumor Foundation (R.H., N.G., C.D.J.), the Matthew Larson Foundation (R.H.), Bear Necessities Pediatric Cancer Foundation (R.H.) and Rally Foundation (R.H.), American Cancer Society IRG-97-150-13 (R.H.), NIH CA157489 (Z.Z.), the Childhood Brain Tumor Foundation (C.P.), Voices against Brain Tumor Foundation (C.P.), and NIH CA164746 (C.P.).
Footnotes
Author Contributions
R.H. designed and conducted all of the experiments and wrote the manuscript. N.A. performed bioinformatics analysis for the data from gene expression and ChIP-seq. Y.I. conducted the in vitro studies and provided valuable discussion. R.L. conducted the cell cycle analysis, flow cytometry analysis, and quantitative RT-PCR. H.G., X.C., and D.F. conducted the ChIP-seq experiments. X.H. provided valuable discussion and intellectual input. M.W.T. contributed to the in vitro studies. V.N. performed microarray analysis. D.S. provided valuable material and intellectual input. S.M. provided clinical information and conceptual advice. P.L.P. supervised microarray analysis and provided valuable discussion and intellectual input. Z.Z. supervised ChIP-seq experiments and provided valuable discussion. C.P. supervised to the in vitro experiments and provided valuable discussion and intellectual input. N.G. provided clinical information and supervised the projects. T.A.W. provided valuable discussion and intellectual input. C.D.J. supervised the projects, provided valuable discussion, and conceptual advice.
References
- 1.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodeling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 3.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender S, et al. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Venneti S, et al. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimm SA, Chamberlain MC. Brainstem glioma: a review. Curr Neurol Neurosci Rep. 2013;13:346. doi: 10.1007/s11910-013-0346-3. [DOI] [PubMed] [Google Scholar]
- 8.Hashizume R, et al. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. J Neurooncol. 2012;110:305–313. doi: 10.1007/s11060-012-0973-6. [DOI] [PubMed] [Google Scholar]
- 9.Mueller S, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol. 2014;16:352–60. doi: 10.1093/neuonc/not220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1943. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 11.Lund AH, van Lohuizen M, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 13.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 15.Aoki Y, et al. An experimental xenograft mouse model of diffuse pontine glioma designed for therapeutic testing. J Neurooncol. 2012;108:29–35. doi: 10.1007/s11060-011-0796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkaria S, et al. Use of an orthotopic xenograft model for assessing the effect of EGFR amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–71. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 17.Bjerke L, et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Disc. 2013;3:512–9. doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.