Abstract
This paper describes a new method for extracting RNA, protein, and lipid mediators from a single tissue specimen. Specifically, mouse bone fracture callus specimens were extracted into a single solution which was processed using three different procedures to measure mRNA levels by RTqPCR, cytokines and growth factors using an xMAP method, and lipid mediators by LC-MS/MS. This method has several advantages because it decreases the number of animals necessary for experimentation, allows the division of the sample from a homogenous mixture which reduces sample variability, and utilizes a solution that protects the integrity of the macromolecules during storage.
Introduction
Bone fractures normally heal by tissue regeneration, which occurs through a temporally and spatially coordinated process [1, 2]. Fractures typically disrupt blood circulation causing hematoma formation and localized tissue hypoxia. Inflammation quickly follows and lipid mediators, growth factors, and cytokines released at the fracture site promote proliferation and chemotaxis of cells to the site. Mesenchymal cells that have proliferated or migrated to the fracture site differentiate into chondrocytes to form a cartilaginous callus around the fracture. Bone replaces the cartilage by endochondral ossification to bridge the fracture. The newly formed bone is remodeled to increase mechanical strength and restore the shape of the bone. While the histological processes that occur during fracture healing are well described, the signaling events that control this tissue regeneration process are poorly understood.
Previous studies showed that cyclooxygenase-2 (COX-2) and 5-lipoxygenase (5-LO) activity regulate bone fracture healing. Pharmacological inhibition or genetic ablation of COX-2 impairs healing [3, 4], while inhibition or ablation of 5-LO accelerates healing [5, 6]. COX-2 and 5-LO catalyze the synthesis of prostaglandins and leukotrienes, respectively, which are lipid mediators that regulate inflammation and other processes, including tissue regeneration [7-10]. Measuring the types and levels of lipid mediator during fracture healing is necessary to understand how COX-2 and 5-LO regulate this tissue regeneration process. This is complicated by the altered dynamics of fracture healing caused by loss of COX-2 or 5-LO activity. Thus, levels of each lipid mediator must be correlated to other cellular processes occurring at that time in the fracture callus in order to understand the role each lipid mediator has during fracture healing. Ideally, lipid mediators would be correlated to protein and mRNA markers of established physiological and cellular processes in order to understand how bone regeneration is regulated by COX-2 or 5-LO. Generally, measurement of mRNA, protein, or lipid mediators is performed with multiple tissue samples prepared separately using extraction methods appropriate for each target molecule [6, 11-14]. The spatial complexity of a fracture callus precludes using this approach since dividing the callus into portions would yield tissue samples with different cellular compositions unless specialized methods such as laser capture micro-dissection were employed [2, 15]. Alternatively, a fracture callus could be used to measure one type of target molecule [6, 16-18]. However, this approach would require using significantly more animals to measure different types of target molecules and would introduce another level of variability into the analysis as levels of the different target molecule types could not be compared from the same specimen.
To overcome these limitations, we developed methods for isolating lipid mediators, proteins, and RNA from the same tissue specimen. The method relies upon RNAlater solution (Ambion, Inc., Austin, TX) to preserve RNA during callus extract preparation and modification of existing methods to measure mRNA, proteins, and lipid mediators from callus extract aliquots using RTqPCR, xMAP, and LC-MS/MS methods, respectively [19].
Materials and Methods
Animal Model
Female ICR mice (Taconic Farms, Germantown, NY) weighing 28.7 ± 2.3 g (mean ± standard deviation) were used in this study. Mice were anesthetized by intraperitoneal injection of ketamine and xylazine (0.1 and 0.01 mg/g body weight, respectively). A closed, diaphyseal fracture was created in the right femur using a custom-made, three-point impactor (BBC Specialty Automotive Center, Linden, NJ) as described previously except the mice were allowed to recover for 7 days between insertion of the intramedullary pin used to stabilize the fracture and production of the fracture [20]. All experimental procedures were approved by the New Jersey Medical School Institutional Animal Care and Use Committee.
Tissue Collection
On day 4 post fracture, mice were injected intraperitoneally with 0.1 ml of 0.5 mg/ml heparin (USB Corp., Cleveland, OH) 30 minutes before euthanization to prevent post-mortem blood clotting. Femurs were resected with surrounding muscle and callus left intact. The proximal and distal epiphyses were removed. The final length of femur diaphysis and surrounding tissue was approximately 1cm and weighed 0.72 ± 0.10 g. The femur was divided at the fracture site to facilitate subsequent tissue homogenization, flash frozen in liquid nitrogen, and stored at −80°C.
Tissue Homogenization
Each femur was homogenized in 3 mls of pre-chilled RNAlater that had been spiked with deuterated eicosanoids to act as internal standards. The deuterated eicosanoids were purchased from Cayman Chemicals (Ann Arbor, MI), dissolved in methanol to 500 ng/ml each, and added to a final concentration of 6.67 ng/ml which would ultimately yield a maximum of 2.67 ng of each deuterated eicosanoid per LC-MS/MS injection (see below). Callus extracts were made with a Precellys 24 Dual Tissue Homogenizer (Bertin Technologies, Washington D.C., MD) using 7 ml tubes containing 2.8 mm diameter zirconium oxide beads. The callus was pulverized twice at 6,500 rpm for 15 seconds. To reduce overheating of the callus extract, samples were placed on dry ice for 1 minute before pulverization and were cooled on ice between pulverization steps. After homogenization, 2 mls of the callus extract was immediately processed for eicosanoid analysis and the remainder was stored at −20°C for protein and RNA analysis.
Protein Extraction
Aliquots of the callus extract were dialyzed to remove RNAlater and solubilize precipitated protein. Protease inhibitors (Sigma-Aldrich P2714 Protease Inhibitor Cocktail) were added to a 0.4 ml aliquot of the callus extract which was then transferred to a dialysis apparatus (Tube-O-DIALYZER, 1kDa Micro, GBiosciences, MO) and dialyzed against M-PER (Mammalian Protein Extraction Reagent, Thermo Fisher Scientific, IL) for 24 hours at 4°C. After the dialysis, insoluble material was removed from the samples by centrifugation (14,000 rpm for 10 minute at 4°C). To remove additional debris, the supernatant was collected and filtered through a 0.2 μm membrane by centrifugation (Nanosep, PALL, Port Washington, NY) at 14,000 rpm and 4°C until all liquid passed through the filter cartridge. The clarified extract was stored at −20°C. Aliquots of the clarified extract were used to measure total protein content using the BCA assay (BCA Protein Assay Reagent, Thermo Fisher Scientific, IL) and to measure target protein amounts using an xMAP method as described below [21, 22]. Protein extract quality was assessed by electrophoresis using 8% polyacrylamide bis-Tris gels in which 15 μg of extract protein was separated in each lane. Protein was detected by Coomassie Blue staining. Precision Plus Protein Standards (BIO-RAD, Berkeley, CA) were used to estimate protein sizes.
xMAP Assay
Growth factors and inflammation-related cytokines were measured in the clarified extracts using a Luminex 100 Multiplexing Instrument (Luminex Corp., Austin, TX) with a Milliplex Map Mouse Cytokine/Chemokine Panel (Millipore Corporation, MA). Thirty two target proteins were analyzed (Table 1). Each clarified extract was measured in duplicate using 25 μl of clarified extract for each assay. Mean fluorescence intensity for each target protein was compared to a standard curve developed using standards contained within the Milliplex Map Mouse Cytokine/Chemokine Panel reagents to determine the amount of each target protein within the clarified extract aliquot. The amount of each target protein was then normalized to the total protein amount used in each assay to yield pg of target protein per mg of soluble protein.
Table 1.
Extract Preparation Effects on Target Protein Yield
| Target Protein |
pg per mg soluble protein
Each sample was measured twice. Single values = mean from 2 samples. B = sample values below detection limit. F = sample failed in assay. |
|||
|---|---|---|---|---|
|
LN-
MPER |
LN-RL | RL-MPER | RL-RL | |
| Eotaxin | 29.0 | 87.7 | 46.0 | 165.2 |
| G-CSF | 1.9 | 9.6 | 2.5 | 7.1 |
| GM-CSF | 2.6 | 4.7 | 2.4 | 5.2 |
| IFN-γ | B, B | 2, B | B, B | B, B |
| Il-1α | 1.6 | 2.8, B | 1.6 | 2, B |
| IL-1β | 16.7 | 6.5 | 4.8 | 8.1 |
| IL-2 | B, B | 1.6 | 0.8 | 1.8, B |
| IL-3 | B, B | B, B | B, B | B, B |
| IL-4 | B, B | B, B | B, B | B, B |
| IL-5 | 1.1 | B, B | B, B | B, B |
| IL-6 | 8.8 | 6.3 | 10.2 | 18.1 |
| IL-7 | 1.2 | 2, B | 1, B | 4.2, B |
| IL-9 | B, B | B, B | B, B | 100, B |
| IL-10 | 4.4 | 7.0 | 2.7 | 4.9 |
| IL-12 (p40) | 3.2, B | 11.6 | 0.6 | 2.1 |
| IL-12 (p70) | 3.4 | 2.8 | B, B | B, B |
| IL-13 | B, B | B, B | B, B | B, B |
| IL-15 | 2.6 | 2.3 | 1.3 | 2.0 |
| IL-17 | 4.6, B | B, B | 1.2 | 3.2, B |
| IP-10 | 30.7 | 73.0 | 35.5 | 140.1 |
| KC | 6.9 | 7.9 | 3.1 | 15.6 |
| LIF | 1.9 | 1.9 | 1.9 | 4.3 |
| LIX | B, B | B, B | B, B | B, B |
| MCP-1 | 9.8 | 7.8 | 2.2 | 15.0 |
| M-CSF | 3.5 | 4.4 | 1.8 | 4.2 |
| MIG | 6.7 | 33.0 | 126.9 | 65.2 |
| MIP-1α | 7.5 | 4.1 | 1.5 | B, B |
| MIP-1β | 1.4 | 1.8 | 0.6 | 1.4, B |
| MIP-2 | 4.9 | 8.0 | 1.9 | 1.9 |
| RANTES | B, B | B, B | B, B | B, B |
| TNF-α | 0.6, F | 1.2, F | B, B | B, B |
| VEGF | 3.1 | 4.2 | 1.0 | 4.9 |
Total RNA Purification
Total RNA was prepared from callus extract aliquots using modified TRIzol extraction and differential filter binding methods in order to eliminate RNAlater from the purified RNA preparation [13]. One hundred μ ls of callus extract was combined with 4 volumes of TRIzol and 4 volumes of water. After mixing, another 6 volumes of TRIzol was added which was mixed gently on a shaker for 30 minutes at 4°C. The aqueous phase was separated by centrifugation (12,000 rpm for 10 mins at room temperature), collected into a new tube, and processed by differential filter binding and elution (RNeasy; Qiagen, Valencia, CA) as follows. The aqueous layer from the TRIzol extraction was mixed with 4 volumes of RLT buffer containing β-mercaptoethanol and 3.5 volumes of methanol. This solution was applied to an RNeasy mini-column. Afterwards, the column was washed with two volumes of RW1 buffer and two volumes of RPE buffer. The RNA was eluted with two washes of 25 μl water. RNA concentration was determined spectrophotometrically with a NanoDrop 1000 (Wilmington, DE). RNA integrity was confirmed by agarose gel electrophoresis and ethidium bromide staining. Only RNA samples with acceptable characteristics, including intact 16S and 28S ribosomal RNA, were used for further analysis.
mRNA Quantification
The reverse transcription-quantitative polymerase chain reaction (RTqPCR) method was used to measure target mRNA levels [23, 24]. cDNA was made from 0.5 μg aliquots of total RNA as described previously with use of an oligo(dT)20 primer, RNase inhibitor (RNAaseOUT; Life Technologies, Grand Island, NY), and MMLV reverse transcriptase (New England BioLabs, Beverly, Massachusetts) [5]. Reactions proceeded for one hour at 42°C and were stopped by incubation at 92°C for ten minutes. cDNA was stored at −20°C until use. Quantitative polymerase chain reaction (qPCR) was performed with Absolute QPCR SYBR Low Rox Mix (Thermo Scientific, Lafayette, CO) and an Applied Biosystems 7500 Real-Time PCR System (Foster City, California). Reactions contained an aliquot of cDNA corresponding to 20 ng of total RNA in a 20 μ l reaction volume. The annealing temperature for the glyceraldehyde-3-phosphate dehydrogenase (Gapdh; forward: 5’-CCTGTGACTT CAACAGCAAC TCC-3’, reverse: 5’-CCACCACCCT GTTGCTGTAG CC-3’), Type II collagen (Col2a1; forward: 5’-TGGGTGTTCT ATTTATTTAT TGTCTTCCT-3’, reverse: 5’-GCGTTGGACT CACACCAGTTAGT-3’) and aggrecan (Acan; forward: 5’-CATGAGAGAG GCGAATGGAA-3’, reverse: 5’-TGATCTCGTA GCGATCTTTC TTCT-3’) mRNA primers was 60°C. Amplified DNA was measured for 40 cycles of polymerase chain reaction (PCR) as SYBR green fluorescence. Target mRNAs were assayed in triplicate for each cDNA preparation. Threshold cycle values (Ct) were determined using the supplied Applied Biosystems software and were manually reviewed for accuracy.
Lipid Mediator Extraction
Eicosanoids and other lipid mediators were extracted from 2 mls of each callus extract. The callus extract was mixed with 9 volumes of ice cold distilled water and insoluble material was removed by centrifugation (14,000 rpm at 4°C for 10 mins using a Sorvall SS-34 rotor). The supernatant was collected and applied to an OASIS HLB 3cc (60mg) Extraction Cartridge (Waters, Milford, MA) which had been previously rinsed with 3 mls of methanol and then equilibrated with 3 mls of water. After sample loading, the cartridge was washed sequentially with 3 mls each of distilled water, 5% methanol, 50% methanol with 0.2% acetic acid, and 50% methanol. After the washes, the cartridge was vacuum dried to remove residual liquid. Eicosanoids were eluted from the cartridge with 1 ml of 97% methyl formate followed by 1 ml of 100% methanol. The combined eluates were dried using a vacuum concentrator (Vacufuge, Eppendorf, Hamburg, Germany).
Mass Spectrometry for Lipid Mediators
Eicosanoids and related lipid mediators were detected by LC-MS/MS and quantified using standard curves. The LC-MS/MS system used included a FAMOS autosampler (LC Packings, San Francisco, CA), an Agilent 1100 degasser and binary pump (Agilent Technologies, Inc., Santa Clara, CA), and an AB SCIEX Qtrap 2000 (AB SCIEX, Redwood City, CA). Eicosanoid samples and standards were separated by reverse-phase liquid chromatography using a Kinetex C18-column (100 × 2.10 mm, 2.6 micron; Phenomenex Inc., Torrance, CA) at room temperature and a flow rate of 300 μl/min. The column eluate was introduced into the Qtrap 2000 using the electro-spray ion source in negative ion mode or in positive ion mode after charge-reversal derivation as described below.
For negative ion mode detection, the dried eluate after SPE purification was reconstituted in 200 μ l of 50% methanol for LC-MS/MS analysis. Injection volume was 50 μ l. For all separations, solvent A was 1% methanol diluted in water and solvent B was 100% methanol. The Kinetex C18-column was developed as outlined in Table 2. The LC eluate was introduced into the Qtrap 2000 using the electro-spray ion source in negative ion mode. Eicosanoids were detected using multiple reaction monitoring (MRM) by measuring the ion pair transition from Q1 to Q3 with parameters listed in Table 3. PGD2 and PGE2 were distinguished by column retention time and co-elution with their cognate deuterated internal standards. The following Qtrap parameters were used: CUR = 20 psi, CAD = 10, IS = −4500 V, TEM = 500°C, GS1 = 50 psi, GS2 = 80 psi, IHE=ON. Lipid standards were dried, reconstituted in 50% methanol, then separated and detected as described above.
Table 2.
Liquid Chromatography Methods for LC-MS/MS Analysis
| Step | Negative Ion Mode | Positive Ion Mode | ||||
|---|---|---|---|---|---|---|
| Time (min) | Buffer A | Buffer B | Time (min) | Buffer A | Buffer B | |
| 0 | 0.0 | 100% | 0% | 0.0 | 100% | 0% |
| 1 | 1.0 | 100% | 0% | 1.0 | 100% | 0% |
| 2 | 1.1 | 100% | 0% | 2.0 | 78% | 22% |
| 3 | 5.0 | 20% | 80% | 8.0 | 74% | 26% |
| 4 | 5.1 | 0% | 100% | 8.1 | 55% | 45% |
| 5 | 10.0 | 0% | 100% | 13.1 | 40% | 60% |
| 6 | 10.1 | 100% | 0% | 14.0 | 0% | 100% |
| 7 | 14.0 | 100% | 0% | 19.0 | 0% | 100% |
| 8 | NA | 21.0 | 100% | 0% | ||
| 9 | NA | 25.1 | 100% | 0% | ||
Table 3.
Mass Spectrometry Parameters for Eicosanoid Detection
| Compound | Negative Ion Mode Parameters | Positive Ion Mode Parameters | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Precursor (m/z) |
Fragment (m/z) |
DP | EP | CE | CXP | Precursor (m/z) |
Fragment (m/z) |
DP | EP | CE | CXP | |
| PGD2 | 351 | 189 | −39 | −7 | −29 | −3 | 519 | 239 | 62 | 11 | 58 | 2 |
| PGD2-d4 | 355 | 193 | −39 | −7 | −29 | −3 | 523 | 311 | 62 | 11 | 58 | 2 |
| PGE2 | 351 | 189 | −41 | −9 | −30 | −2 | 519 | 239 | 62 | 11 | 58 | 2 |
| PGE2-d4 | 355 | 193 | −41 | −9 | −30 | −2 | 523 | 241 | 62 | 11 | 58 | 2 |
| PGF2α | 353 | 193 | −41 | −5 | −30 | −2 | 521 | 239 | 90 | 5 | 61 | 2 |
| PGF2α-d4 | 357 | 197 | −41 | −5 | −30 | −2 | 525 | 241 | 90 | 5 | 61 | 2 |
| 6-Keto-PGF1α | 369 | 207 | −41 | −5 | −30 | −2 | 537 | 239 | 90 | 6 | 74 | 2 |
| 6-Keto-PGF1α-d4 | 373 | 211 | −41 | −5 | −30 | −2 | 541 | 241 | 90 | 6 | 74 | 2 |
| TXB2 | NA | 537 | 337 | 72 | 10 | 56 | 5 | |||||
| TXB2-d4 | NA | 541 | 341 | 72 | 10 | 56 | 5 | |||||
| 15-deoxy- Δ12,14-PGJ2 |
NA | 482 | 254 | 80 | 10 | 30 | 2 | |||||
| 15-deoxy- Δ12,14-PGJ2-d4 |
NA | 486 | 241 | 80 | 10 | 30 | 2 | |||||
| LTB4 | NA | 503 | 323 | 45 | 10 | 45 | 3 | |||||
| LTB4-d4 | NA | 507 | 325 | 45 | 10 | 45 | 3 | |||||
| 5(S)-HETE | NA | 487 | 283 | 65 | 10 | 54 | 3.5 | |||||
| 5(S)-HETE-d8 | NA | 495 | 284 | 65 | 10 | 54 | 3.5 | |||||
| Lipoxin A4 | NA | 519 | 283 | 110 | 11 | 55 | 2 | |||||
| Lipoxin A4-d5 | NA | 524 | 283 | 110 | 11 | 55 | 2 | |||||
| 5-oxo-ETE | NA | 485 | 239 | 120 | 10 | 60 | 0 | |||||
| 5-oxo-ETE-d7 | NA | 492 | 239 | 120 | 10 | 60 | 0 | |||||
DP = Declustering Potential; EP = Entrance Potential; CE = Collision Energy; CXP = Collision Cell Exit Potential
For quantification in positive ion mode, eicosanoids and related lipid mediators in the dried elutes were converted into positively charged molecules by attaching N-(4-aminomethylphenyl)pyridinium through an amide bond at the carboxylic acid of each lipid as described previously [25]. This was accomplished using an AMP+ Mass Spectrometry Kit (Cayman Chemicals) following the manufacturer’s instructions except hydroxybenzotriazole was replaced with 20 mM 1-hydroxy-7-azabenzotriazole (Santa Cruz, Biotechnology, Inc., Dallas, TX) to prevent ion suppression of some eicosanoids. The AMP+ derived samples were diluted to a final volume of 500 μ l with water prior to LC-MS/MS analysis. Lipid standards (Cayman Chemicals) were dried and then derived as described above. The derivatized eicosanoid samples and standards were separated by reverse-phase liquid chromatography using a Kinetex C18-column (100 × 2.10 mm, 2.6 micron) at room temperature and a flow rate of 300 μl/min. Injection volume was 100 μ l. For all separations, solvent A was 1% acetic acid diluted in water and solvent B was 1% acetic acid in acetonitrile. The Kinetex C18-column was developed as outlined in Table 2. The LC eluate was introduced into the Qtrap 2000 using the electro-spray ion source in positive ion mode. Eicosanoids were detected using multiple reaction monitoring (MRM) by measuring the ion pair transition from Q1 to Q3 with parameters listed in Table 3. PGD2 and PGE2 were also distinguished by column retention time and co-elution with their cognate deuterated internal standards. The following Qtrap parameters were used: CUR = 40 psi, CAD = 5, IS = 5500 V, TEM = 400°C, GS1 = 50 psi, GS2 = 80 psi, IHE=ON.
Lipid Mediator Data Analysis
Eicosanoids and other lipid mediators were quantified using a standard curve for each lipid with Analyst Quantification Software (AB SCIEX). LC-MS/MS chromatogram peak areas were determined for each lipid using the Analyst Software. Each peak was manually reviewed for accuracy. For each lipid standard, 6 values ranging from 0 to 4 ng were analyzed in triplicate and the peak areas were fitted to a quadratic standard curve. These standard curves were used to calculate the amount of each eicosanoid or other lipid mediator present in each injected sample. The amount of recovered deuterated eicosanoid that had been spiked into the callus extract was used to determine the percent yield for each eicosanoid in each sample and data were corrected based upon the yields. Quality control standards and blanks were used periodically throughout the analysis to ensure accuracy. The statistical analysis was performed using SigmaPlot 12.5 software (Systat Software Inc., Chicago, IL).
Results and Discussion
An outline of the optimized procedures for extract preparation and processing to measure protein, mRNA, and lipid mediators can be found in the Supplemental Materials.
Callus Extract Preparation
Because fracture calluses contain bone, a robust tissue homogenization process is necessary to prepare callus extracts. We chose to use a bead pulverization method using the Precellys 24 Dual Tissue Homogenizer to promote reproducibility in future experiments. Pulverization was performed using different Precellys parameters to optimize callus homogenization. Lower rpm was used initially to prevent an increase in extract temperature but resulted in incomplete callus breakage. Pulverization was also tried in 2 ml rather than 7 ml tubes but resulted in excessive foam formation and increased extract temperature. Pulverization with metal rather than zirconium oxide beads caused extract temperature to increase above 40°C. Pulverizing the callus in 3 ml of buffer with 2 cycles at 6500 rpm for 15 seconds and with cooling before and after each cycle gave complete callus disruption without extract temperature rising above room temperature.
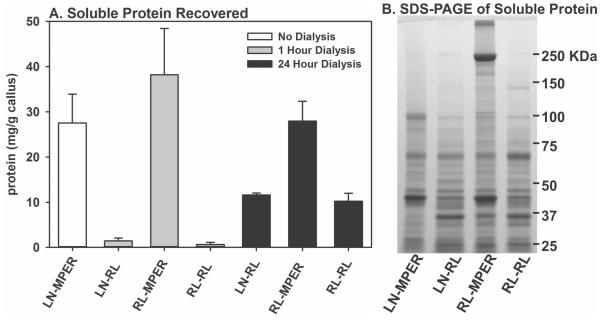
Common methods for extracting RNA from tissues using phenol or guanidinium thiocyanate are not compatible with obtaining proteins in their native state [14]. Thus, we had to develop a procedure that would allow for callus disruption into a buffer that would be compatible with downstream mRNA, native protein, and lipid mediator isolation. We chose to test for native protein isolation using RNAlater as the extract buffer because (a) RNAlater should preserve the RNA, (b) most proteins precipitated by the ammonium sulfate in RNAlater should be solubilized by dialysis, and (c) we did not expect the RNAlater to affect the lipid mediators (see below). Extract preparation in RNAlater was compared to M-PER buffer (Mammalian-Protein Extraction Reagent, Pierce Biotechnology, Rockford, IL) which was developed for isolating native proteins from mammalian cells and tissues. Four processing methods were tested: (LN-MPER) callus flash-frozen in liquid nitrogen and pulverized in M-PER reagent, (LN-RL) callus flash-frozen in liquid nitrogen and pulverized in RNAlater, (RL-MPER) callus stored in RNAlater and pulverized in M-PER reagent, and (RL-RL) callus stored in RNAlater and pulverized in RNAlater. All samples were stored at −80°C before pulverization. We found that calluses stored in RNAlater were more difficult to pulverize. Following pulverization, aliquots of each callus extract that had been stored in or pulverized in RNAlater were dialyzed against M-PER for 1 hour, clarified by centrifugation, and soluble protein measured (Figure 1A). The samples pulverized in RNAlater had less protein (0.2 mg/ml) than those pulverized in M-PER, suggesting that the 1 hour dialysis was not sufficient. When dialysis was extended to 24 hours, soluble protein yield substantially increased for the LN-RL (2.2 mg/ml) and RL-RL (2.0 mg/ml) extract methods though yields were less than extracts prepared with M-PER (LN-MPER, 5.0 mg/ml and RL-MPER, 5.3 mg/ml) (Figure 1A).
Figure 1. Callus Extract Protein Yields and Quality.
Panel A: Soluble protein levels were measure in callus extracts after 1 hour or 24 hours of dialysis to remove RNAlater. Samples sizes were LN-MPER = 12 (not dialyzed); LN-RL 1 hour = 8; RL-MPER 1 hour = 4; RL-RL 1 hour = 4; LN-RL 24 hour = 3; RL-MPER 24 hours = 3; RL-RL 24 hours = 2. Error bars represent standard errors of the mean. Panel B: Aliquots of soluble protein from extracts prepared using each method were separated by SDS-PAGE and detected by Coomassie Blue staining. Positions for protein standards are shown.
Protein composition of extracts dialyzed for 24 hours and the LN-MPER extract were determined by SDS-PAGE (Figure 1B). The RL-MPER extract appeared to have the largest number of different proteins, particularly those greater than 100 kDa with a very abundant protein of approximately 220 kDa. For proteins below 100 kDa, the protein composition between extraction methods appeared to be similar except for increased abundance of an approximately 42 kDa protein in the LN-MPER and RL-MPER extracts and reduced amounts of an approximately 35 kDa protein in the LN-MPER extract (Figure 1B). Since cytokines and growth factors are generally below 100 kDa in molecular weight, all extraction methods appeared to be suitable for protein extract preparation.
Since RNAlater precipitates protein, we also collected precipitated protein directly from the LN-RL and RL-RL crude extracts by centrifugation using a Beckman TL-100 Ultracentrifuge and TLA100 rotor (80,000 rpm for 30 mins at 4°C). Protein in the pellet and supernatant were determined. Ninety percent of the total protein was found in the precipitated fraction. Resuspending the precipitated protein collected by centrifugation of the RL-RL or LN-RL extracts in MPER buffer produced amounts of soluble protein similar to directly dialyzing the crude extracts (data not shown) and so collecting the precipitated protein by centrifugation was omitted.
xMAP Analysis
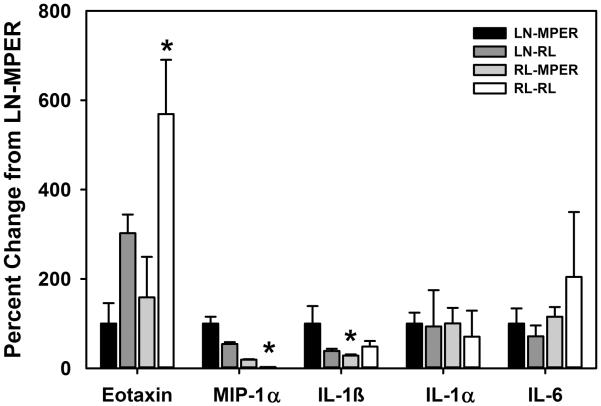
Callus extracts prepared using the different pulverization and dialysis procedures were assayed for cytokine and growth factor levels to determine which method gave the best overall yield of potential target proteins. LN-RL, RL-MPER, RL-RL samples dialyzed for 1 or 24 hours were compared to LN-MPER samples using an xMAP assay for 32 different target proteins (Milliplex Map Mouse Cytokine/Chemokine Panel). The majority of target protein readings (mean fluorescence intensity) from the LN-RL, RL-MPER, or RL-RL which had been dialyzed for 1 hour failed or fell below the detectable limits of the assay (data not shown). When dialysis time was increased to 24 hours, cytokines could be readily detected in the LN-RL, RL-MPER, and RL-RL samples (Table 1 and Figure 2). Cytokine levels in the RL-MPER samples were comparable to those in the LN-MPER samples, except IL-1β, which was significantly lower in the RL-MPER samples (Figure 2). In RL-RL samples, eotaxin levels were significantly higher while MIP-1α level were significantly lower when compared to LN-MPER (Figure 2). Cytokines IL-3, IL-4, IL-13, LIX, and RANTES were under the detectable limit in all samples (Table 1). For the remaining 27 cytokines, the number of reactions that failed or were below the detection limit was determined from 2 replicate assays of 2 samples for each callus extract preparation method with 24 hour dialysis (Table 1). Cytokine and growth factor measurements for 9 of the LN-MPER, 10 of the LN-RL, 11 of the RL-MPER, and 16 of the RL-RL samples failed or were below the detection limit (Table 1). Although the RL-RL samples were dialyzed against M-PER buffer to remove RNAlater, improper solvent flow through the Luminex 100 assay plate resulted in variable or undetectable target protein levels in the RL-RL samples. Development of xMAP methods that use magnetic particles may permit use of the RL-RL callus extract preparation method but this was not tested. Based upon these results, we chose to use the LN-RL callus extract preparation method because callus pulverization was more easily accomplished, extract prepared in RNAlater should stabilize RNA, soluble protein yield was sufficient for measurement of target proteins, xMAP results were similar to the LN-MPER method (which is comparable to a standard protein extraction method), and the RNAlater is compatible with lipid mediator extraction (see below).
Figure 2. Variation in Cytokine Recovery using Different Extraction Methods.
Selected cytokine protein levels from LN-MPER, LN-RL, RL-MPER, and RL-RL samples were normalized against the mean value for the LN-MPER samples as the percent change. These data are shown as mean values (± SEM). Significant differences were determined using One Way ANOVA or Kruskall Wallis tests. Post-hoc tests was used to determine the differences between groups. Dunnett post-hoc method was used when sample sizes were the same and Dunn’s post-hoc method was used when sample sizes were different in each group. Differences were considered to be significant at P < 0.05. The statistical analysis was performed using SigmaPlot 12.5 software (Systat Software Inc.).
RNA Extraction
After successful detection and quantitation of proteins in LN-RL callus extracts, we developed a procedure for isolation of total RNA from LN-RL callus extracts. We had previously found that sequentially performing a phenol-quanidinium extraction (TRIzol) followed by differential filter binding (RNeasy) was necessary to provide high quality total RNA from fracture callus specimens, likely because of the large amount of extracellular proteins present in these samples [13]. However, the large amount of ammonium sulfate in the RNAlater solution necessitated modification of the previously used TRIzol extraction and RNeasy filter binding purification method. The large salt concentration in the RNAlater solution enabled complete solvation of TRIzol which prevented separation of aqueous and organic phases after chloroform addition. This problem was overcome by diluting the LN-RL callus extract aliquot with 4 volumes of water before mixing in 10 volumes of TRIzol. After mixing, chloroform was added to 10%, mixed vigorously, and phases separated by centrifugation at 4°C at 12,000 rpm for 10 mins. The aqueous phase was collected for final RNA purification by differential filter binding using RNeasy mini-columns as follows. Buffer RLT was prepared as described by the manufacturer except ethanol was replaced with methanol. Use of ethanol in the RLT buffer caused salt precipitation, which reduced RNA yield and purity. The RNeasy column was then washed with 6 mls of RW1 wash buffer and with 6 mls RPE buffer. These additional washes were necessary to prevent salt contamination of the RNA. Total RNA was then eluted from the RNeasy column with water.
Total RNA was prepared from 3 fracture callus extracts collected 4 days after fracture. RNA was quantified by UV absorbance, checked for quality by agarose gel electrophoresis (not shown), and analyzed for GAPDH, Type II collagen (Col2a1), and aggrecan (Acan) mRNA levels by RT-qPCR (Table 4). The amount of contaminating genomic DNA was not determined. The average A260/A280 ratio for the purified RNA was 1.84 ± 0.15. RTqPCR detected GAPDH, Type II collagen, and aggrecan mRNA in all callus extract RNA preparations showing that the modified RNA isolation procedure was successful (Table 4). To assess RNA stability using the different extraction methods, total RNA was extracted from mouse liver using the 4 different extraction methods and the crude extracts were incubated at room temperature (data not shown). Intact total RNA was evident using all extraction methods when RNA was immediately extracted after pulverization. RNA yields from the RL-RL extract were low. Within 30 minutes, significant RNA degradation was evident in the LN-MPER and RL-MPER extracts, while no RNA degradation was evident after 3 hours in the LN-RL extract.
Table 4.
RNA Yields from 100 μl of LN-RL Callus Extract
| Sample Size |
Range | Mean | Std. Dev. |
|
|---|---|---|---|---|
| RNA Yield (μg) | 3 | 0.29-0.62 | 0.40 | 0.19 |
| Gapdh mRNA (Ct value) | 3 | 18.43-19.27 | 18.97 | 0.47 |
| Col2a1 mRNA (Ct value) | 3 | 5.96-9.47 | 8.04 | 1.84 |
| Acan mRNA (Ct value) | 3 | 6.14-9.73 | 8.31 | 1.91 |
Lipid Mediator Extraction
Before using RNAlater for callus extract preparation, we compared recovery of PGE2, PGF2α, 6-keto-PGF1α (the initial breakdown product of PGI2), and TXB2 (the initial breakdown product of thromboxane A2) from RNAlater, M-PER, and water. Prostaglandins were added to water, M-PER, or RNAlater and then recovered by solid-phase extraction (SPE) using Oasis HLB columns. The recovered prostaglandins were analyzed by LC-MS/MS in negative ion mode and compared to the starting amount of each prostaglandin as a percentage (Table 5). The best recovery of the tested prostaglandins occurred with RNAlater which likely reflects enhanced hydrophobic interaction between the prostaglandins and the SPE matrix promoted by the high salt concentration of RNAlater. Other SPE matrices were tested including C18, weak cation exchanger, weak anion exchanger, and hydrophobic polydivinylbenzene matrices but the Oasis HLB matrix gave the best prostaglandin yield and was used in all subsequent experiments (data not shown).
Table 5.
Prostaglandin Recovery following SPE after Dilution in Different Solvents
| Prostaglandin | Percent Recovery from: | ||
|---|---|---|---|
| Water | M-PER Buffer | RNAlater | |
| PGE2 | 60 | 100 | 100 |
| PGF2α | 38 | 40 | 100 |
| 6-keto-PGF1α | 51 | 72 | 90 |
| TXB2 | 15 | 22 | 77 |
We noticed that some of the salt from the RNAlater also eluted with the prostaglandins from the Oasis HLB column. Consequently, SPE column loading and washing steps were optimized to reduce ammonium sulfate carry-over while maintaining prostaglandin recovery. The RNAlater and prostaglandin mixture was diluted with 9 volumes of water immediately before applying to the Oasis HLB column. The SPE column was then thoroughly washed sequentially using 1 ml each of water, 5% methanol, 50% methanol with 0.1% acetic acid, and 50% methanol. Washes using methanol concentrations greater than 50% decreased prostaglandin yield. Similarly, excessive washing with 50% methanol solutions also reduced the overall yield of prostaglandins (data not shown). Lipids were eluted from the SPE column with 1 ml methyl formate followed by 1 ml of methanol. The eluates were combined and dried in a vacuum centrifuge. Sample heating during drying decreased overall lipid yield and was avoided. Dried samples were stored at −20°C until analysis.
LC-MS/MS Analysis of Callus Extract Lipid Mediators
To initial optimize LC methods, eicosanoid standards were detected using negative ion mode, electrospray ionization. A variety of solvents, gradients, and columns were tested based upon prior work [25-35]. We found that a Kinetex C18 (100 × 2.1 mm) column developed using a multi-step gradient with water and acetonitrile, each containing 1% acetic acid, gave satisfactory separation of the target eicosanoids in our system. We also found that the eicosanoids were more stable in 1% acetic acid than in 1% formic acid (data not shown). Initial experiments using lipids prepared from callus extracts found that the AB SCIEX Qtrap 2000 had sufficient sensitivity to detect only the most abundant prostaglandins in negative ion mode. For instance, when all of the lipids prepared from 2 mls of callus extract were used in a single separation, only PGE2 and TXB2 could be readily detected. Since our goal is to survey lipid mediators present during fracture healing, we sought methods to increase the sensitivity of the LC-MS/MS assay. Mass spectrometers with better ionization devices and ion detectors with much greater sensitivity than the Qtrap 2000 could obviate the necessity for the following procedures.
Multiple methods have been developed to derivatize prostaglandins and other fatty acids in order to enhance detection of these lipids. Most derivation methods target the carboxyl group of the prostaglandins or other fatty acids in order to add a moiety for fluorescence detection or to convert the carboxyl group into an amine derivative that will promote positive ion formation for detection by mass spectrometry [25, 30-38]. Since formation of positive ions by electrospray ionization is more efficient than formation of negative ions, we chose to use a charge reversal derivatization method on the callus extract lipid preparations in order to enhance sensitivity. The method developed by Bollinger et al. adds N-(4-aminomethylphenyl)pyridinium (AMP) at the carboxylic acid of fatty acids through an amide linkage [25]. The AMP moiety is resistant to fragmentation in the mass spectrometer and so provides a constant increase of 168 amu to each prostaglandin. We also modified the AMP derivation method by using 1-hydroxy-7-azabenzotriazole instead of hydroxybenzotriazole in the derivation reaction because use of hydroxybenzotriazole caused ion suppression of LTB4 (data not shown and personal communication from James Bollinger and Michael Gelb, U. of Washington, Seattle, WA).
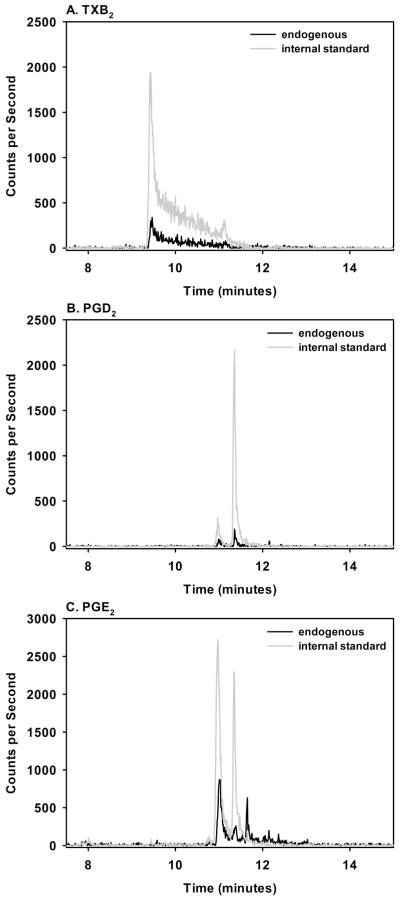
Using the optimized methods for callus extraction, lipid mediator purification, and derivation, we assayed for 10 different eicosanoids and their deuterated standards in the lipids purified from 2 mls of callus extract. Characteristic MS/MS ion pairs and eicosanoid peak retention times were used to identify each lipid mediator (Figure 3). Only eicosanoid MS/MS curves with peak values 10X greater than baseline were used for quantification. Consequently, identification and quantification of the eicosanoids was highly reproducible. The limit of quantification for each eicosanoid are noted into Table 6. Average percent recovery of the deuterated standards ranged from 9.5% for 6-keto-PGF1alpha to 62% for 5(S)-HETE (Table 6). We were able to detect and quantify all 10 eicosanoids in the day 4 fracture callus extracts (Table 6).
Figure 3. Representative Chromatograms from LC-MS/MS Measurements.
Lipids extracted from a mouse femur fracture callus frozen in liquid nitrogen and pulverized in RNAlater were derivatized and then separated on a reverse phase column. Endogenous eicosanoids (black lines) and deuterated internal standards (gray lines) were detected by MS/MS. Shown are chromatograms for TXB2 (A), PGD2 (B), and PGE2 (C) from a single fracture callus.
Table 6.
Lipid Mediator Yields: LN-RL Extraction of Day 4 Fracture Calluses
| Lipid Mediator | Sample Size |
Deuterated Standards: | Fracture Callus Eicosanoids: | |||||
|---|---|---|---|---|---|---|---|---|
| Percent Recovery | ng per mg protein | LOQ (ng) |
||||||
| Range | Mean | SD | Range | Mean | SD | |||
| PGD2 | 7 | 17-42 | 29.50 | 12.82 | 0.72-1.11 | 0.91 | 0.16 | 0.115 |
| PGE2 | 7 | 7-49 | 19.00 | 14.27 | 1.41-2.54 | 1.89 | 0.43 | 0.110 |
| PGF2α | 7 | 20-39 | 27.00 | 8.52 | 0.35-0.84 | 0.58 | 0.18 | 0.120 |
| TXB2 | 7 | 9-36 | 20.57 | 10.29 | 1.00-1.78 | 1.44 | 0.28 | 0.100 |
| 6-keto-PGF1α | 6 | 4-18 | 9.50 | 5.72 | 0.79-1.53 | 1.19 | 0.28 | 0.130 |
| 15-deoxy-Δ12,14-PGJ2 | 3 | 46-78 | 56.67 | 18.48 | 12.29-14.11 | 13.22 | 0.91 | 0.265 |
| LTB4 | 6 | 19-100 | 46.00 | 30.59 | 0.0-3.7 | 1.62 | 1.60 | 0.100 |
| 5(S)-HETE | 5 | 49-100 | 62.15 | 18.82 | 0.42-1.53 | 0.97 | 0.42 | 0.100 |
| Lipoxin A4 | 4 | 17-52 | 31.50 | 14.98 | 0.38-0.71 | 0.56 | 0.14 | 0.140 |
| 5-oxo-ETE | 7 | 23-100 | 52.43 | 31.89 | 0.35-1.58 | 0.82 | 0.43 | 0.240 |
Conclusions
For this project, our goal was to isolate lipid mediators, protein, and RNA from the same bone tissue sample. A variety of methods exist for extracting proteins [39-41], lipid mediators [6, 18], and RNA [13, 42] from bone. Unfortunately, these methods typically employ different initial extraction methods. Using the same initial extraction procedure as described here, eliminates the need to divide the initial tissue sample for each separate extraction procedure or use tissues from multiple animals in order to measure all 3 molecular classes. This method successfully extracted lipid mediators, proteins, and RNA simultaneously from a single bone callus tissue.
Supplementary Material
Acknowledgments
This project was supported by Award Number R01DE019926 from the National Institute of Dental & Craniofacial Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental & Craniofacial Research or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Claes L, Recknagel S, Ignatius A. Fracture healing under healthy and inflammatory conditions. Nature reviews. Rheumatology. 2012;8:133–143. doi: 10.1038/nrrheum.2012.1. [DOI] [PubMed] [Google Scholar]
- [2].Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. Journal of cellular biochemistry. 2003;88:873–884. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- [3].Simon AM, Manigrasso MB, O'Connor JP. Cyclo-oxygenase 2 function is essential for bone fracture healing. Journal of Bone and Mineral Research. 2002;17:963–976. doi: 10.1359/jbmr.2002.17.6.963. [DOI] [PubMed] [Google Scholar]
- [4].Simon AM, O'Connor JP. Dose and time-dependent effects of cyclooxygenase-2 inhibition on fracture-healing. The Journal of bone and joint surgery. American volume. 2007;89:500–511. doi: 10.2106/JBJS.F.00127. [DOI] [PubMed] [Google Scholar]
- [5].Cottrell JA, O'Connor JP. Pharmacological inhibition of 5-lipoxygenase accelerates and enhances fracture-healing. The Journal of bone and joint surgery. American volume. 2009;91:2653–2665. doi: 10.2106/JBJS.H.01844. [DOI] [PubMed] [Google Scholar]
- [6].Manigrasso MB, O'Connor JP. Accelerated fracture healing in mice lacking the 5-lipoxygenase gene. Acta orthopaedica. 2010;81:748–755. doi: 10.3109/17453674.2010.533931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haeggstrom JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- [8].Smith WL, Urade Y, Jakobsson PJ. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem Rev. 2011;111:5821–5865. doi: 10.1021/cr2002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kalish BT, Kieran MW, Puder M, Panigrahy D. The growing role of eicosanoids in tissue regeneration, repair, and wound healing. Prostaglandins & other lipid mediators. 2013;104-105:130–138. doi: 10.1016/j.prostaglandins.2013.05.002. [DOI] [PubMed] [Google Scholar]
- [10].Stables MJ, Gilroy DW. Old and new generation lipid mediators in acute inflammation and resolution. Progress in lipid research. 2011;50:35–51. doi: 10.1016/j.plipres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- [11].Barclay D, Zamora R, Torres A, Namas R, Steed D, Vodovotz Y. A simple, rapid, and convenient Luminex-compatible method of tissue isolation. J Clin Lab Anal. 2008;22:278–281. doi: 10.1002/jcla.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Welsh TN, Hubbard S, Mitchell CM, Mesiano S, Zarzycki PK, Zakar T. Optimization of a solid phase extraction procedure for prostaglandin E2, F2 alpha and their tissue metabolites. Prostaglandins & other lipid mediators. 2007;83:304–310. doi: 10.1016/j.prostaglandins.2007.02.004. [DOI] [PubMed] [Google Scholar]
- [13].Balaburski G, O'Connor JP. Determination of variations in gene expression during fracture healing. Acta orthopaedica Scandinavica. 2003;74:22–30. doi: 10.1080/00016470310013608. [DOI] [PubMed] [Google Scholar]
- [14].Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- [15].Lin H-N, O’Connor JP. Immunohistochemical Localization of Key Arachidonic Acid Metabolism Enzymes during Fracture Healing in Mice. PloS one. 2014;9:e88423. doi: 10.1371/journal.pone.0088423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hadjiargyrou M, Lombardo F, Zhao S, Ahrens W, Joo J, Ahn H, Jurman M, White DW, Rubin CT. Transcriptional profiling of bone regeneration. Insight into the molecular complexity of wound repair. The Journal of biological chemistry. 2002;277:30177–30182. doi: 10.1074/jbc.M203171200. [DOI] [PubMed] [Google Scholar]
- [17].Coords M, Breitbart E, Paglia D, Kappy N, Gandhi A, Cottrell J, Cedeno N, Pounder N, O'Connor JP, Lin SS. The effects of low-intensity pulsed ultrasound upon diabetic fracture healing. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2011;29:181–188. doi: 10.1002/jor.21223. [DOI] [PubMed] [Google Scholar]
- [18].Dekel S, Lenthall G, Francis MJ. Release of prostaglandins from bone and muscle after tibial fracture. An experimental study in rabbits. The Journal of bone and joint surgery. 1981;63-B:185–189. doi: 10.1302/0301-620X.63B2.7217139. British volume. [DOI] [PubMed] [Google Scholar]
- [19].Lader ES. Methods and reagents for preserving RNA in cell and tissue samples. 2001. Google Patents.
- [20].Manigrasso MB, O'Connor JP. Characterization of a closed femur fracture model in mice. Journal of orthopaedic trauma. 2004;18:687–695. doi: 10.1097/00005131-200411000-00006. [DOI] [PubMed] [Google Scholar]
- [21].Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Analytical biochemistry. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- [22].Vignali DA. Multiplexed particle-based flow cytometric assays. Journal of immunological methods. 2000;243:243–255. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- [23].Kawasaki ES, Clark SS, Coyne MY, Smith SD, Champlin R, Witte ON, McCormick FP. Diagnosis of chronic myeloid and acute lymphocytic leukemias by detection of leukemia-specific mRNA sequences amplified in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5698–5702. doi: 10.1073/pnas.85.15.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM, Moorman AF. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Analytical biochemistry. 2002;307:63–69. doi: 10.1016/s0003-2697(02)00021-0. [DOI] [PubMed] [Google Scholar]
- [25].Bollinger JG, Thompson W, Lai Y, Oslund RC, Hallstrand TS, Sadilek M, Turecek F, Gelb MH. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Analytical chemistry. 2010;82:6790–6796. doi: 10.1021/ac100720p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Deems R, Buczynski MW, Bowers-Gentry R, Harkewicz R, Dennis EA. Detection and quantitation of eicosanoids via high performance liquid chromatography-electrospray ionization-mass spectrometry. Methods in enzymology. 2007;432:59–82. doi: 10.1016/S0076-6879(07)32003-X. [DOI] [PubMed] [Google Scholar]
- [27].Surette ME, Odeimat A, Palmantier R, Marleau S, Poubelle PE, Borgeat P. Reverse-phase high-performance liquid chromatography analysis of arachidonic acid metabolites in plasma after stimulation of whole blood ex vivo. Analytical biochemistry. 1994;216:392–400. doi: 10.1006/abio.1994.1057. [DOI] [PubMed] [Google Scholar]
- [28].Blewett AJ, Varma D, Gilles T, Libonati JR, Jansen SA. Development and validation of a high-performance liquid chromatography-electrospray mass spectrometry method for the simultaneous determination of 23 eicosanoids. J Pharm Biomed Anal. 2008;46:653–662. doi: 10.1016/j.jpba.2007.11.047. [DOI] [PubMed] [Google Scholar]
- [29].Eberhard J, Jepsen S, Albers HK, Acil Y. Quantitation of arachidonic acid metabolites in small tissue biopsies by reversed-phase high-performance liquid chromatography. Analytical biochemistry. 2000;280:258–263. doi: 10.1006/abio.2000.4540. [DOI] [PubMed] [Google Scholar]
- [30].Wickramasinghe AJ, Shaw RS. An electron-capture gas-liquid-chromatographic method for the determination of prostaglandin F2alpha in biological fluids. The Biochemical journal. 1974;141:179–187. doi: 10.1042/bj1410179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee SH, Williams MV, DuBois RN, Blair IA. Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid communications in mass spectrometry : RCM. 2003;17:2168–2176. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- [32].Higashi T, Ichikawa T, Inagaki S, Min JZ, Fukushima T, Toyo'oka T. Simple and practical derivatization procedure for enhanced detection of carboxylic acids in liquid chromatography-electrospray ionization-tandem mass spectrometry. J Pharm Biomed Anal. 2010;52:809–818. doi: 10.1016/j.jpba.2010.03.001. [DOI] [PubMed] [Google Scholar]
- [33].Iwasaki Y, Nakano Y, Mochizuki K, Nomoto M, Takahashi Y, Ito R, Saito K, Nakazawa H. A new strategy for ionization enhancement by derivatization for mass spectrometry, Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2011;879:1159–1165. doi: 10.1016/j.jchromb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- [34].Bollinger JG, Rohan G, Sadilek M, Gelb MH. LC/ESI-MS/MS detection of FAs by charge reversal derivatization with more than four orders of magnitude improvement in sensitivity. J Lipid Res. 2013;54:3523–3530. doi: 10.1194/jlr.D040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Leng J, Wang H, Zhang L, Zhang J, Wang H, Guo Y. A highly sensitive isotope-coded derivatization method and its application for the mass spectrometric analysis of analytes containing the carboxyl group. Anal Chim Acta. 2013;758:114–121. doi: 10.1016/j.aca.2012.11.008. [DOI] [PubMed] [Google Scholar]
- [36].Turk J, Weiss SJ, Davis JE, Needleman P. Fluorescent derivatives of prostaglandins and thromboxanes for liquid chromatography. Prostaglandins. 1978;16:291–309. doi: 10.1016/0090-6980(78)90031-x. [DOI] [PubMed] [Google Scholar]
- [37].Wintersteiger R, Juan H. Prostaglandin determination with fluorescent reagents. Prostaglandins Leukot Med. 1984;14:25–40. doi: 10.1016/0262-1746(84)90021-0. [DOI] [PubMed] [Google Scholar]
- [38].Yue H, Strauss KI, Borenstein MR, Barbe MF, Rossi LJ, Jansen SA. Determination of bioactive eicosanoids in brain tissue by a sensitive reversed-phase liquid chromatographic method with fluorescence detection. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2004;803:267–277. doi: 10.1016/j.jchromb.2003.12.027. [DOI] [PubMed] [Google Scholar]
- [39].Urist MR, Strates BS. Bone morphogenetic protein. Journal of dental research. 1971;50:1392–1406. doi: 10.1177/00220345710500060601. [DOI] [PubMed] [Google Scholar]
- [40].Einhorn TA, Majeska RJ, Rush EB, Levine PM, Horowitz MC. The expression of cytokine activity by fracture callus. Journal of Bone and Mineral Research. 1995;10:1272–1281. doi: 10.1002/jbmr.5650100818. [DOI] [PubMed] [Google Scholar]
- [41].Jiang X, Ye M, Jiang X, Liu G, Feng S, Cui L, Zou H. Method development of efficient protein extraction in bone tissue for proteome analysis. J Proteome Res. 2007;6:2287–2294. doi: 10.1021/pr070056t. [DOI] [PubMed] [Google Scholar]
- [42].Cho T-J, Gerstenfeld LC, Einhorn TA. Differential temporal expression of members of the transforming growth factor β superfamily during murine fracture healing. Journal of Bone and Mineral Research. 2002;17:513–520. doi: 10.1359/jbmr.2002.17.3.513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.