Abstract
Background
We previously developed a prognostic classifier using the expression levels of BRCA1, HIF1A, DLC1, and XPO1 that identified stage I lung adenocarcinoma patients with a high risk of relapse. That study evaluated patients in five independent cohorts from various regions of the world. In an attempt to further validate the classifier, we have used a meta-analysis based approach to study 12 cohorts consisting of 1069 TNM stage I lung adenocarcinoma patients from every suitable, publically available dataset.
Materials and Methods
Cohorts were obtained through a systematic search of public gene expression datasets. These data were used to calculate the risk score using the previously published 4-gene risk model. A fixed effects meta-analysis model was used to generate a pooled estimate for all cohorts.
Results
The classifier was associated with prognosis in ten of the twelve cohorts (p<0.05). This association was highly consistent regardless of the ethnic diversity or microarray platform. The pooled estimate demonstrated that patients classified as high risk had worse overall survival for all stage I (Hazard Ratio [HR], 2.66; 95% Confidence Interval [CI], 1.93-3.67; P<0.0001) patients and in stratified analyses of stage IA (HR, 2.69; 95%CI, 1.66-4.35; P<0.0001) and stage IB (HR, 2.69; 95%CI, 1.74-4.16; P<0.0001) patients.
Conclusions
The -4-gene classifier provides independent prognostic stratification of stage IA and stage IB patients beyond conventional clinical factors
Impact
Our results suggest that the 4-gene classifier may assist clinicians in decisions regarding postoperative management of early stage lung adenocarcinoma patients.
Introduction
Lung cancer is the leading cause of cancer-death in the world, accounting for more than one-fourth of all cancer-deaths (1). Approximately 85% of lung cancers are non-small cell lung cancer (NSCLC). The most common histology for NSCLC is adenocarcinoma (ADC), followed by squamous cell carcinoma (SQC) and large cell carcinoma. Despite therapeutic advances, prognosis remains considerably poor relative to other solid cancers, even in early stage patients (1). Thus, more refined treatment strategies are needed.
TNM staging is the best prognostic factor for NSCLC. TNM staging is used by clinicians to guide treatment options for NSCLC. Early stage patients, including TNM stage I and II, are typically approached with curative surgery as the optimal treatment. Among such patients with completely resected NSCLC, adjuvant chemotherapy is recommended only for stage II patients based on several randomized trials that demonstrated survival benefit of platinum-based chemotherapy (2–4). By contrast, clinical trials have revealed no survival advantage and potential deleterious side-effects of adjuvant chemotherapy for stage IA patients (2, 5). With regard to stage IB patients, the evidence supporting routine use of adjuvant chemotherapy is controversial (2, 6,7). A more detailed histological subtyping of lung cancer may improve on TNM classification system. For example, the presences of the micropapillary histologic subtype has been found to be associated with cancer recurrence after limited resection of peripheral lung ADC and my help guide treatment strategies (8).
Approximately 30% of stage I lung cancer patients will relapse and ultimately die of this disease. The majority of these patients are being treated by surgery alone because of the lack of clear evidence of benefit from adjuvant chemotherapy. Consequently, 5-year overall survival rates for pathological stage IA and IB are 73% and 58%, respectively, based on the recently-revised, 7th edition of TNM staging (9). One simple and critical question is how clinicians can distinguish the approximately 30% of stage I patients who have higher risk of relapse from the other 70% of patients who have excellent prognosis. High-risk patients might have undetectable micrometastases at the time of surgery. Hence their outcome could potentially be improved by postoperative systemic therapy with the primary goal of eliminating residual occult metastases that lead to disease recurrence. There is a substantial need to identify stage IB patients who are unlikely to benefit from adjuvant chemotherapy and/or immunotherapy as well as stage IA patients who have the highest risk of relapse. In view of that, it is vital to develop prognostic biomarkers that can help clinicians determine appropriate postoperative management for each individual patient. The demand for such clinical prognostic tests is now undoubtedly increasing, as the extensive use of computed tomography (CT) screening becomes widely accepted, in which the majority of patients are diagnosed at stage I (10).
Numerous studies have identified prognostic biomarkers for NSCLC based on multigene expression by using qRT-PCR and/or microarray technology (11–26). However, associations reported in single studies often failed to provide sufficient validation in additional populations (12, 26,27). A recent review criticized prognostic gene signatures for their unspecified clinical utility as well as the lack of reproducibility, and suggested that no lung cancer signatures are ready for clinical application (27). Taking into account the guidelines suggested in that review, we started to develop a gene expression-based prognostic signature that was intended to be used for early stage lung ADCs, especially for stage I patients. Our goal was to make a classifier based on a few key genes that would be a simple and robust classifier for prognosis of stage I lung cancer patients. A similar strategy based on analyzing important 31 cell proliferation genes has shown to be a robust prognostic classifier for lung cancer (28). Our resulting signature, namely the 4-gene classifier, was found to be highly robust in all 5 cohorts that we analyzed. Its prognostic significance was independent of other clinical factors, including age, gender, TNM stage and smoking status (29). These results suggest that the 4-gene classifier may be useful in guiding therapeutic decisions for early stage lung cancer patients. We have now set out to test the 4-gene classifier in as many independent populations as we could identify from publically available gene expression data. As a result, we have evaluated more than 1000 stage I ADC patients from 12 independent cohorts using different gene expression platforms. We use a meta-analytic approach to measure the association of the 4-gene classifier with prognosis and evaluate its reproducibility across those cohorts. We focus on stage IA and IB patients separately to further evaluate the clinical usefulness of this classifier.
Materials and Methods
Selection of studies
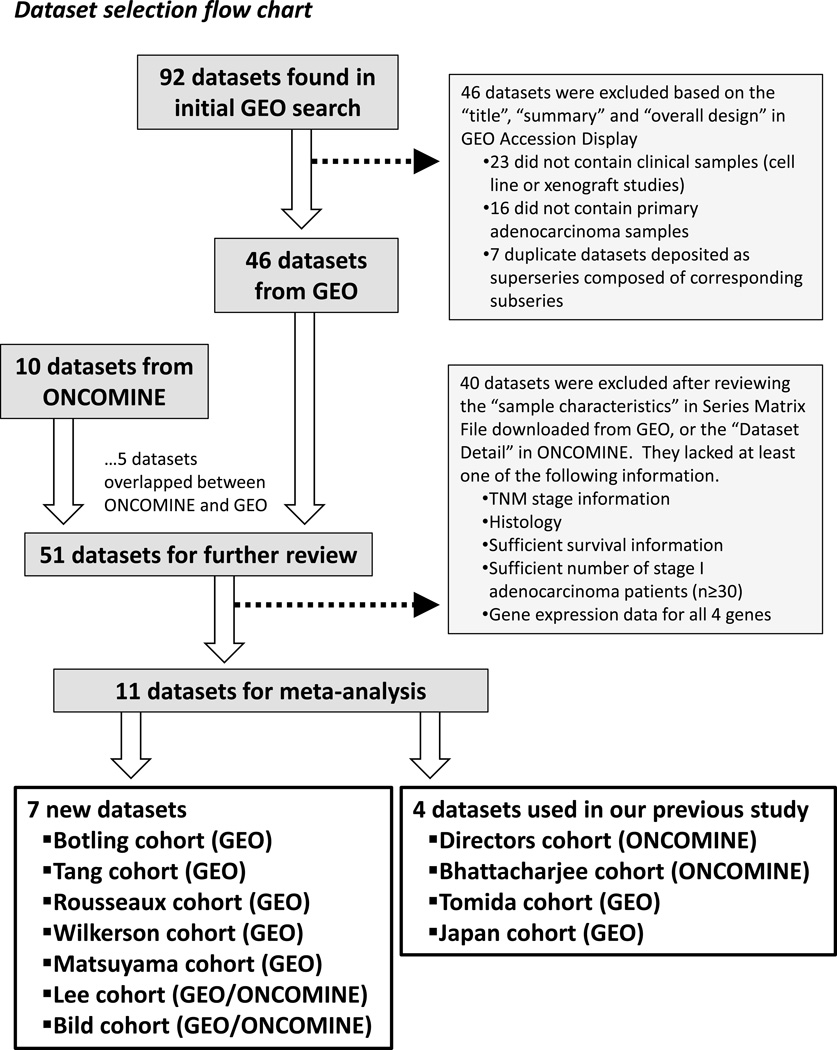
We searched GEO (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/) in June 2013 with the search terms “lung cancer”, “non-small cell lung cancer”, “lung adenocarcinoma”, “lung adenocarcinomas” and “NSCLC”. The retrieved GEO series were filtered by Organism (Homo Sapiens) and Series Type (Expression Profiling by Array) as well as sorted by the number of samples (series that have at least 30 samples). Ninety-two GEO series identified by the initial GEO search were screened on the basis of their Title, Summary and Overall Design as described in GEO Accession Display. Datasets were excluded if they analyzed only cell lines/xenograft samples, only non-tumor specimens (e.g., bronchial epithelial cells, blood, fluid), or contained no primary ADC tumors. Also, several superseries that consisted of one or more subseries were excluded (due to duplicate data) and the corresponding subseries with gene expression data were retrieved, leaving 46 GEO datasets of lung cancer-related clinical studies. In parallel with this search, we used ONCOMINE (Compendia Bioscience, Ann Arbor, MI; http://www.oncomine.com) to identify public microarray datasets that had ADC patients with survival status. ONCOMINE search identified 10 datasets, 5 of which were not deposited in GEO. The resulting 51 datasets containing primary ADC samples were further reviewed based on the Sample Characteristics in Series Matrix File, or the Dataset Detail in ONCOMINE. Selection criteria for all publicly available datasets required each dataset to include survival information for more than 30 TNM stage I patients of ADC and have expression data for BRCA1, HIF1A, DLC1 and XPO1. After removing 40 datasets that did not fit our criteria, we found 11 independent microarray datasets, including the Botling (GSE37745) (30), Tang (GSE42127) (24), Rousseaux (GSE30219) (31), Matsuyama (GSE11969) (32), Wilkerson (GSE26939) (33), Lee (GSE8894/ONCOMINE) (17), Bild (GSE3141/ONCOMINE) (34) cohorts as well as the Bhattacharjee (ONCOMINE) (35), Directors (ONCOMINE) (12), Japan (GSE31210) (36), Tomida (GSE13213) (16) cohorts. Among them, the former 7 cohorts were newly obtained from GEO or ONCOMINE (if available) for this present study, whereas the latter 4 datasets were the original cohorts that we analyzed in our initial study (29). The selection flowchart and the list of retrieved datasets are presented in Figure 1 and Supplementary Table 1. All of the data from the stage I lung adenocarinoma cohorts are available in the supplemental data.
Figure 1.
Dataset selection flow chart. A total of 92 datasets from GEO and 10 datasets from ONCOMINE were evaluated. A total of 11 datasets were selected to be included in this meta-analysis.
For the 4-coding gene analyses in SQC patients, we used multiple cohorts of stage I SQC. Six cohorts, including the Botling, Rousseaux, Tang, Matsuyama, Lee and Bild datasets among the ADC datasets mentioned above, were included, as these cohorts also contained expression data for SQC patients with survival information. We obtained one SQC dataset from GEO (GSE17710) deposited by Wilkerson et al. (37), separated from their ADC data (GSE26939, the Wilkerson ADC cohort) (33). Additionally, among 3 SQC datasets with survival information which were found in ONCOMINE, including the Raponi (SQC only) (38), Larsen (SQC only) (39) and Zhu (ADC and SQC) (11) cohort, the Raponi and the Zhu cohorts were included in SQC analyses. For the Zhu cohort, only SQC patients were analyzed, while ADC patients (n=14, Stage I) were disregarded, since considerable number of ADC patients were already used within (CAN/DF) of the Directors cohort (11). The Larsen cohort was excluded because BRCA1 gene was not available in their platform.
Gene expression data analysis
In this study, we focused only on stage I patients. The 4-coding gene analysis of five original cohorts used AJCC TNM 6th edition as described previously (29). Concerning 7 new cohorts, although the TNM edition was not specified as either 6th or 7th in each of original papers, we assumed that they were based on AJCC TNM 6th edition, since most tumors were collected before the development of TNM 7th edition in 2009. For the Rousseaux cohort, T1N0 tumors were defined as stage IA, while T2N0 tumors were defined as stage IB, according to the provided TNM classification for each patient. Among all available stage I cases obtained from the public datasets, 2 ADCs and 1 SQC in the Tang cohort, 3 ADCs and 4 SQCs in the Lee cohort were excluded from the analysis, since survival information was not provided for those cases.
For all analyses, the normalized expression values were obtained from each dataset and were not processed further. We then generated criteria to select the most reliable, informative probes. In brief, if there were multiple probes to a single gene, pairwise correlation of each probe was analyzed in each cohort. Probes were removed if they showed no correlation (R<0.5) between any of the other probes for that gene. If there were only 2 probes to a single gene that didn't correlate with each other, the probe with the highest expression was selected. Probes that were selected are shown in Supplementary Table 2. If more than one probe was selected, they were averaged and no further processing was performed. The 4-coding gene classifier [(0.104 × BRCA1) + (0.133 × HIF1A) + (−0.246 × DLC1) + (0.378 × XPO1)] was applied to all newly-obtained cohorts using microarray expression data, and the resulting classifier score was categorized as low, medium, or high based on tertiles within each cohort separately. The within cohort categorization was performed to standarize risk scores across all cohorts. This will compensate for the fact that each study used different methodologies to measure the expression of each of the four genes and these expression values are not directly comparable to one another across cohorts. The association between the 4-gene classifier and survival was assessed by the Kaplan–Meier log-rank test for trend using Graphpad Prism v5.0 (Graphpad Software Inc). Cox regression analyses were carried out using SPSS 11.0 (SPSS Inc), and all univariate and multivariate models were adjusted for cohort membership where appropriate. Forest plot analyses and calculations for the nomograms were performed using Stata 11.2 (Stata-Corp LP). Heterogeneity test for the combined HR was carried out using the I-squared statistics (40). Nomograms were developed based on coefficients from multivariable Cox regression models on 5-year overall survival using all variables that were significantly associated with patient outcome.
Results
Identification of eligible studies
Since the purpose of this gene expression-based classifier is to identify high-risk, stage I ADC patients who may benefit from additional intervention after surgery, we limited all the analyses in this study to stage I primary ADC tumors. The systematic search identified 11 microarray datasets consisting each of more than 30 cases of stage I ADC patients with sufficient survival information with gene expression data for all 4 genes, including BRCA1, HIF1A, DLC1 and XPO1, as described in Figure 1. Four of the 11 datasets were previously analyzed for our initial paper describing the signature, in which 5 independent cohorts of stage I ADCs were each analyzed by qRT-PCR and/or microarrays (29). Hence, 7 independent cohorts were newly obtained through this systematic search and a total of 12 cohorts were included in this study. The characteristics of the 12 cohorts are summarized in Table 1. This analysis includes 1069 patients in total, consisting of 546 stage IA and 518 stage IB cases (5 cases were not specified as stage IA or IB). These cohorts were derived from 6 different countries, including Japan, Norway, Sweden, France, South Korea, as well as at least 8 different institutions in the United States. Nine of 12 cohorts reported overall survival information, 2 cohorts reported relapse-free survival and 1 cohort reported cancer-specific survival. In each cohort, RNA samples were isolated from frozen tumor specimens and were subjected to gene expression analysis based on various platforms, including qRT-PCR and Affymetrix, Illumina, or Agilent microarrays. (Table 1)
Table 1.
Twelve independent cohorts of stage I, lung adenocarcinoma patients
| Cohorts | Country | n | TNM Stage |
Age |
Gender |
Smoker (%) | Postoperative Therapy |
Outcome | Platform | GEO ID | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IA | IB | IAor IB |
Mean | M | F | CT /RT |
None | Unknown | |||||||
| Five cohorts | |||||||||||||||
| Japan | Japan | 149 | 100 | 49 | 0 | 59.7 | 66 | 83 | 45.6 | 0 | 149 | 0 | RFS | qRT-PCR | - |
| US/Norway | USA (UMD), Norway | 67 | 29 | 38 | 0 | 64.6 | 37 | 30 | 96.9 | 4 | 43 | 20 | CSS | qRT-PCR | - |
| Directors | USA (MSK,HLM,CAN/DF,UM) | 276 | 114 | 162 | 0 | 64.4 | 131 | 145 | NA | 46 | 157 | 73 | OSa | Affymetrix U133A | NAb |
| Bhattacharjee | US (Harvard) | 76 | 35 | 40 | 1 | 64.2 | 32 | 44 | 90.8 | 0 | 0 | 76 | OSa | Affymetrix U95A | NAb |
| Tomida | Japan | 79 | 42 | 37 | 0 | 61.4 | 41 | 38 | 50.6 | 0 | 79 | 0 | OSa | Agilent 44K | GSE13213 |
| Seven new cohorts | |||||||||||||||
| Tang | USA (MD Anderson) | 87 | 32 | 55 | 0 | 64.1 | 37 | 50 | NA | 22 | 65 | 0 | OSa | Illumina WG6 V3 | GSE42127 |
| Rousseaux | France | 81 | 73 | 8 | 0 | 61.8 | 65 | 16 | NA | 0 | 0 | 81 | OSa | Affymetrix U133+2 | GSE30219 |
| Botling | Sweden | 70 | 28 | 42 | 0 | 63.5 | 31 | 39 | NA | 5 | 31 | 34 | OSa | Affymetrix U133+2 | GSE37745 |
| Wilkerson | USA (UNC) | 62 | 31 | 31 | 0 | 65.6 | 26 | 36 | 88.0 | 0 | 0 | 62 | OSa | Agilent 44K custom | GSE26939 |
| Matsuyama | Japan | 52 | 28 | 24 | 0 | 62.3 | 28 | 24 | 46.2 | 0 | 0 | 52 | OSa | Agilent 21.6K custom | GSE11969 |
| Lee | Korea | 36 | 13 | 23 | 0 | 61.4 | 16 | 20 | 38.9 | 0 | 0 | 36 | RFS | Affymetrix U133+2 | GSE8894b |
| Bild | USA (Duke) | 34 | 21 | 9 | 4 | 64.8 | 17 | 17 | NA | 0 | 0 | 34 | OSa | Affymetrix U133+2 | GSE3141b |
| Total | 1069 | 546 | 518 | 5 | 63.1 | 527 | 542 | 77 | 524 | 468 | |||||
NOTE:
Nine cohorts with overall survial information were used in the combined analysis (n=817).
Data were obtained from ONCOMINE.
Abbreviation: CT/RT, chemotherapy and/or radiotherapy; NA, not available; RFS, relapse-free survival; CSS, cancer-specific survival; OS, overall survival.
The 4-gene classifier is tested in 12 independent cohorts
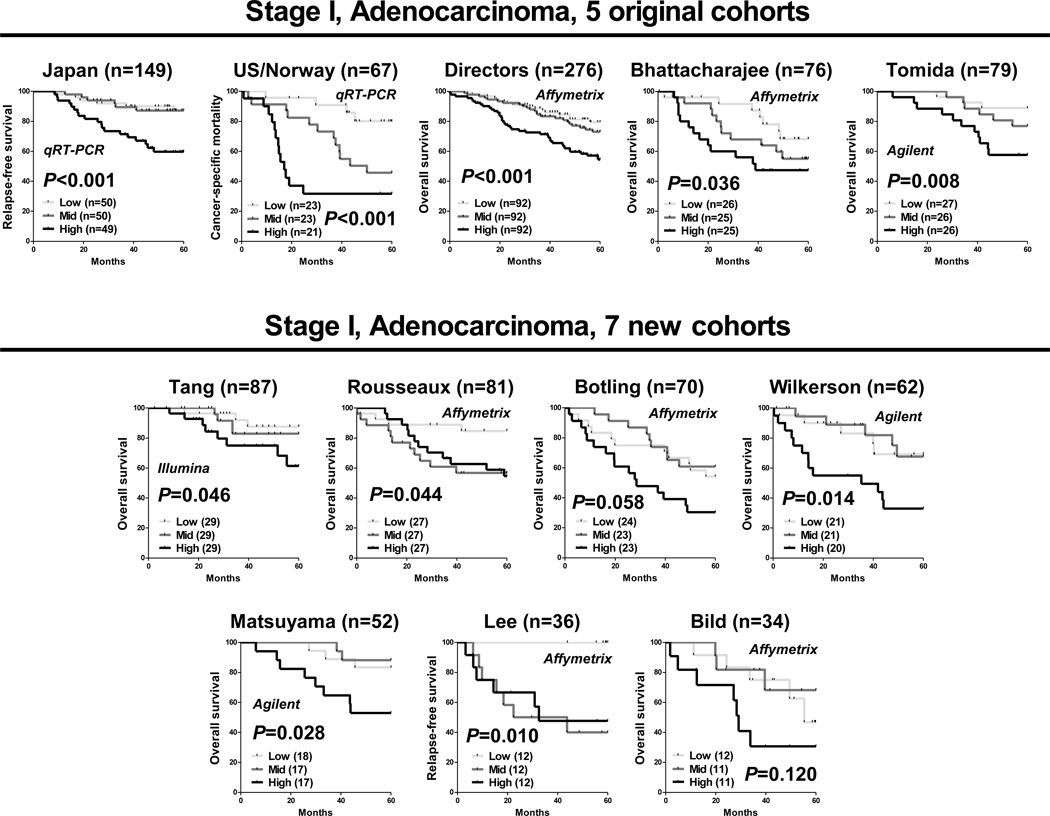
The 4-gene classifier was applied to each of the 7 newly-obtained cohorts using microarray expression data for 4 genes, and then cases were categorized as high, medium or low, based on tertiles for each cohort separately. Similar to our previous results, highly concordant associations were found between the 4-gene classifier and prognosis in all 7 newly-obtained cohorts, including the Tang (p=0.046), Rousseaux (p=0.044), Wilkerson (p=0.014), Matsuyama (p=0.028), Lee (p=0.010), Botling (p=0.058) and Bild (p=0.120) cohorts by the Kaplan-Meier analysis (Figure 2). Overall, the 4-gene classifier was significantly associated with prognosis in 10 cohorts, while the remaining 2 cohorts had marginal associations in the proper direction.
Figure 2.
The performance of the 4-coding gene classifier in 12 independent cohorts of stage I lung adenocarcinoma patients. For each cohort, cases were categorized as high, medium or low based on tertiles. P-values were obtained by the log-rank test for trend.
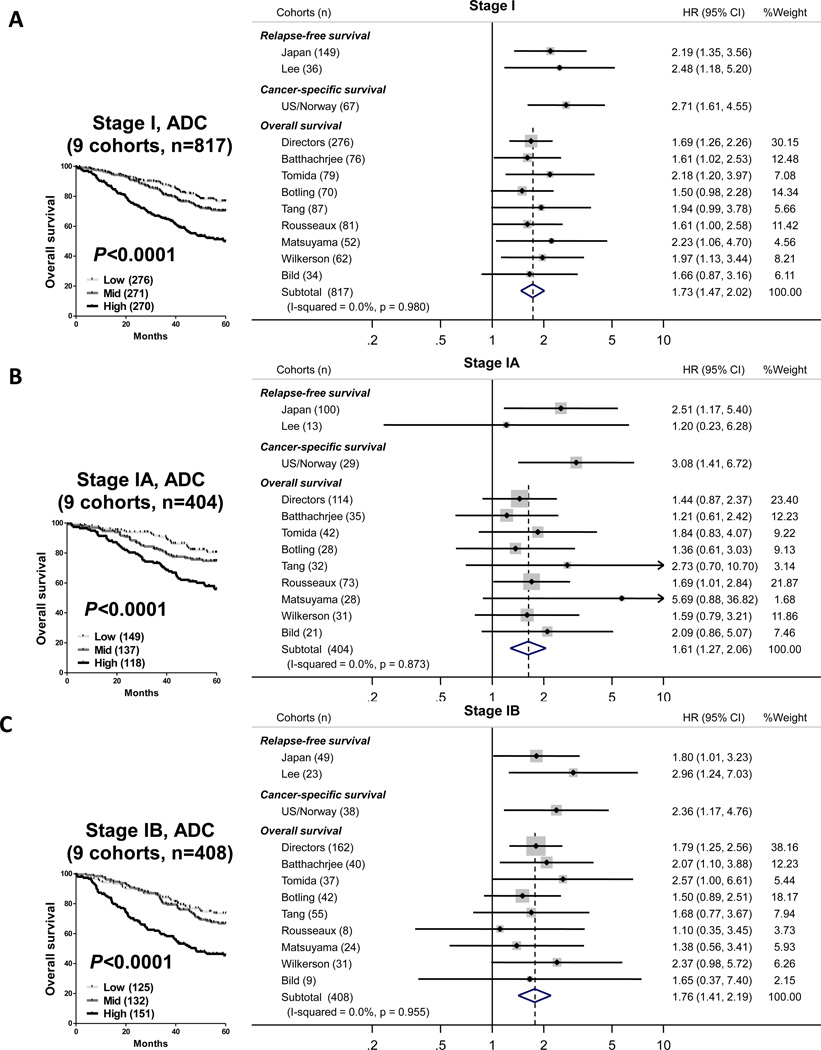
The 9 cohorts that had overall survival information were analyzed in a fixed effects meta-analysis model, which included 817 stage I cases. There was no evidence for heterogeneity or inconsistency across multiple cohorts (I-squared=0.0%, p=0.980), suggesting that these results are representative of most lung ADCs and not a result of selection bias (Figure 3). Patients that were classified as high risk had significantly worse overall survival (HR, 1.73; 95%CI, 1.47-2.02) in stage I analysis (Figure 3A). The corresponding Kaplan-Meier analysis for the combined stage I patients demonstrated a significant association with overall survival and the 4-gene classifier (p<0.0001; Figure 3A). Furthermore, stratified analyses were performed for stage IA and IB separately, to address the prognostic impact of this classifier in these subgroups. Significant associations between the 4-gene classifier and overall survival were found in both stage IA (HR, 1.61; 95%CI, 1.27-2.06) and stage IB (HR, 1.76; 95%CI, 1.41-2.19) analyses, respectively (Figure 3B and C).
Figure 3.
Forest plot of the prognostic impact of the 4-coding gene classifier in 12 independent cohorts of stage I lung adenocarcinoma. A) Meta-analysis of all patients with TNM stage IA or IB lung adenocarcinoma. B) Meta-analysis of all patients with TNM stage IA lung adenocarcinoma. C) Meta-analysis of all patients with TNM stage IB lung adenocarcinoma.
The 4-gene classifier is an independent prognostic biomarker for stage IA as well as stage IB patients
Given that the classifier is significantly associated with survival in stage IA and IB subgroups, Cox regression analysis was conducted using the combined cohort with respect to each stage (Table 2). All univariate and multivariate Cox analyses were adjusted for cohort membership and multivariate models were adjusted for age, gender and TNM stage. Since most of the public datasets did not provide complete clinical information, we could not apply other parameters, such as smoking status or adjuvant chemotherapy to the Cox analysis. In univariate analysis, older age, male gender, TNM stage IB and the 4-gene classifier were each significantly associated with worse outcome. Multivariate models revealed that the 4-gene classifier was significantly associated with poor overall survival, independent of other parameters, in stage I patients (HR, 2.66; 95%CI, 1.93-3.67; P<0.0001) and in stratified analyses of stage IA (HR, 2.69; 95%CI, 1.66-4.35; P<0.0001) and stage IB (HR, 2.69; 95%CI, 1.74-4.16; P<0.0001) patients. We have also performed an analysis of the risk score as a linear variable (rather than a ordered categorical variable) to demonstrate these associations are highly robust and do not rely on using tertiles as cutpoints for the data (Supplementary Table 3)
Table 2.
Univariable and Multivariable Cox regression of the 4-coding gene classifier in the combined cohorta of Stage I, adenocarcinoma patients
| Variable (n) | Univariable Analysis b |
Multivariable Analysis b |
|||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| TNM Stage I (n=817) | |||||
| 4 gene classifier c | Low (276) | Reference | NA | Reference | NA |
| Medium (271) | 1.34 (0.95 ― 1.89) | 0.101 | 1.27 (0.89 ―1.80) | 0.183 | |
| High (270) | 2.83 (2.07 ― 3.86) | <0.0001 | 2.66 (1.93 ― 3.67) | <0.0001 | |
| Trend P< 0.0001 | Trend P< 0.0001 | ||||
| Staged | IB (408) / IA (404) | 1.68 (1.29 ― 2.19) | 0.0001 | 1.55 (1.19 ― 2.03) | 0.001 |
| Age | Continous | 1.03 (1.02 ― 1.05) | <0.0001 | 1.04 (1.02 ― 1.05) | <0.0001 |
| Gender | Female (409) / Male (408) | 0.67 (0.52 ― 0.87) | 0.002 | 0.78 (0.60 ― 1.01) | 0.062 |
| TNM Stage IA (n=404) | |||||
| 4 gene classifier c | Low (149) | Reference | NA | Reference | NA |
| Medium (137) | 1.47 (0.87 ― 2.49) | 0.151 | 1.42 (0.84 ―2.40) | 0.191 | |
| High (118) | 2.69 (1.67 ― 4.34) | <0.0001 | 2.69 (1.66 ― 4.35) | <0.0001 | |
| Trend P< 0.0001 | Trend P< 0.0001 | ||||
| Age | Continous | 1.03 (1.01 ― 1.06) | 0.002 | 1.04 (1.02 ― 1.06) | 0.0007 |
| Gender | Female (205) / Male (199) | 0.61 (0.40 ― 0.91) | 0.016 | 0.65 (0.43 ― 0.99) | 0.043 |
| TNM Stage IB (n=408) | |||||
| 4 gene classifier c | Low (125) | Reference | NA | Reference | NA |
| Medium (132) | 1.20 (0.74 ― 1.93) | 0.456 | 1.14 (0.71 ― 1.84) | 0.586 | |
| High (151) | 2.88 (1.88 ― 4.43) | <0.0001 | 2.69 (1.74 ― 4.16) | <0.0001 | |
| Trend P< 0.0001 | Trend P< 0.0001 | ||||
| Age | Continous | 1.04 (1.02 ― 1.05) | <0.0001 | 1.03 (1.02 ― 1.05) | <0.0001 |
| Gender | Female (203) / Male (205) | 0.75 (0.54 ― 1.06) | 0.102 | 0.90 (0.64 ― 1.26) | 0.533 |
The combined cohort consists of 9 publicly availble, independent microarray datasets of stage I patients with overall survival information, including the Directors (276), Bhattacharjee (76), Tomida (79), Botling (70), Tang (87), Rousseaux (81), Matsuyama (52), Wilkerson (62), and Bild (34) cohorts.
The univariable models was adjusted for cohort membership and the multivariable model included the 4 gene classifier, cohort membership, age, gender, and TMN staging.
The 4-coding gene classifier was categorized based on tertiles of Stage I patients for each cohort.
There were a total of 5 stage I cases in the Bhattachrjee (1) and Bild (4) cohorts for which stage IB/IA infromation is not available. These are included in univariate analyses and excluded in multivariate analyses.
The potential use of the 4-gene classifier to predict prognosis for stage I lung adenocarcinoma
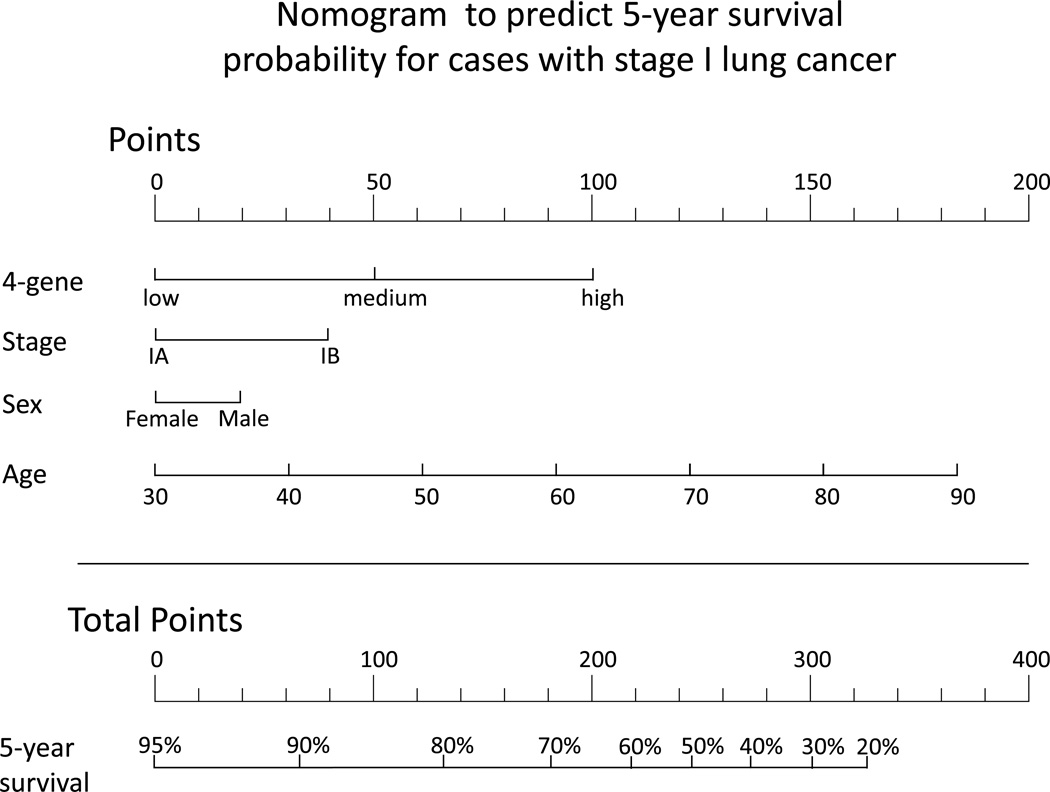
In order for a prognostic classifier to be clinically useful, it has to provide actionable information to the physician. To demonstrate this potential for the 4-gene classifier, we developed a nomogram to predict 5-year survival rates in patients diagnosed with stage I lung ADC (Figure 4). This nomogram is based on the 9 cohorts with overall survival data and includes all variables that were significantly associated with 5-year overall survival. The points assigned to each variable are weighted based on Cox regression coefficients. This nomogram demonstrates that the 4-gene classifier could be used with clinical staging and other clinical parameters to predict the probability of 5-year survival. Subgroup analysis within stage is important to demonstrate to show clinical utility, therefore we created nomograms stratified by TNM stage IA and IB separately as another example of how this classifier can be integrated with TNM staging to help determine patient prognosis (Supplementary Figure 1).
Figure 4.
Nomogram to predict 5-year survival rates for stage I lung adenocarcinoma. Each clinical variable (4-gene score, TNM Stage, sex and age) is assigned a point value. The sum of those points can then be used to estimate probability of survival for 5-years. For example, the sum of the points is 300, a patient has approximately a 30% 5-year survival probability.
The 4-gene classifier is only applicable to adenocarcinoma patients
Up to now we have focused only on lung cancer patients with ADC histology. To determine if this association could be observed across different histologies, we examined the 4-gene classifier in SQC, which is another major histological type of NSCLC. Nine independent cohorts, consisting of 337 stage I SQC patients, were obtained and the 4-gene classifier was applied to each cohort (Supplementary Table 4). In a combined analysis, the 8 cohorts with overall survival information were combined (n=292). However, no significant association was found in any of the SQC analyses (Supplementary Figure 2), indicating that the 4-gene classifier is specific to ADC. This was not completely unexpected because we built this classifier using ADC gene expression data only. SQC and ADC are also considered to be molecularly distinct entities (41). Therefore, this classifier appears to be only useful for lung ADC and not SQC. Sufficient numbers were not available to examine other histologies.
Discussion
Translating prognostic gene signatures into clinical use is a major challenge in the field of lung cancer research. There is little doubt that prognostic tests are necessary for stage I lung cancer patients after complete resection. In breast cancer, it is striking to note that several multigene assays are already commercially available and are currently used by clinical oncologists and supported by the NCCN and other guidelines (42, 43). As for lung cancer, no prognostic biomarkers have been incorporated into the current guidelines despite a large number of published gene signatures. This may be at least in part due to insufficient reproducibility as well as the lack of large-scale validation (27). Also, it has been suggested that many signatures were developed without clear focus on specific clinical contexts (27). In order to address those issues, our study has set out to test whether the 4-gene classifier that we previously identified is a robust prognostic classifier for stage I lung ADC using every publically available dataset. The 4-gene classifier was a robust classifier for over 1000 TNM Stage I lung ADC cases from 12 cohorts regardless of ethnic difference in the genetic background of the patients. When each cohort was separately analyzed, the 4-gene classifier showed highly consistent results for its association with survival. The classifier was highly reproducible across multiple platforms for gene expression measurement, including qRT-PCR and commercial/custom microarrays from Affymetrix, Agilent and Illumina. There is no evidence of selection bias in any of the 12 cohorts, which suggests these results presented here are representative of stage I lung ADC. However, the classifier had no prognostic impact on SQC patients, indicating its limited utility only to lung ADCs.
The 4-gene classifier has potential to be used as a prognostic biomarker for the management of stage IA and stage IB ADC patients in the current clinical setting. The pooled estimate revealed significant association between the 4-gene classifier and prognosis in stage IA and IB lung ADC subgroups, independent of other clinical variables. This suggests that the classifier can add additional discriminative value to identify high-risk patients beyond conventional clinical characteristics. Hence, “low-risk” stage IB patients identified by the classifier and predicted to have excellent survival probabilities may be recommended to forgo adjuvant therapy. Likewise, the classifier may also identify “high-risk” stage IA patients for whom intensive postoperative intervention is considered. Future work should explore and identify the optimal cutpoint for this assay that should distinguish "high-risk" and "low-risk patients". For convenience, we have used tertile as a cutpoint for our studies to distinguish between high/medium/low risk. It is likely that optimal cutpoints can be found that will improve the clinical utility of this classifier.
Many published multigene signatures for NSCLC had utilized dozens to hundreds, or even thousands of genes along with complex classification models that are difficult to understand. We believe that this is an obstacle to the rapid development and feasibility of the clinical tests. By contrast, the 4-gene classifier uses the expression values of only 4 genes, potentially providing an opportunity to develop a simple and practical laboratory test. The classifier is composed of biologically relevant genes that are mechanistically important and are each significantly associated with prognosis in early stage lung ADC (29). We consider that our strategy of focusing on only biologically relevant genes have improved the chances of developing a robust classifier that will be generalizable to lung ADC.
A potential limitation for this analysis is that there was incomplete data on smoking status and types of chemotherapy that were retrieved for several of the cohorts used for the meta-analysis. Therefore, we were not able to include these covariates in the models. Smoking history was considered as a covariate within the discovery cohorts and found to not contribute to the risk association model (29). Adjuvant chemotherapy is not recommended for stage I patients, thus the majority of the patients included in our analysis would not have received any. In fact, only 4% of patients in the Japan discovery cohort received adjuvant chemotherapy (29). Still, these factors are important for survival after lung cancer surgery and it is possible that their inclusion may modulate the association between the 4-gene classifier and prognosis.
We have shown that the classifier is robust and that using qRT-PCR data and microarray data from a variety of labs provides similar results. In order to turn the 4-gene classifier into a clinical test, future work should focus on developing a standardized assay which will include developing methods for measuring each of the four genes and recommending methods for tissue handling, processing and RNA isolation. Assays will have to be designed to minimize the potential batch effects and inter-laboratory differences. Another future possibility is the use of RNA samples extracted from formalin-fixed paraffin-embedded (FFPE) tissues. This could extend the practical utility of the classifier to readily-available archived specimens. Furthermore, our previous study demonstrated an improved prognostic association by combining multiple, validated classifiers, namely the combination of the 4-coding gene classifier with non-coding microRNA-21 (miR-21) classifier for stage I lung ADC (29). In addition to the present results, recent meta-analysis studies demonstrating miR-21 as a promising prognostic biomarker for NSCLC may be supportive to the potential combined use of the 4-gene and miR-21 classifiers as validated biomarkers (44, 45). This demonstrates that the integration of multiple, independent classifiers can further improve prognostic predictions and suggests that these classifiers can be combined with additional biomarkers and histological subtyping data to improve decision making capabilities for early stage lung cancer.
In conclusion, the 4-gene classifier that we recently developed was rigorously validated in large-scale, multiple cohorts with a meta-analytic approach consisting of more than 1000 stage I patients. To our knowledge, this is the first report of an RNA-based classifier in lung ADC to be tested and validated this extensively. The reproducibility of its performance was clearly demonstrated in the intended clinical context based on unbiased approaches. Particularly, the classifier provides additional prognostic stratification beyond the current risk factors, namely, stage IA and stage IB subgroups, highlighting the potential of this classifier in personalized management for early stage patients. These results support the development of standardized tests for the 4-gene assays and the incorporation of these assays into prospective studies.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute, NIH and a Department of Defense Congressionally Directed Medical Research Program Grant PR093793
Footnotes
Conflict of Interest Statement:
The authors have no conflicts of interest to declare.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 3.NCCN Clinical Practice Guidelines in Oncology. http://www.nccn.org.
- 4.Howington JA, Blum MG, Chang AC, Balekian AA, Murthy SC. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e278S–e313S. doi: 10.1378/chest.12-2359. [DOI] [PubMed] [Google Scholar]
- 5.Arriagada R, Auperin A, Burdett S, Higgins JP, Johnson DH, Le Chevalier T, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 2010;375:1267–1277. doi: 10.1016/S0140-6736(10)60059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butts CA, Ding K, Seymour L, Twumasi-Ankrah P, Graham B, Gandara D, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: updated survival analysis of JBR-10. J Clin Oncol. 2010;28:29–34. doi: 10.1200/JCO.2009.24.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss GM, Herndon JE, 2nd, Maddaus MA, Johnstone DW, Johnson EA, Harpole DH, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitadori J, Bograd AJ, Kadota K, Sima CS, Rizk NP, Morales EA, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. Journal of the National Cancer Institute. 2013;105:1212–1220. doi: 10.1093/jnci/djt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 10.Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan F, et al. Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med. 2013;368:1980–1991. doi: 10.1056/NEJMoa1209120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 14.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 15.Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu CQ, Strumpf D, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 16.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 17.Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14:7397–7404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 18.Boutros PC, Lau SK, Pintilie M, Liu N, Shepherd FA, Der SD, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106:2824–2828. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–3444. doi: 10.1172/JCI32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raz DJ, Ray MR, Kim JY, He B, Taron M, Skrzypski M, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res. 2008;14:5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26:877–883. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
- 23.Roepman P, Jassem J, Smit EF, Muley T, Niklinski J, van de Velde T, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15:284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen DT, Hsu YL, Fulp WJ, Coppola D, Haura EB, Yeatman TJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst. 2011;103:1859–1870. doi: 10.1093/jnci/djr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo NL, Wan YW, Tosun K, Lin H, Msiska Z, Flynn DC, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wistuba II, Behrens C, Lombardi F, Wagner S, Fujimoto J, Raso MG, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res. 2013;19:6261–6271. doi: 10.1158/1078-0432.CCR-13-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akagi I, Okayama H, Schetter AJ, Robles AI, Kohno T, Bowman ED, et al. Combination of Protein Coding and Noncoding Gene Expression as a Robust Prognostic Classifier in Stage I Lung Adenocarcinoma. Cancer Res. 2013;73:3821–3832. doi: 10.1158/0008-5472.CAN-13-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 31.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama Y, Suzuki M, Arima C, Huang QM, Tomida S, Takeuchi T, et al. Proteasomal non-catalytic subunit PSMD2 as a potential therapeutic target in association with various clinicopathologic features in lung adenocarcinomas. Mol Carcinog. 2011;50:301–309. doi: 10.1002/mc.20632. [DOI] [PubMed] [Google Scholar]
- 33.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7:e36530. doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of Genes Up-regulated in ALK-positive and EGFR/KRAS/ALK-negative Lung Adenocarcinomas. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 37.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 39.Larsen JE, Pavey SJ, Passmore LH, Bowman R, Clarke BE, Hayward NK, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28:760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- 40.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goncalves R, Bose R. Using multigene tests to select treatment for early-stage breast cancer. J Natl Compr Canc Netw. 2013;11:174–182. doi: 10.6004/jnccn.2013.0025. quiz 82. [DOI] [PubMed] [Google Scholar]
- 43.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 44.Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2012;49:604–615. doi: 10.1016/j.ejca.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu K, et al. The Prognostic Value of miR-21 and miR-155 in Non-small-cell Lung Cancer: A Meta-analysis. Jpn J Clin Oncol. 2013 doi: 10.1093/jjco/hyt084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.