Abstract
Novel strategies to directly thwart malaria transmission are needed to maintain the gains achieved by current control measures. Transmission-blocking interventions (TBIs), namely vaccines and drugs targeting parasite or mosquito molecules required for vector-stage parasite development, have been recognized as promising approaches for preventing malaria transmission. However, the number of TBI targets is limited and their degree of conservation among the major vector-parasite systems causing human disease is unclear. Therefore, discovery and characterization of novel proteins involved in vector-stage parasite development of Plasmodium falciparum and Plasmodium vivax is paramount. We mined the recent Anopheles gambiae midgut lipid raft proteome for putative mosquito-derived TBI targets and characterized a secreted glycoconjugate of unknown function, AgSGU. We analyzed molecular variation in this protein among a range of anopheline mosquitoes, determined its transcriptomic and proteomic profiles, and conducted both standard and direct membrane feeding assays with P. falciparum (lab/field) and P. vivax (field) in An. gambiae and Anopheles dirus. We observed that α-AgSGU antibodies significantly reduced midgut infection intensity for both lab and field isolates of P. falciparum in An. gambiae and An. dirus. However, no transmission-reducing effects were noted when comparable concentrations of antibodies were included in P. vivax-infected blood meals. Although antibodies against AgSGU exhibit transmission-reducing activity, the high antibody titer required for achieving 80% reduction in oocyst intensity precludes its consideration as a malaria mosquito-based TBI candidate. However, our results suggest that P. falciparum and P. vivax ookinetes use a different repertoire of midgut surface glycoproteins for invasion and that α-AgSGU antibodies, as well as antibodies to other mosquito-midgut microvillar surface proteins, may prove useful as tools for interrogating Plasmodium-mosquito interactions.
Keywords: Malaria, Anopheles, transmission-blocking, Plasmodium vivax, Plasmodium falciparum, midgut
1. Introduction
Over the last decade, more than 200 million cases of malaria have been reported annually, with an estimated 473,000 – 789,000 people succumbing to the disease in 2012 (WHO, 2013). These statistics, though eye opening, fail to capture the tremendous social and economic burdens that the disease imposes on the developing world. Although recent financial and political commitments to malaria control and elimination have led to significant declines in disease transmission, progress is imminently threatened by mosquito resistance to insecticides and delayed parasite clearance times following treatment with Artemisinin (Alonso et al., 2011; Dondorp et al., 2009; Enayati and Hemingway, 2010; Phyo et al., 2012). Novel strategies to curb transmission are critical to ensuring continual progress towards malaria elimination and eradication.
The etiologic agents of human malaria are Apicomplexan parasites in the genus Plasmodium. For mosquito-to-human transmission to occur, a mosquito must first take a blood meal from an infected individual with gametocytes in peripheral circulation. Once inside the lumen of the mosquito midgut, gametocytes egress from parasitized red blood cells and develop into male or female gametes. These stages undergo sexual reproduction to form zygotes that transform into motile ookinetes, which attach to and invade the mosquito midgut epithelium. Upon successful invasion, ookinetes traverse the cell to reach the basal lamina of the basement membrane, where they develop into oocysts. Oocysts mature over a 10 – 14 day period during which sporogony occurs, resulting in the production of hundreds to thousands of sporozoites. Upon oocyst maturation and rupture, these sporozoites are released into the open circulatory system of the hemocoel, a portion of which make their way into the mosquito salivary glands. In subsequent blood meals, salivary-gland sporozoites are injected into the capillary bed, thereby transmitting parasites to a new host. Midgut invasion by ookinetes represents a critical bottleneck in the life cycle of Plasmodium and offers a unique opportunity to interrupt the parasite’s life cycle (Dinglasan and Jacobs-Lorena, 2008).
One promising approach to combating malaria is the use of transmission-blocking interventions (TBIs), namely vaccines or drugs that target parasite stages in the blood meal and therefore prevent developmental steps in the mosquito vector required for subsequent transmission to human hosts. The rationale is that if a susceptible population is adequately treated with a TBI, the average blood meal ingested by a mosquito will contain antibodies (vaccine) or small molecules (drug) that target parasite sexual stages ingested in the blood (gametocytes) and/or those that develop in the midgut after feeding (e.g., macrogametes, microgametes, zygotes, ookinetes). TBIs may either kill the parasites or interfere with molecular interactions necessary for specific developmental steps to occur (e.g., fertilization, ookinete invasion of the midgut epithelium) (Dinglasan and Jacobs-Lorena, 2008; Mathias et al., 2013). Strategies aiming to prevent Plasmodium invasion of the mosquito midgut may target surface antigens on either the ookinete or the midgut epithelium, and several such TBIs have shown encouraging results (Armistead et al., 2014; Mathias et al., 2012, 2013; Shimp et al., 2013; Miyata et al., 2010). However, the various mechanisms through which an ookinete invades a midgut epithelial cell remain poorly understood. Recent studies have suggested that Plasmodium ookinetes use multiple ligands on the apical midgut plasma membrane during the invasion process, which may be the result of multiple pathways of midgut invasion (Angrisano et al., 2012; Parish et al., 2011; Vega-Rodriguez et al., 2013). As such, an integral understanding of the parasite’s entire invasion process will contribute greatly to improving current strategies to interrupt parasite transmission with TBIs.
Lipid-raft microdomains play a fundamental role in the invasion pathways of a diverse array of pathogens (Riethmuller et al., 2006). Lipid rafts are dynamic, ordered structures of proteins and lipids, rich in cholesterol and sphingolipids, in the plasma membrane of eukaryotes. These microdomains can fuse together to form platforms that facilitate key cellular functions including cell signal transduction, membrane trafficking and pathogen invasion (reviewed by Simons and Gerl, 2010). Recent evidence suggests that lipid rafts may be an important component of ookinete invasion of the midgut epithelium. Six out of the seven known ookinete-interacting proteins, including the recently reported enolase binding protein (EBP) (Vega-Rodríguez et al., 2014) and the two mosquito-based TBI antigens Anopheles alanyl aminopeptidase N (AnAPN1) (Dinglasan et al., 2007) and Anopheles gambiae carboxypeptidase B (CPBAg1) (Lavazec et al., 2007), were found associated with apical midgut-microvilli detergent resistant membranes (DRM), which are enriched in lipid rafts (Parish et al., 2011). We argue that mining the midgut DRM proteome will likely result in the identification of novel TBI candidates and thus, provide insight into invasion models proposed in the literature, a stance validated by the work on EBP.
The Anopheles gambiae protein AGAP000570, a secreted glycoconjugate of unknown function, which we refer to as AgSGU, was consistently identified in replicate DRM or lipid raft preparations from An. gambiae midguts (Parish et al., 2011) and was among the most highly abundant proteins in one of the replicates. Interestingly, AgSGU was also the second most abundant protein in the An. gambiae peritrophic matrix (PM) proteome (Dinglasan et al., 2009). The presence of AgSGU in the DRM fraction suggests it is partitioned into the same raft structures as the ookinete-interacting proteins mentioned above. Given the presence of AgSGU in midgut DRMs, we hypothesized that this protein plays a role in ookinete invasion of the mosquito midgut. Because these rafts may act as platforms for ookinete invasion, AgSGU may interact either directly with Plasmodium ookinetes (transacting) or with other important ligands within midgut lipid-raft structures (cis-acting) during the invasion process. As such, characterization of this protein may be relevant to the molecular biology underlying Plasmodium transmission and therefore yield new insights into the midgut invasion pathway(s) of Plasmodium species.
2. Methods
2.1. Homology Searches and Evolutionary Analyses of AgSGU and Putative Orthologs
To investigate homology and potential functions of AgSGU, its amino acid sequence was searched against the Conserved Domain Database (www.ncbi.nlm.nih.gov/cdd/) using RPS (Reverse Position Specific)-BLAST. To identify putative orthologs among anophelines, the AgSGU amino acid sequence was searched against the assembled genome sequences from all anophelines in VectorBase (Release VB-2014-02) using BLASTp (peptide vs. peptide). Where putative orthologs were not found or gene sets were not yet available, all additional datasets were searched using tBLASTn (peptide vs. translated nucleotide), including genome scaffolds, contigs, assembled trascriptomes, and sets of EST sequences. To identify homologs outside of Anophelinae, the amino acid sequence of AgSGU was searched against the NCBI non-redundant database using BLASTp.
Protein sequences for 15 anophelines were selected for further analyses based on confidence of orthology. The signal peptide for each was identified using the SignalP 4.1 server (Petersen et al., 2011; www.cbs.dtu.dk/services/SignalP/) and then removed manually. All sequences were then aligned with TCOFFEE (Notredame et al., 2000; http://tcoffee.crg.cat/) and that alignment was used to align the corresponding DNA sequences while taking the reading frame into account with the program TranslatorX (Abascal et al., 2010; http://translatorx.co.uk/). The DNA alignment was then uploaded to Datamonkey (Delport et al., 2010; Pond and Frost, 2005a; www.datamonkey.org/), the web-based server of the HyPhy package (Pond et al., 2005), and used to construct a phylogenetic tree (neighbor joining method) and select an appropriate model of nucleotide substitution. The latter process identified model 012302 (LRT = 9.435, AIC = 8579.799, p = 0.00893), a time-reversible model of nucleotide substitution that accounts for biases in substitution rates inherent in the sequences analyzed (e.g., transition/transversion biases). In subsequent analyses of selection, this model was used to estimate ω, or dN/dS, the ratio of the number of non-synonymous substitutions per non-synonymous site to the number of synonymous substitutions per synonymous site. Estimates of ω were generated across AgSGU using three separate methods in HyPhy, single-likelihood ancestor counting (SLAC), random effects likelihood (REL), and fixed effects likelihood (FEL),
Predictions of secondary structure and putative protein-binding residues were made using the PredictProtein server (Rost et al., 2004; www.predictprotein.org) with the amino acid sequence from An. gambiae minus the signal peptide.
2.2. Expression, Purification, and Analysis of AgSGU
A 510 bp fragment of AgSGU, representing the complete exon, minus the predicted signal peptide and transmembrane domain, was amplified from An. gambiae midgut cDNA with the gene-specific primers AgSGU F1 5′-GACCCCGTACTGCACGTTTC-3′ and AgSGU R1 5′-GAACCCGTAGATGGCAGC-3′, cloned into the pBAD/Thio-TOPO® vector (Life Technologies, Carlsbad, CA) with a C-terminal 6XHis tag, and expressed in E. coli BL21 (DE3) cells. Cells were grown at 37°C to an optical density (O.D.) of ~0.4 and induced with 0.02% arabinose per liter for 5–6 hours. Soluble protein was extracted from the final cell pellet with BugBuster™ Protein Extraction Reagent (Novagen, Madison, WI), loaded onto a Ni-NTA agarose column (Qiagen, Venlo, Netherlands) and eluted with 300mM imidazole. Elutions were concentrated using Amicon Ultra 3kD centrifugal filters (Millipore, Billerica, MA), and purity was assessed by SDS-PAGE using 12% Tris-glycine gels and SimplyBlue™ SafeStain (Life Technologies).
2.3. Antibody Purification and Fab Preparation
Rabbit polyclonal antibody (PAb) generation was outsourced and produced against the purified recombinant protein (Washington Biotechnology, Baltimore, MD). AgSGU-specific antibody was purified from serum by affinity chromatography using AminoLink Plus Coupling Resin (Thermo Fisher, Rockford, IL) to which recombinant AgSGU was immobilized. To generate Fab fragments AgSGU-specific antibody, normal rabbit IgG (Life Technologies), and normal rabbit IgM (Southern Biotech, Birmingham, AL) was digested with papain (Thermo Fisher) overnight at 37°C.
2.4. SDS-PAGE and Immunoblot Analysis
Mosquito (An. gambiae, Anopheles dirus, Anopheles stephensi, or Aedes aegypti) midgut, salivary gland, abdomen (minus midguts), or thorax (minus salivary glands) lysates were loaded (~5 μg per well) into 4–20% Tris-glycine gels. Proteins were separated under reducing conditions and transferred to a nitrocellulose membrane. The membranes were blocked in a 1:1 solution of Odyssey blocking buffer (LI-COR, Lincoln, NE) and PBS plus tween-20 (0.1%) (PBST), probed with rabbit anti-AgSGU antibodies (20 μg/mL), and detected with donkey anti-rabbit IgG secondary antibodies labeled with IRDye 680RD (LI-COR) and diluted 1:50,000 in a 1:1 solution of blocking buffer and PBST. For loading controls, the same membranes were also probed with mouse anti-α-tubulin (Developmental Studies Hybridoma Bank, MAb AA4.3) (10 μg/mL) and detected with goat anti-mouse IgG secondary antibodies labeled with IRDye 800CW (LI-COR) diluted as above. Quantitative immunoblotting of An. gambiae and An. stephensi midgut AgSGU was performed as above using 4 midgut equivalents per lane from 3, 10, 16, and 27 day old mosquitoes. Expression levels were determined relative to An. gambiae lactate dehydrogenase (AgLDH; AGAP004880), which was detected by rabbit α-LDH antiserum (1:200) and donkey anti-rabbit 680 IRDye labeled secondary antibodies diluted 1:50,000 in a 1:1 solution of blocking buffer and PBST. All immunoblots were imaged using the LI-COR Odyssey infrared imaging system (LI-COR).
2.5. qRT-PCR
Total RNA was extracted from An. gambiae midguts (sugar fed and blood fed) using TRIzol reagent (Life Technologies), and cDNA was synthesized with the RevertAid First Strand cDNA Synthesis Kit using random hexamers (Fermentas, Waltham, MA). Real-time quantitative PCR (qRT-PCR) was performed using SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA) and the StepOnePlus Real-Time PCR System (Applied Biosystems). AgSGU was amplified with gene-specific primers AgSGU F2 5′-GTCGC-GGTTACGTCGTGT-3′ and AgSGU R2 5′-GTAGATGGCAGCAGTGTAGCTG-3′. Anopheles gambiae ribosomal protein L32 (AgRpL32; AGAP002122) was amplified with the primers AgRpL32 F 5′-GCCGAAGATTGTGAAGAAGC-3′ and AgRpL32 R 5′-GACGTTGTGGACCAGGAACT-3′ to serve as an internal control for normalization of cDNA templates. All qRT-PCR reactions were performed in triplicate.
2.6. Immunofluorescence Microscopy
Paraformaldehyde (4%)-fixed, sucrose (20%)-protected An. gambiae KEELE midguts (sugar-fed and blood-fed) were embedded in Tissue Tek OCT compound (Sakura Finetek USA, Torrence, CA) and flash frozen in a butanol bath on ethanol-dry ice. Prior to immunohistochemistry, slides of 7 μm cryosections were allowed to equilibrate to room temperature for 15 minutes and hydrated 3 times with PBS for 5 minutes. Cryosections were blocked for 1 hour with PBS-BSA (3%) and probed with rabbit α-AgSGU PAbs (1:500) or normal rabbit IgG (controls) for 2 hrs at 37°C in a humidified chamber. Slides were washed 3 times with PBS for 5 minutes and probed with Alexa Fluor 488-conjugated α-rabbit IgG secondary antibodies (Invitrogen, Carlsbad, CA) in 0.02% Evans blue in blocking buffer. Slides were washed as above and imaged under oil immersion at 1000x total magnification using a Nikon Eclipse E800 epifluorescence microscope and SPOT imaging software (version 4.9).
2.7. Transmission-blocking Assays
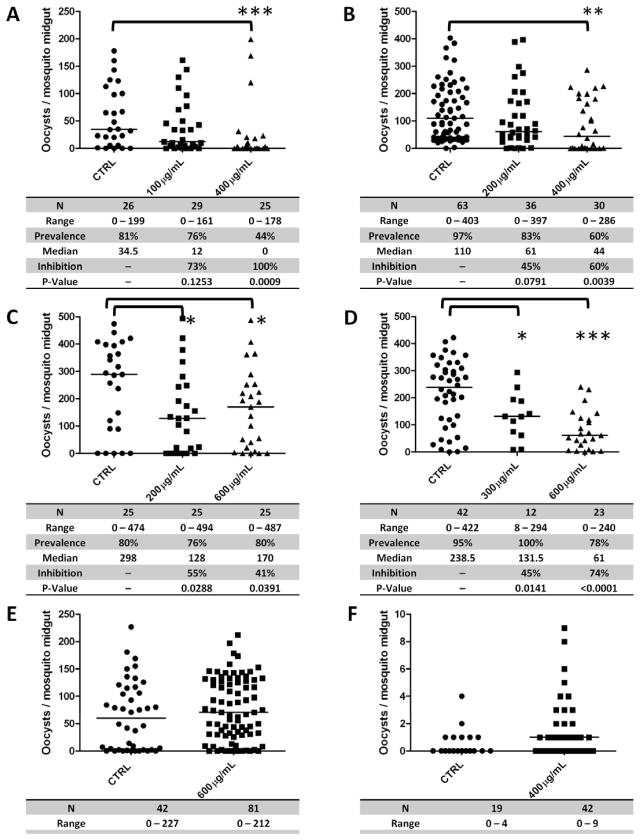
For standard membrane feeding assays (SMFAs), approximately 50 female An. gambiae (Keele strain) mosquitoes per treatment group (5–6 days old) were allocated to pint-sized cup cages and starved for ~ 16 hours prior to each assay. P. falciparum (NF54) gametocyte cultures (15–19 days post-initiation) were pelleted and diluted to 0.3% gametocytemia with human blood that had been washed with RPMI and brought up to 40–50% hematocrit with normal AB serum (Interstate Blood Bank, Inc., Memphis, TN). Gametocytemic blood was mixed with pre-immune rabbit IgG (control), rabbit α-AgSGU whole antibody (0.1, 0.2, 0.4, or 0.6 mg/ml) or Fab fragments (0.6 or 0.9 mg/ml) and then delivered to water-jacketed membrane feeders at 37°C. Fifty mosquitoes per treatment (starved overnight prior to experiment) were allowed to feed for 30–60 minutes. Midguts were dissected 8 days post- blood feeding, stained with 0.2% mercurochrome for 15 minutes, and scored for oocysts with a compound microscope at 200x total magnification. Note that in one SMFA experiment (reported in figure 4B), two pre-immune replicates were included and the oocyst counts for each were pooled prior to making comparisons with α-AgSGU treatments.
Figure 4.
α-AgSGU antibodies inhibit parasite invasion in laboratory and field Plasmodium falciparum isolates but not Plasmodium vivax.
(A–C) α-AgSGU PAbs inhibit Plasmodium falciparum laboratory isolates in three independent standard membrane feeding assays (SMFAs). Each panel shows P. falciparum oocyst counts in Anopheles gambiae mosquitoes fed on gametocytemic blood along with CTRL (pre-immune IgG) or α-AgSGU antigen-specific antibodies. (A) α-AgSGU inhibits parasite invasion at 100 and 400 μg/mL. (B) α-AgSGU inhibits parasite invasion at 200 and 400 μg/mL. (C) α-AgSGU inhibits parasite invasion at 200 and 600 μg/mL. (D) α-AgSGU inhibits parasite invasion in P. falciparum field isolates. SMFA showing P. falciparum oocyst counts in An. dirus mosquitoes fed on gametocytemic blood with CTRL (human AB serum) or α-AgSGU antibodies (300 and 600 μg/mL). (E, F) α-AgSGU does not block parasite invasion in P. vivax field isolates. SMFA showing P. vivax oocyst counts in An. dirus mosquitoes fed gametocytemic blood with CTRL (human AB Serum) or α-AgSGU antibodies (400 and 600 μg/mL). N: numbers of mosquitoes dissected per group (Sample Size); Range: range of oocyst numbers per midgut observed for each group; Prevalence: percent of blood fed mosquitoes in which at least one oocyst developed; Inhibition: reduction in median oocyst intensity in the treatment group relative to the control reported as the % inhibition, calculated as follows: . P-values were determined by the non-parametric Mann Whitney U Test (α = 0.05).
Direct membrane feeding assays (DMFAs) were performed using blood from gametocytemic volunteers infected with P. falciparum or P. vivax collected from patients in the Bongti, Thasao, and Lumsum malaria clinics in Kanchanaburi Province, Thailand. The protocol for the use of parasite-infected blood from volunteers (TMEC 11-033) was approved by the Ethics Committee of the Faculty of Tropical Medicine of Mahidol University, Thailand (Approval # MUTM 2011-040-01) and by the Johns Hopkins Bloomberg School of Public Health Institutional Review Board (protocol #IRB00001629). Infected blood-cell pellets were washed with RPMI and resuspended with human AB serum alone or mixed with rabbit α-AgSGU antibodies (0.3, 0.4, or 0.6 mg/ml) at 40% hematocrit, and fed to ~100 An. dirus mosquitoes per treatment (starved overnight prior to experiment) for 30 minutes using water- jacketed membrane feeders at 37°C. Mosquito midguts were dissected 7 (P. vivax) or 9 (P. falciparum) days after blood feeding, stained with 0.2% mercurochrome for 15 minutes, and scored for oocysts with a compound microscope at 200x total magnification. Due to low numbers of malaria cases at the field clinics, only one isolate of P. falciparum and four isolates of Plasmodium vivax from infected individuals were used in DMFA experiments. Two of the P. vivax experiments failed to generate infections in mosquitoes, so data from only two of those experiments are reported in the Results. Differences in oocyst intensity between control and α-AgSGU groups for both SMFA and DMFA experiments were analyzed by nonparametric statistical analysis using the Mann-Whitney U test (α= 0.05, one tailed) using GraphPad Prism (v5) software.
2.8. Enzyme-linked Immunosorbent Assay (ELISA)
The presence of IgG and IgM was detected in rabbit α-AgSGU whole antibody and Fab fragments by enzyme-linked immunosorbent assay (ELISA). Maxisorp 96-well ELISA plates (Nunc, Fisher Scientific) were incubated overnight at 4°C with recombinant AgSGU at 1 μg/mlin 100 mM Na2CO3/100 mM NaHCO3 buffer and washed three times with PBS. Plates were blocked for 1 h at room temperature with 1:1 solution of 5% milk and 1% gelatin and probed with rabbit α-AgSGU antibody (10 μg/ml) or Fab fragments (10 μg/ml) diluted in blocking buffer for 1 hour at room temperature. Plates were washed five times with PBS plus Tween-20 (0.05%) (PBST20), followed by incubation with 100 μl per well of horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MD) at 1:1000 in blocking buffer, or mouse anti-rabbit IgM (BD Biosciences, San Jose, CA) at 1:3000 followed by HRP-conjugated goat anti-mouse IgG (KPL) at 1:5000. Plates were washed five times with PBST20 and developed by adding 100 μl of TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase substrate (KPL) to each well. Development was stopped after 5 min by the addition of 100 μl of TMB Stop solution (KPL), and plates were read at 450 nm using a SpectraMax 384 Plus plate reader (Molecular Devices, Sunnyvale, CA) and analyzed by SOFTmax Pro 5.3 software. Anti-AgSGU absorbance readings were normalized against that of normal rabbit IgG, IgM, or Fab isotype controls.
3. Results and Discussion
3.1. In silico Analyses Suggest Conservation of AgSGU among Mosquitoes and Dipterans with Notable Exceptions
AgSGU is a relatively short, intronless gene (507 bases) on the X chromosome of An. gambiae coding for an 18.1 kDa protein highly expressed in the midgut epithelium (Parish et al., 2011) and found in the peritrophic matrix (Dinglasan et al., 2009). The molecular function of the protein is unknown and a search of its amino acid sequence against the Conserved Domain Database using RPS-BLAST failed to identify a single domain with known function. The protein has identifiable homologs in other mosquitoes and insects in the order Diptera (Tables 1, S1), a diverse group of insects that includes fruit flies and taxa of medical and veterinary importance such as mosquitoes, sand flies, biting midges, and black flies. Mosquitoes belong to the family Culicidae, which is divided into two monophyletic subfamilies, Culicinae and Anophelinae. Only the latter includes vectors of human malaria, all of which are found in the genus Anopheles. Currently, Anopheles is divided into seven subgenera, although the major malaria vectors are restricted to just four, Cellia (Old World), Anopheles (Cosmopolitan), Nyssorhynchus (Neotropics), and Kerteszia (Neotropics) (Harbach, 2013). An. gambiae and its sibling species belong to the subgenus Cellia, and BLAST searches of AgSGU against the genomic and/or transcriptomic sequences from other anopheline mosquitoes included in VectorBase returned single putative orthologs for all species with the exception of those in the subgenus Nyssorhynchus, represented only by two species Anopheles darlingi and Anopheles albimanus. Percent identity at the amino acid level among putative orthologs ranged from 59% – 98% with the greatest similarity in the An. gambiae species complex as expected (Table 1). For the two Nyssorhynchus species, An. darlingi and An. albimanus, the top BLAST hits were only 37.1% and 26.7% identical at the amino acid level, with low alignment scores (≤79) and poor E values (E > 0.06) in both cases. Reciprocal BLAST searches of these proteins against the An. gambiae genome returned different genes with greater percent identity than AgSGU indicating that orthology is unlikely. Moreover, neither a transcriptomic (Martinez-Barnetche et al., 2012) nor a proteomic study (Ubaida-Mohien et al., 2012) of the An. albimanus midgut identified putative orthologs of this protein, suggesting that the AgSGU ortholog in the Nyssorhynchus lineage was either lost or diversified beyond recognition following divergence from the Anopheles-Cellia lineage, which according to mtDNA data occurred approximately 79 million years ago (Moreno et al., 2010). However, caveats that must be considered are that these analyses were based only on two species of the Nyssorhynchus subgenus and that transcriptomic and proteomic data were from colonies that may not fully reflect genomes occurring in nature.
Table 1. Putative orthologs of AgSGU (AGAP000570) from select anopheline mosquitoes.
The protein sequence of AgSGU was searched against gene sets in VectorBase (Release VB-2014-02) using BLASTp or against DNA contigs, scaffolds, and transcriptomes using tBLASTn. Proteins are ordered by amino-acid percent identity.
| VectorBase ID | % Identity | Mr (kDa) | Species | Subgenus | Region | E-value |
|---|---|---|---|---|---|---|
| AGAP000570 | 100 | 18.1 | An. gambiae | Cellia | Afrotropical | 1E-117 |
| AQUA010799 | 98.2 | 18.1 | An. quadriannulatus A | Cellia | Afrotropical | 9E-101 |
| AARA010474 | 97.6 | 18.0 | An. arabiensis | Cellia | Afrotropical | 2E-99 |
| AEPI009084 | 77.5 | 17.5 | An. epiroticus | Cellia | Asian (Oriental) | 2E-62 |
| AFUN010154a | 65.9 | 17.7 | An. funestus | Cellia | Afrotropical | 3E-54 |
| ACUA014121 | 65.5 | 17.8 | An. culicifacies A | Cellia | Asian (Oriental) | 4E-62 |
| AMIN005326 | 64.9 | 18.0 | An. minimus A | Cellia | Asian (Oriental) | 4E-59 |
| ASTE001198 | 64.5 | 17.9 | An. stephensi | Cellia | Middle East, India | 1E-61 |
| AMAM022625 | 63.9 | 17.9 | An. maculatus B | Cellia | Asian (Oriental) | 8E-62 |
| KI421895b | 63.6 | 17.1 | An. atroparvus | Anopheles | Holarctic | 7E-43 |
| SRS008483_4015c | 63.0 | 16.7d | An. quadrimaculatus | Anopheles | Nearctic | 1E-46 |
| SRS008433_3269c | 62.9 | 18.2 | An. dirus A | Cellia | Nearctic | 2E-53 |
| AFAF004320 | 61.7 | 17.8 | An. farauti | Cellia | Australasian | 5E-52 |
| ASIS013596e | 60.2 | 19.1 | An. sinensis | Anopheles | Asian (Oriental) | 2E-45 |
| SRS008481_11032c | 59.9 | 17.2 | An. freeborni | Anopheles | Nearctic | 7E-53 |
This entry in the database (347 amino acids) likely combines two distinct open reading frames into a single annotation. The predicted ortholog in An. gambiae is AGAP013413, but only aligns with the C-terminal half of the protein (amino acids 201 – 347). A BLASTp search of the N-terminal half (amino acids 1 – 169) against the An. gambiae genome results in AGAP000570 with the predicted molecular weight reported above.
Supercontig identifier. The sequence corresponding to the BLAST hit aligned to bases 3,892,853 – 3,893,284 in the reverse strand.
Transcript identifier from Hittinger et al. (2010). The transcript data are searchable on VectorBase.
The sequence is likely incomplete. There is no start codon and appears to be 9 residues shorter than the sequence for An. gambiae.
This entry in the database (8 exons, 745 amino acids) likely combines multiple open reading frames into a single annotation. The predicted ortholog in An. gambiae is AGAP000569, but only aligns with a portion of the protein (no alignment with the last 2 exons, which code for 187 amino acids). A BLASTp search of the final 2 exons against the An. gambiae genome results in AGAP000570 with the predicted molecular weight reported above.
Outside of Anophelinae, homologs were identified in the culicine mosquitoes Aedes aegypti and Culex quinquefasciatus, as well as in the biting midge Culicoides sonorensis, the sand flies Phlebotomus duboscqi, Phlebotomus papatasi, and Lutzomyia longipalpis, and in numerous fruit flies (Table S1). Taken together, the analysis of homology suggests that AgSGU has descended from an ancient protein conserved across the order Diptera, including families of the suborder Nematocera, such as Culicidae (mosquitoes), Ceratopogonidae (biting midges), and Psychodidae (sand flies), as well as at least one lineage of the suborder Brachycera, the family Drosophilidae (fruit flies).
3.2. Evolutionary Analyses of AgSGU and Putative Orthologs Indicate a Prominent Role for Purifying Selection During Anopheline Diversification
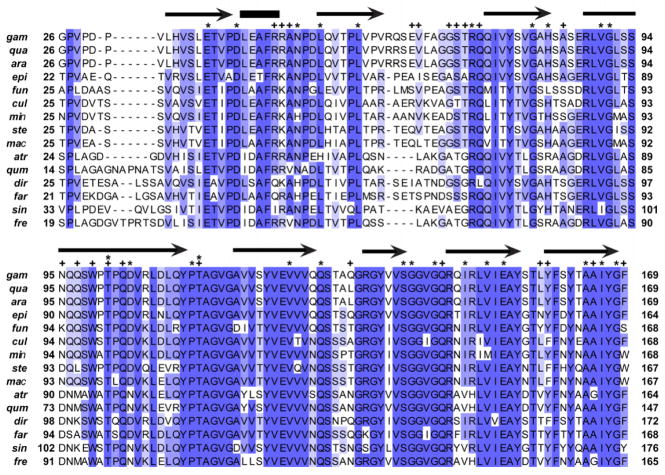
Compared to other mosquito-midgut proteins implicated in Plasmodium ookinete invasion in the literature, the degree of divergence among anophelines is greater than expected for AgSGU (Table 1). Thus, to further analyze the evolution of AgSGU and its putative orthologs among anophelines, we used Datamonkey, the web-based server (www.datamonkey.org) of the HyPhy package (Delport et al., 2010; Pond et al., 2005; Pond and Frost, 2005a), to investigate signatures of selection over the protein’s evolutionary history by estimating ω, or dN/dS, the ratio of the number of non-synonymous substitutions per non-synonymous site to the number of synonymous substitutions per synonymous site. When estimated on a site-by-site basis, this ratio provides insight into the selection pressure experienced by each codon in a gene of interest over its evolutionary history (Pond and Frost, 2005b; Yang and Bielawski, 2000). Ratios equal to 1 indicate neutral evolution, those greater than 1 indicate positive selection, and those less than 1 indicate negative, or purifying, selection (Yang and Bielawski, 2000). Estimates of ω were generated across AgSGU using three separate methods, single-likelihood ancestor counting (SLAC), random effects likelihood (REL), and fixed effects likelihood (FEL). Gene-wide estimates of ω ranged from 0.216835 – 0.244641, suggesting that negative selection has played a prominent role in AgSGU evolution. On a finer scale, REL failed to identify specific codons under negative selection, while SLAC and FEL identified 25 and 50 such codons (α = 0.01), respectively. The alignment in Figure 1 suggests that 52 out of 152 amino acids (34.2%) are invariant among anophelines, of which 18 correspond to codons with ω significantly less than 1 identified by both SLAC and FEL (Table 2). Eighteen additional amino acids are conserved among more than 80% (> 12/15) of the anophelines in the alignment, and 3 of these are also under negative selection (Table 2).
Figure 1. Amino acid alignment of AgSGU with putative orthologs from a sample of anopheline mosquitoes.
Shading corresponds to different levels of amino acid identity: dark, 13–15 out of 15 conserved (> 80% identity); intermediate, 10–12 out of 15 conserved (66.7%–80% identity); light = 7–9 out of 15 conserved (46.7%–66.6% identity). Above the alignment, arrows and bars represent annotations based on predictions of secondary structure, where arrows are predicted beta strands and bars are predicted alpha helices. Stars represent sites under negative selection identified by two separate methods (SLAC and FEL), while pluses indicate amino acids predicted to take part in protein binding. All species are in the genus Anopheles with abbreviations of the species names as follows: gam, gambiae; qua, quadriannulatus A; ara, arabiensis; epi, epiroticus; fun, funestus; cul, culicifacies A; min, minimus A; ste, stephensi; mac, maculatus B; atr, atroparvus; qum, quadrimaculatus; dir, dirus A; far, farauti; sin, sinensis; fre, freeborni.
Table 2. Codons under negative selection identified by both SLAC and FEL analyses.
Numbers for codon position correspond to amino acid numbers for An. gambiae as indicated in the alignment in Figure 1. Codon positions and amino acids predicted to be involved in protein binding are in bold.
| Codon Position | Amino Acidsa | Categoryb | ω SLAC | P-value | ω FEL | P-value |
|---|---|---|---|---|---|---|
| 38 | E15 | charged | 0 | 0.006603 | 0 | 0.000768 |
| 42 | D15 | charged | 0 | 0.001918 | 0 | 0.000102 |
| 50 | N12, H3 | polar | 0.063 | 0.000755 | 0.034 | 0.000160 |
| 53 | L14, H1 | hydrophobic | 0.099 | 0.009734 | 0.080 | 0.004253 |
| 58 | L15 | hydrophobic | 0 | 0.002737 | 0 | 0.000036 |
| 73 | R15 | charged | 0 | 0.008455 | 0 | 0.000972 |
| 81 | G15 | hydrophobic | 0 | 0.000476 | 0 | 0.000027 |
| 83 | H11, R3, L1 | polar | 0.112 | 0.003022 | 0.012 | 0.000027 |
| 90 | V14, I1 | hydrophobic | 0.070 | 0.002433 | 0.014 | 0.000043 |
| 91 | G15 | hydrophobic | 0 | 0.004328 | 0 | 0.000302 |
| 101 | T15 | polar | 0 | 0.004328 | 0 | 0.000085 |
| 104 | D11, N4 | charged | 0.037 | 0.001294 | 0.026 | 0.000764 |
| 112 | P15 | hydrophobic | 0 | 0.004115 | 0 | 0.000082 |
| 113 | T15 | polar | 0 | 0.004115 | 0 | 0.000049 |
| 125 | V15 | hydrophobic | 0 | 0.004115 | 0 | 0.001721 |
| 129 | Q15 | polar | 0 | 0.000303 | 0 | 0.000011 |
| 140 | S15 | polar | 0 | 0.004818 | 0 | 0.000034 |
| 141 | G15 | hydrophobic | 0 | 0.004186 | 0 | 0.000197 |
| 144 | G15 | hydrophobic | 0 | 0.004276 | 0 | 0.000544 |
| 148 | I11, V4 | hydrophobic | 0.060 | 0.003162 | 0.038 | 0.000294 |
| 151 | V14, M1 | hydrophobic | 0.082 | 0.006518 | 0.048 | 0.001644 |
| 153 | E15 | charged | 0 | 0.006568 | 0 | 0.001085 |
| 164 | A15 | hydrophobic | 0 | 0.000457 | 0 | 0.000021 |
| 166 | I15 | polar | 0 | 0.000119 | 0 | 0.000001 |
| 168 | G15 | hydrophobic | 0 | 0.004611 | 0 | 0.000405 |
For each codon position, all amino acids present in the alignment are provided with the frequency indicated by the subscript.
Amino acids were categorized into three broad groups based on physicochemical properties of side chains (Wolfenden et al., 1981): hydrophobic - A, F, G, I, L, P, V, W; polar - C, H, M, N, Q, S, T, Y; charged - D, E, K, R.
Considering that structural details and the molecular function of the AgSGU remain unknown, the nature of the highly conserved and negatively selected amino acids may provide important clues about AgSGU structure and function. In the secreted version of the protein (i.e., no signal peptide), hydrophobic amino acids are the most common residues, collectively comprising ~53% with valine being the most frequent (23/144, ~16%). Polar amino acids make up one third of the protein (47/144, ~33%), while the remaining residues are charged (20/144, ~14%) and likely to be on the protein surface. Of these amino acid categories, negative selection appears to be operating on 13 hydrophobic, 7 polar, and 5 charged residues (Table 2). Predictions of secondary structure and protein-binding residues using the PredictProtein server (Rost et al., 2004) suggest that AgSGU consists of 8 beta sheets and a single alpha helix (Figure 1). Twenty-two amino acids within the peptide sequence were predicted to take part in protein-protein interactions, of which only 2 were under negative selection (Figure 1, Table 2). Both residues were threonines and were invariant among the anophelines investigated. Although not predicted to be glycosylated (see section 3.3), it is noteworthy that both threonines are flanked by at least one proline in the majority of species in the alignment, which increases the likelihood of O-linked, or mucin-type, glycosylation in Drosophila (Gerken et al., 2008). Thus, as an alternative to protein binding, conservation at these two residues may instead be due to important sites of post-translational modification. Interestingly, of the remaining amino acids predicted to take part in protein binding, 14 of 20 were relatively less conserved among the anophelines investigated (Figure 1) and were mostly polar or charged. These data, along with evidence for negative selection predominantly occurring on hydrophobic residues, suggest that the protein’s interactive motifs have been more evolutionarily flexible while purifying selection has maintained the protein’s general structure.
3.3. AgSGU Expression Data Suggest Conserved Roles in Physiology of Blood feeding
Published microarray data indicated that AgSGU transcription in An. gambiae adult females was down-regulated 29.9 fold at 3 hours post-blood feeding relative to the non-blood fed condition (p < 0.001) (Marinotti et al., 2006). However, at 24 hours post-blood feeding, transcription partially rebounded and was up-regulated 9.8 fold (p < 0.001) relative to the previous time point. At 48 hours the upward trend continued with a 2.2 fold increase (p = 0.0019), but transcription plateaued thereafter with no significant change at 72 or 96 hours post-blood feeding, and a small but significant increase of 1.7 fold 15 days after taking a blood meal (p = 0.016) (Marinotti et al., 2006). Moreover, in a comprehensive analysis of the adult tissues in An. gambiae, AgSGU was most highly expressed in the female midgut (Baker et al., 2011), and was also the second most abundant protein detected in the peritrophic matrix (PM) proteome (Dinglasan et al., 2009), which forms after the mosquito has taken a blood meal. This indicates that despite a reduction in transcript, this protein is highly abundant in the midgut following the ingestion of blood. Together, these data suggest that the transcript is translated immediately in response to blood feeding, which is in line with the association of AgSGU with the PM. Transcription is then likely initiated within 24 hours after blood feeding to replenish mRNA transcript in preparation for subsequent blood meals. An increase in transcription levels of AgSGU was also found in larval salivary glands relative to the whole larvae with the glands removed (Neira et al., 2009). These data, as well as homology with salivary-gland proteins empirically identified in sand flies (Kato et al., 2006), suggests a role for AgSGU in the salivary glands or saliva, in addition to the midgut.
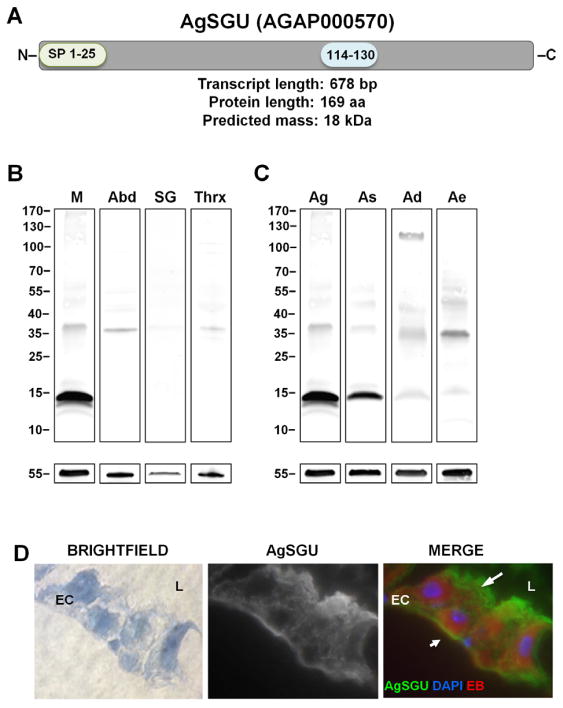
Although AgSGU lacks a transmembrane domain, it is predicted to have a GPI-anchor according to both algorithm-derived (GPISOM) and empirical data, as it was identified in two preparations of the GPI-anchored An. gambiae midgut proteome (<1% False Discovery Rate, 2 unique peptides) (Parish et al., 2011). A GPI-anchor on AgSGU provides further support for its presence in lipid rafts, since GPI-anchored proteins are enriched in lipid rafts (Simons et al., 1997). Finally, AgSGU is predicted to lack either N- or O-linked glycans as determined by the glycosylation prediction programs NetOGlyc (Steentoft et al., 2013; www.cbs.dtu.dk/services/), ISOGlyP (http://isoglyp.utep.edu/index.php), and NetNGly (www.cbs.dtu.dk/services/NetNGlyc/) (Figure 2A). Given the above, AgSGU cannot be called a glycoprotein since the GPI anchor appears to be the only empirically supported glycosylation on this protein. Following convention, it is therefore referred to as a glycoconjugate.
Figure 2. Anopheles gambiae AgSGU.
(A) Schematic of native AgSGU indicating functional characteristics of the protein (www.vectorbase.org); SP, Signal peptide (aa1-25); Arrow indicates predicted GPI-anchor site (aa27-29). Low complexity region, blue boxed area (aa114-130). (B) AgSGU is expressed in An. gambiae midgut. Immunoblot with α-AgSGU PAbs of female An. gambiae midgut lysate (‘M’, lane 1), abdomen lysate minus midguts (‘Abd’, lane 2), salivary gland lysate (‘SG’, lane 3), and thorax lysate minus salivary glands(‘Thrx’, lane 4). Bands for a loading control, α-tubulin, are shown beneath each lane. (C) Conservation of AgSGU among Old World anopheline and aedine mosquitoes. Immunoblot of midgut lysates from An. gambiae (Ag, lane 1), An. stephensi (As, lane 2), An. dirus (Ad, lane 4), and Ae. aegypti (Ae, lane 4) probed with α-AgSGU PAbs. As in panel (B), bands for a loading control, α-tubulin, are shown beneath each lane. (D) AgSGU is present on the midgut surface. Immunofluorescence assay of sugar-fed An. gambiae mosquito-midgut cryosections counterstained with Evans blue (EB); (left panel) Brightfield, (middle panel) α-AgSGU, and (right panel) merged image of α-AgSGU (green), DAPI nuclei stain (blue) and EB protein counterstain (red). α-AgSGU (green) preferentially stains the luminal compartment (top larger arrow) of the midgut, however partial staining of the basal lamina is also observed (bottom smaller arrow). L, lumen of the midgut. EC, epithelial cell. Images were acquired at 400X total magnification.
3.4. Empirical Evidence for AgSGU Expression in the An. gambiae Midgut
To confirm the presence of AgSGU in the midgut and begin investigating its function, midgut proteins were resolved by SDS-PAGE and antibodies against full-length recombinant AgSGU were used for immunoblotting. A distinct protein doublet with a more prominent lower band was observed at an apparent Mr of ~ 14 – 16 kDa under reducing conditions (Figures 2B, 2C), corresponding to the Mr of the predicted size of AgSGU without a signal sequence (15.5 kDa). These data provide additional support for the presence of a GPI-anchor, since the signal peptide is cleaved upon GPI attachment (Englund, 1993). Some GPI-anchored proteins undergo shifts in mobility causing either an apparent increase or, in some cases, a decrease in Mr compared to what is predicted due to the lipid anchor altering protein conformation or binding to SDS (Eugster et al., 2007; Li et al., 2007; Luders et al., 2003). Since AgSGU is aglycosylated except for the anchor, we suspect that the ~16 kDa band in the SDS-PAGE is AgSGU with a GPI- anchor, while the lower band is without anchor. This could be due to a simple loss of the GPI anchor or its targeted removal for the secondary release of the secreted glycoconjugate from the midgut surface.
Contrary to what we observed in An. gambiae midgut lysates, only a faint single band at ~35 kDa was observed in the salivary gland lysate (Figure 2B). Because similarly faint bands were seen in both abdomen (minus midguts) and thorax (minus salivary glands) lysates, the salivary-gland band is likely due to cross reactivity. Thus, we find no definitive evidence for protein expression of AgSGU in adult female salivary glands, results congruent with a published proteome for this tissue in An. gambiae (Kalume et al., 2005) despite its transcription in the salivary glands of larvae (Neira et al., 2009).
A comparative immunoblot analysis of midgut lysates from An. gambiae, Anopheles stephensi, Anopheles dirus and Aedes aegypti support homology as predicted by in silico analyses. AgSGU appears as a protein doublet in both An. gambiae and An. stephensi at ~14–16 kDa with the lower band being much more prominent (Figure 2C). For An. dirus sample, a ~15 kDa band is present but very faint. However, across all samples, faint bands are consistently observed from~ 35–55 kDa, which may indicate AgSGU SDS-resistant complexes or simply the presence of cross-reactive proteins. It is noteworthy that the signal for An. dirus was not as strong as it was for An. stephensi despite similar percent amino acid identities with An. gambiae, suggesting differences in protein abundance between species or variation in the sequence of the epitope recognized by the antibodies. As described above no ortholog to AgSGU was found in the midgut transcriptome (Martinez-Barnetche et al., 2012) or proteome (Ubaida-Mohien et al., 2012) of the neotropical mosquito, An. albimanus (subgenus Nyssorhynchus). Nevertheless, the confirmed presence of AgSGU in the midguts of Old World malaria vectors from the subgenus Cellia (Figure 2C), as well as predicted conservation among other vectors from subgenera Cellia and Anopheles, suggests an important role for the protein in mosquito physiology and perhaps in vector-parasite interactions.
AgSGU shows a considerable degree of conservation with four Ae. aegypti proteins: AAEL009985 and AAEL013885 (Table S1). The full-length homologs of Ae. aegypti, AAEL009985, AAEL013885, AAEL004809, and AAEL013895, are 17.12 kDa, 17.15 kDa, 16.44 kDa, and 16.55 kDa, respectively (Table S1). In the immunoblot we observed a faint low Mr band in Ae. aegypti close to the predicted Mr of all four homologs, as well as more prominent bands at 35 and 45 kDa (Figure 2C). Although the higher molecular weight bands may also indicate multi-protein complexes in Aedes, we again cannot rule the fact that the antibody is simply cross reacting with other proteins. Further characterization of these homologs in the biology of Aedes mosquitoes may help elucidate the function of AgSGU in Anopheles gambiae.
Secretion to the membrane surface is one function of the GPI post-translational modification (Englund, 1993), but it is unclear if AgSGU is further secreted following the cleavage of the GPI-anchor or if it remains associated with the extracellular matrix through interactions with surface glycoproteins. A bona fide GPI anchor would indicate localization of AgSGU to the midgut microvilli (MV) on the luminal surface. Indeed, AgSGU was found to be present in the An. gambiae midgut luminal brush border microvilli (BBMV) proteome (Ubaida-Mohien, et al. 2012). Immunofluorescence microscopy of sugar-fed An. gambiae midgut cryosections indicates the presence of AgSGU on the MV and surprisingly, also on the midgut basal lamina (Figure 2D). As expected from our previous work, normal rabbit IgG does not stain midgut sections (Figure S1). Therefore, several orthogonal lines of evidence supports the presence of AgSGU on the apical membrane and its association with lipid rafts, which primarily localize to the apical plasma membrane of polarized cells. Additionally, the presence of AgSGU on the basal side of the midgut suggests a different, albeit unknown function on the basal lamina extracellular matrix.
3.5. AgSGU Expression Throughout Mosquito Development
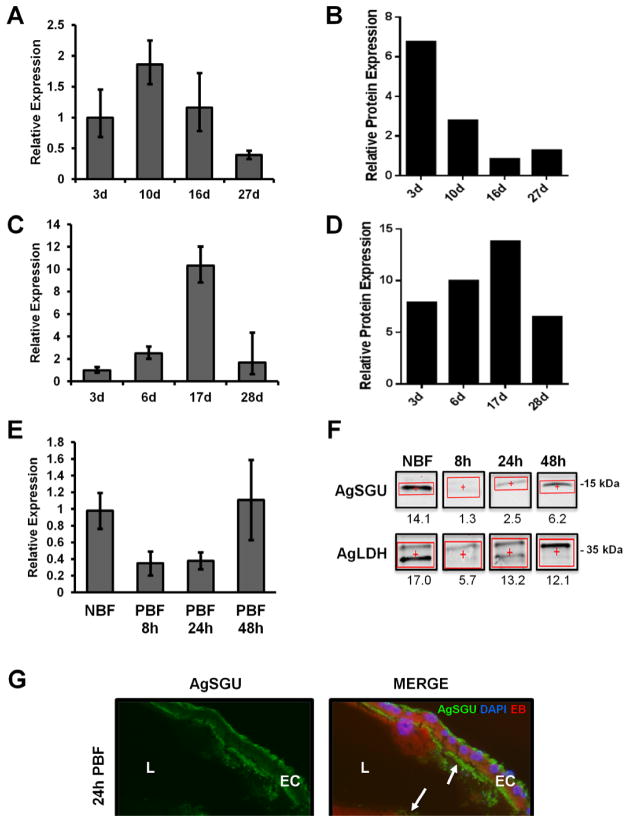
To further characterize AgSGU expression, both immunoblot and quantitative RT-PCR (qRT-PCR) analyses of anopheline midgut lysates at various life stages, and adult ages were performed (Figure 3). AgSGU transcript expression in sugar-fed mosquitoes appears to peak at ~10 days post-eclosion, and then decreases incrementally (Figure 3A). Protein expression, however, is highest at the beginning of the female mosquito’s adult life and decreases until 16 days post-emergence, when it appears to stay relatively stable for its remaining life span (Figure 3B). This discordance in transcript and protein expression in An. gambiae suggests that transcription occurs either in the pupal stage or within the first two days following eclosion. The protein is then expressed in the emerged adult, perhaps in preparation for the first blood meal. In the absence of blood feeding, transcripts likely accumulate in the midgut, while protein levels slowly decrease over time. In contrast, protein expression in An. stephensi midguts is relatively stable with little variation over the adult life span (Figure 3D), although transcript expression peaks later at greater relative levels than An. gambiae, near day 17 (Figure 3C). Interestingly, relative protein expression is similar at day 3 for each species but quite distinct thereafter. These differences in relative transcript and protein expression between An. gambiae and An. stephensi may indicate differences in AgSGU function between the two species, or may reflect variation in expression of this protein during senescence.
Figure 3. AgSGU expression throughout the adult mosquito lifespan.
(A) Quantitative RT-PCR expression levels of AgSGU transcript in An. gambiae sugar-fed midguts from 3, 10, 16 and 27 day old mosquitoes relative to the An. gambiae ribosomal protein RpL32 (AgRpL32). (B) Quantitative immunoblot using α-AgSGU or α-An. gambiae lactate dehydrogenase (LDH) rabbit PAbs to probe An. gambiae midgut lysates from 3, 10, 16 and 27 day old mosquitoes maintained on sugar only. Four midgut equivalents were loaded per lane for the immunoblot. The relative protein expression levels for AgSGU are presented and are normalized to AgLDH expression. (C) Quantitative RT-PCR expression levels of AgSGU transcript in An. stephensi sugar-fed midguts from 3, 6, 17 and 28 day old mosquitoes relative to AgRpL32. (D) Quantitative immunoblot using α-AgSGU and α-LDH PAbs to probe An. stephensi midgut lysates from 3, 6, 17 and 28 day old mosquitoes maintained on sugar only. The amount of protein loaded per lane and normalization as in (B). (E) Quantitative RT-PCR expression levels of AgSGU transcript in sugar-fed (NBF) An. gambiae mosquito midguts, and at 8, 24, and 48 hours post-blood feeding (PBF) relative to AgRpL32. (F) Quantitative immunoblot with α-AgSGU or α-AgLDH PAbs in sugar-fed (NBF) An. gambiae mosquito midgut lysates, and 8, 24, and 48 hours post-blood feeding (PBF) (10 μg per lane). The relative protein expression levels for AgSGU are presented and are normalized to AgLDH expression. (G) Immunofluorescence assay of An. gambiae mosquito midgut cryosections 24 hours after blood feeding with α-AgSGU (green), and merged with images of the Evans blue (EB) protein counterstained midgut (red) and nuclear staining with DAPI (blue). Arrows identify staining both in the peritrophic matrix (in the lumen) and along the apical-membrane surface of midgut epithelial cells (EC). BM, blood meal; L, lumen.
Quantitative RT-PCR (Figure 3E) and quantitative immunoblots (Figure 3F) of AgSGU pre- and post-blood feeding (8h, 24h and 48h) show that levels of both RNA and protein are comparatively higher prior to blood feeding, diminish immediately after the blood meal, and then slowly rise back to pre-blood fed levels as digestion progresses. These data support the previously reported transcriptomic data discussed above (Marinotti et al., 2006), and in combination with data from the peritrophic matrix (PM) proteome, suggest that AgSGU has a role in midgut physiology related to blood feeding. In the PM study, AgSGU was identified as the second most abundant protein present in all fractions of the PM (Dinglasan et al., 2009), suggesting the presence of AgSGU during blood feeding, and a possible direct or indirect role in PM structure. This is not unexpected, since many midgut-surface proteins associated with the PM in blood-fed females are apparently sheared off the midgut microvillar surface and then regenerated after blood feeding, indicating that the majority of AgSGU may be purged with the digested blood meal (Klowden, 2007; Parish et al., 2011). Importantly, data from quantitative immunoblots in Figure 3F are from blood fed midguts with the blood meal and PM removed, and thus represents protein that is present only in the midgut tissues themselves. Following a blood meal, it is therefore unlikely to find abundant levels of AgSGU in the midgut tissue until its epithelial surface is regenerated.
To confirm this hypothesis the localization of AgSGU in blood fed midguts was examined by immunofluorescence microscopy on midgut cryosections from mosquitoes dissected 24 hrs post-blood feeding (Figure 3G). AgSGU is present on the apical MVsurface, on the midgut basal lamina and at the interface of the blood meal and the apical midgut epithelium, which roughly corresponds to the PM interface. As with cryosections from sugar-fed midguts, normal rabbit IgG does not stain sections from blood-fed mosquitoes (Figure S1). These staining patterns are consistent with those observed for the localization of AgAPN1 (Dinglasan et al., 2007) and AgMUC1 (Devenport et al., 2005), both of which are bona fide apical microvillar surface proteins, as well as Ag-Aper-14 (Devenport et al., 2005), which was independently confirmed to be a PM protein (Dinglasan et al., 2009).
These data demonstrate that AgSGU is present and accessible during ookinete invasion, and is consistently expressed throughout the mosquito life span, although it is unclear whether it is bound to the PM or not. The PM is thickest at 24 hrs (Figure 3G) and while there is previous proteomic evidence for its presence, AgSGU may simply be intercalated into the PM and not necessarily attached to it.
3.6. Inhibition of P. falciparum Oocyst Formation by α-AgSGU Polyclonal Antibodies (PAbs)
Due to the presence of AgSGU in lipid rafts on the mosquito midgut, and its putative role in blood feeding, we hypothesized that if AgSGU has a role in ookinete attachment to the midgut, then α-AgSGU PAbs may prevent Plasmodium ookinete invasion. Concentrations of 100, 200, 400 and 600 μg/mL of antigen-specific PAbs significantly reduced P. falciparum oocyst formation in An. gambiae mosquitoes, resulting in 40 – 100% inhibition of median oocyst intensity (P <0.05) with reductions in mosquito infection prevalence of ~20–42% across groups (Figures 4A–C) and no apparent reductions in vector survivorship at the time of dissection (8 days post-blood feeding). Inhibition by PAbs implies either a direct or indirect role for AgSGU in Plasmodium ookinete invasion. High concentrations necessary for inhibition may be indicative of high levels of AgSGU present in the midgut that neutralize or sequester PAbs prior to reaching lipid rafts, or an abundance of the AgSGU on the midgut surface during ookinete invasion.
Inhibition of ookinete invasion by α-AgSGU PAbs could occur through various mechanisms. For example, AgSGU may interact with a portion of ookinete-interacting proteins on lipid rafts, such as AgAPN1 (Mathias et al., 2012; Ubadai-Mohien et al., 2012), and prevent ookinete inhibition by steric hindrance of the ookinete-interacting protein; or the PAbs may efficiently diffuse through the blood meal and bind to AgSGU on lipid rafts and prevent lipid raft clustering.
Immunizing mice with mosquito-midgut lysate elicits a repertoire of antibodies that have transmission-blocking or reducing activity against murine and human malaria parasites (Almeida and Billingsley, 2002; Lal et al., 2001). Midgut proteins that are recognized by mouse antisera (Almeida and Billingsley, 2002) or the monoclonal antibody MG25E (Lal et al., 2001) span a large spectrum of molecular masses from ~15 kDa – 200 kDa, but to date the identities of these proteins remain unknown. It should be noted that MG25E exhibited a multi-banding profile by immunoblot, suggesting that like α-AgSGU and α-AgAPN1 (Dinglasan et al., 2007), MG25E may also see a multimer of the same protein. Only seven bona fide ookinete-interacting, apical midgut-surface proteins have been identified to date (Parish et al., 2011; Vega-Rodríguez et al., 2014) and of these, only two proteins from An. gambiae have thus far been shown to be potent transmission-blocking vaccine antigens: carboxypeptidase B, CPBAg1 (Lavec et al., 2007) and alanyl aminopeptidase N, AgAPN1 (Dinglasan et al., 2007; Mathias et al., 2012; Armistead et al., 2014).
Since P. falciparum NF54 parasites maintained in the laboratory do not represent naturally circulating Plasmodium populations worldwide, we conducted a limited study to determine whether α-AgSGU PAbs would block field isolates of P. falciparum equivalently to our laboratory observations. Although the numbers of cases were low in the field site (Kanchanaburi, Thailand) we were able to test one human isolate of P. falciparum and two isolates of Plasmodium vivax from infected individuals in An. dirus mosquitoes, a primary Plasmodium vector in Southeast Asia. PAbs significantly inhibited oocyst development of local Thai P. falciparum at concentrations of 300 and 600 μg/mL (45–74% inhibition, P<0.05 and P<0.0001 respectively) (Figure 4D), corroborating our laboratory data. Interestingly, α-AgSGU PAbs did not inhibit P. vivax oocyst development in An. dirus (Figures 4E, F). These data suggest different invasion strategies by P. falciparum and P. vivax in the anopheline midgut although our sample size was limited.
One potential explanation for this difference is the speed of ookinete invasion following blood meals. Plasmodium vivax ookinetes invade within 18–24 hours, while P. falciparum invasion begins around 24 hours post- blood feeding (Baton and Ranford-Cartwright, 2004; Zollner et al., 2006). In An. gambiae, AgSGU levels appear lowest in midgut tissue at 8 hours post-blood feeding (Figures 3E, F). Therefore, discordant AgSGU expression during that same time frame may eliminate its potential as a transmission-blocking vaccine candidate for P. vivax. At 24 hours, AgSGU is clearly present on the midgut surface in addition to the lumen (Figures 3E–G), and can thus affect P. falciparum invasion (Figures 4A–D). Moreover, if our hypothesis above is correct, it implicates the role of AgSGU in ookinete invasion as a bystander through its lipid raft association with other ookinete-interacting proteins, rather than as a critical component to ookinete invasion. The presence of α-AgSGU IgM in our antiserum (Figure S2) and its potential to cross-link proteins on the midgut surface or influence ookinete invasion by non-specific steric hindrance provides some support to the possible bystander inhibition phenomenon. However, we generated Fab fragments from the AgSGU IgG and IgM and observed a significant reduction in P. falciparum oocyst numbers in mosquitoes, albeit only at the 900 μg/ml concentration of Fab (Figure S2).
4. Conclusions
We have shown that AgSGU is a GPI-anchored midgut protein conserved among many of the major vectors of malaria with the likely exception of some in the neotropics (i.e., subgenus Nyssorhynchus). The evolutionary history of AgSGU and its putative orthologs has been governed predominantly by purifying selection during the diversification of the subfamily Anophelinae. Anti-AgSGU PAbs bind to sugar fed and blood fed midguts of An. gambiae and An. dirus and can inhibit the development of both lab and field-derived P. falciparum oocysts, suggesting a direct or indirect role in ookinete invasion. However, the same antibodies did not inhibit oocyst development of P. vivax, suggesting that invasion pathways of the two major human malaria parasites vary. At present, the exact mechanism of inhibition in P. falciparum is unclear, but the data provide additional support for the role of lipid-raft glycoproteins found on the mosquito-midgut surface in ookinete-midgut interactions. However, from a practical perspective, we note that the observed requirement for very high concentrations of PAbs to confer >80% transmission-blocking activity would likely preclude AgSGU for consideration as a mosquito-based malaria transmission-blocking vaccine candidate. For comparison, 10–100 μg/mL concentrations of α-AgAPN1 specific IgG can confer between 80–100% inhibition of oocyst development in both An. stephensi and An. gambiae (Armistead, et al., 2014), which at least meets the minimum requirements for the TBV target product profile (Dinglasan et al., 2013; MalERA Consultative Group on Vaccines, 2011). Regardless, further studies are needed to elucidate the mechanism of AgSGU-mediated inhibition of P. falciparum oocyst development, especially given the inability of α-AgSGU PAbs to reduce P. vivax oocyst infection intensity.
Currently, our understanding of P. vivax ookinete-anopheline midgut interactions falls well short of the body of knowledge underlying the interface of P. falciparum-An. gambiae, and P. falciparum-An. stephensi. The presence of AgSGU after blood feeding, its association with lipid rafts (Parish et al., 2011), and its presence on the surface of the midgut epithelium suggests a possibility for (i) direct interaction between AgSGU and P. falciparum ookinete surface, (ii) indirect interaction with plasmodial micronemal proteins secreted during the invasion process, or (iii) interacting with other MV surface glycoconjugates nearby, e.g., AgAPN1. Exploring these possibilities will require additional reagent and method development as well as interdisciplinary approaches that fall outside of the scope of this report. However, such studies are needed to shed light on key biological differences in the transmission biology of the two major human malaria parasites, especially in light of the present interest in disease elimination and eradication.
Supplementary Material
The amino acid sequence of AgSGU was searched against the NCBI non-redundant database using BLASTp. Proteins are listed by BLAST E-value.
Immunofluorescence assay control using normal rabbit IgG (NRIgG) with midgut cryosections from sugar-fed An. gambiae counterstained with Evans blue (EB); (left panel) Brightfield, (middle panel) NRIgG, and (right panel) merged image of NRIgG (green), DAPI nuclei stain (blue) and EB protein counterstain (red). (B) Same as panel (A) but with midgut cryosections from An. gambiae 24 hours after blood feeding.
(A) Plates coated with recombinant AgSGU (1 μg/ml) and probed with AgSGU-specific whole IgG (10 μg/ml), or Fab (10 μg/ml). IgM was detected with either Mouse anti-Rabbit IgM (mu chain)-Biotin at 10 μg/ml (IgG1 Ab) followed by Goat anti-mouse IgG (H+L) HRP at 0.2 μg/ml, or Goat anti-Rabbit IgM HRP at 1:4000. IgG was detected with Goat anti-Rabbit IgG (gamma chain) HRP at 1:40,000. Absorbance readings were normalized to that of pre-immune rabbit serum, normal IgG, IgM, or Fab. (B) Standard membrane feeding assay using P. falciparum NF54 and An. gambiae mosquitoes.
Acknowledgments
The authors thank Hilary Hurd and Paul Eggleston for the Anopheles gambiae KEELE strain and Travis van Warmerdam for excellent technical assistance. This work was funded in part by the National Institutes of Health (NIH) (grants K22AI077707, R01AI082587 to RRD; and T32 to LAP) and the NIH National Center for Research Resources (grant UL1 RR 025005). We are grateful to the staff at the malaria clinics in Kanchanaburi, Thailand, for their support during patient recruitment. We also acknowledge support from the Bloomberg Family Foundation and the Johns Hopkins Malaria Research Institute (JHMRI) to RRD, a JHMRI Pre-doctoral Fellowship to JSA, the Calvin A. and Helen L. Lang postdoctoral fellowship to DKM, as well as the Swiss National Science Foundation (grant PBZHP3_141430) for postdoctoral fellowship support to HVT.
Footnotes
Competing interests
The authors declare that no competing interests exist such as those described under the guidelines or that might be perceived to influence the results and discussion reported in this paper.
Authors’ contributions
DKM, JGJ, LAP, and HVT carried out the molecular biology studies and analysis of data. DKM performed the analyses of molecular evolution and generated the sequence alignment. JGJ, DKM, LAP, JSA, HVT and CK performed standard and direct membrane feeding assays. JSA performed immunoassays, the preparation and provision of critical reagents and analysis of data. JGJ, RRD, and DKM performed the microscopy. JGJ, LAP and RRD participated in the design of the study and performed the statistical analysis. JGJ and JS participated in the design, coordination and execution of field studies in Thailand. RRD conceived of the study, and participated in its design and coordination. All authors drafted the manuscript with final edits by DKM and RRD.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Derrick K. Mathias, Email: dmathia2@jhu.edu.
Juliette G. Jardim, Email: juliettejardim@gmail.com.
Lindsay A. Parish, Email: lindsayannparish@gmail.com.
Jennifer S. Armistead, Email: armistead.j@wehi.edu.au.
Hung V. Trinh, Email: hungvtrinh@gmail.com.
Chalermpon Khumpitak, Email: chalermpon.kum@mahidol.ac.th.
Jetsumon Sattabongkot, Email: jetsumon@hotmail.com.
Rhoel R. Dinglasan, Email: rdingla2@jhu.edu.
References
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida AP, Billingsley PF. Induced immunity against the mosquito Anopheles stephensi (Diptera: Culicidae): effects of cell fraction antigens on survival, fecundity, and Plasmodium berghei (Eucoccidiida: Plasmodiidae) transmission. J Med Entomol. 2002;39:207–214. doi: 10.1603/0022-2585-39.1.207. [DOI] [PubMed] [Google Scholar]
- Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, Doumbo OK, Greenwood B, Hall BF, Levine MM, Mendis K, Newman RD, Plowe CV, Rodríguez MH, Sinden R, Slutsker L, Tanner M. A research agenda to underpin malaria eradication. PLoS Med. 2011;8:e1000406. doi: 10.1371/journal.pmed.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrisano F, Tan Y, Sturm A, McFadden GI, Baum J. Malaria parasite colonisation of the mosquito midgut - placing the Plasmodium ookinete centre stage. Int J Parasitol. 2012;42(6):519–527. doi: 10.1016/j.ijpara.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Armistead JA, Morlais I, Mathias DK, Jardim J, Joy J, Fridman A, Finnefrock A, Bagchi A, Plebanski M, Scorpio D, Churcher T, Borg NA, Sattabongkot J, Dinglasan RR. Antibodies to a single, conserved epitope in Anopheles APN1 inhibit universal transmission of Plasmodium falciparum and Plasmodium vivax malaria. Infect Immun. 2014;82(2):818–829. doi: 10.1128/IAI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baton LA, Ranford-Cartwright LC. Plasmodium falciparum ookinete invasion of the midgut epithelium of Anopheles stephensi is consistent with the Time Bomb model. Parasitology. 2004;129:663–676. doi: 10.1017/s0031182004005979. [DOI] [PubMed] [Google Scholar]
- Chandre F, Darrier F, Manga L, Akobgeto M, Faye O, Mouchet J, Guillet P. Status of pyrethroid resistance in Anopheles gambiae sensu lato. Bull World Health Organ. 1999;77:230–234. [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26(19):2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport M, Fujioka H, Donnelly-Doman M, Shen Z, Jacobs-Lorena M. Storage and secretion of Ag-Aper14, a novel peritrophic matrix protein, and Ag-Muc1 from the mosquito Anopheles gambiae. Cell Tissue Res. 2005;320:175–185. doi: 10.1007/s00441-004-1067-3. [DOI] [PubMed] [Google Scholar]
- Dinglasan RR, Armistead JA, Nyland JF, Jiang X, Mao HQ. Single-dose microparticle delivery of a malaria transmission-blocking vaccine elicits a long-lasting functional antibody response. Curr Mol Med. 2013;13(4):479–487. doi: 10.2174/1566524011313040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinglasan RR, Devenport M, Florens L, Johnson JR, McHugh CA, Donnelly-Doman M, Carucci DJ, Yates JR, 3rd, Jacobs-Lorena M. The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem Mol Biol. 2009;39:125–134. doi: 10.1016/j.ibmb.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinglasan RR, Jacobs-Lorena M. Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 2008;24:364–370. doi: 10.1016/j.pt.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinglasan RR, Jacobs-Lorena M. Insight into a conserved a lifestyle: protein-carbohydrate adhesion strategies of vector-borne pathogens. Infect Immun. 2005;73:7797–7807. doi: 10.1128/IAI.73.12.7797-7807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinglasan RR, Kalume DE, Kanzok SM, Ghosh AK, Muratova O, Pandey A, Jacobs-Lorena M. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc Natl Acad Sci USA. 2007;104:13461–13466. doi: 10.1073/pnas.0702239104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Eng J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayati A, Hemingway J. Malaria management: past, present, and future. Annu Rev Entomol. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- Englund PT. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- Eugster C, Panakova D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 2008;18(11):861–870. doi: 10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach RE. Anopheles. Mosquito Taxonomic Inventory; 2013. [accessed on March 15, 2014]. http://mosquito-taxonomic-inventory.info/simpletaxonomy/term/6047. [Google Scholar]
- Kalume DE, Okulate M, Zhong J, Reddy R, Suresh S, Deshpande N, Kumar N, Pandey A. A proteomic analysis of salivary glands of female Anopheles gambiae mosquito. Proteomics. 2005;5(14):3765–3777. doi: 10.1002/pmic.200401210. [DOI] [PubMed] [Google Scholar]
- Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, Sigutova L, Volf P, Doumbia S, Valenzuela JG. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya) BMC Genomics. 2006;7:226. doi: 10.1186/1471-2164-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klowden MJ. Physiological Systems in Insects. 2. Elsevier; California: 2007. [Google Scholar]
- Lal AA, Patterson PS, Sacci JB, Vaughan JA, Paul C, Collins WE, Wirtz RA, Azad AF. Anti-mosquito midgut antibodies block development of Plasmodium falciparum and Plasmodium vivax in multiple species of Anopheles mosquitoes and reduce vector fecundity and survivorship. Proc Natl Acad Sci USA. 2001;98:5228–5233. doi: 10.1073/pnas.091447398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Boudin C, Lacroix R, Bonnet S, Diop A, Thiberge S, Boisson B, Tahar R, Bourgouin C. Carboxypeptidases B of Anopheles gambiae as targets for a Plasmodium falciparum transmission-blocking vaccine. Infect Immun. 2007;75:1635–1642. doi: 10.1128/IAI.00864-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Bourgouin C. Mosquito-based transmission blocking vaccines for interrupting Plasmodium development. Microbes Infect. 2008;10:845–849. doi: 10.1016/j.micinf.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Li X, Kaloyanova D, van Eijk M, Eerland R, van der Goot G, Oorschot V, Klumperman J, Lottspeich F, Starkuviene V, Wieland FT, Helms JB. Involvement of a Golgi-resident GPI-anchored protein in maintenance of the Golgi structure. Mol Biol Cell. 2007;18:1261–1271. doi: 10.1091/mbc.E06-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Pyrowolakis G, Jentsch S. The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP. EMBO Rep. 2003;4:1169–1174. doi: 10.1038/sj.embor.7400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Calvo E, Ngyuyen QK, Dissanayake S, Ribeiro JM, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Barnetche J, Gomez-Barreto RE, Ovilla-Munoz M, Tellez-Sosa J, Garcia-Lopez DE, Dinglasan RR, Ubaida Mohien C, Maccallum RM, Redmond SN, Gibbons JG, Rokas A, Machado CM, Cazares-Raga F, Gonzalez-Ceron L, Hernandez-Martinez S, Rodriguez-Lopez MH. Transcriptome of the adult female malaria mosquito vector Anopheles albimanus. BMC Genomics. 2012;13:207. doi: 10.1186/1471-2164-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias DK, Pastrana-Mena RP, Ranucci E, Tao D, Ferruti P, Ortega C, Staples GO, Zaia J, Takashima E, Tsuboi T, Borg NA, Verotta L, Dinglasan RR. A small molecule glycosaminoglycan mimetic blocks Plasmodium invasion of the mosquito midgut. PLoS Pathog. 2013;9(11):e1003757. doi: 10.1371/journal.ppat.1003757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias DK, Plieskatt JL, Armistead JS, Bethony JM, Abdul-Majid KB, McMillan A, Angov E, Aryee MJ, Zhan B, Gillespie P, Keegan B, Jariwala AR, Rezende W, Bottazzi ME, Scorpio DG, Hotez PJ, Dinglasan RR. Expression, immunogenicity, histopathology, and potency of a mosquito-based malaria transmission-blocking recombinant vaccine. Infect Immun. 2012;80:1606–1614. doi: 10.1128/IAI.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Harakuni T, Tsuboi T, Sattabongkot J, Kohama H, Tachibana M, Matsuzaki G, Torii M, Arakawa T. Plasmodium vivax ookinete surface protein Pvs25 linked to cholera toxin B subunit induces potent transmission-blocking immunity by intranasal as well as subcutaneous immunization. Infect Immun. 2010;78(9):3773–3782. doi: 10.1128/IAI.00306-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira Oviedo M, Ribeiro JMC, Heyland A, VanEkeris L, Moroz T, Linser PJ. The salivary transcriptome of Anopheles gambiae (Diptera:Culicidae) larvae: a microarray-based analysis. Insect Biochem Mol Biol. 2009;39:382–394. doi: 10.1016/j.ibmb.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Parish LA, Colquhoun DR, Mohien CU, Lyashkov AE, Graham DR, Dinglasan RR. Ookinete-interacting proteins on the microvillar surface are partitioned into detergent resistant membranes of Anopheles gambiae midguts. J Prot Res. 2011;10:5150–5162. doi: 10.1021/pr2006268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NPJ, White NJ, Anderson TJC, Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005a;21(10):2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005b;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–6709. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Ramalho-Ortigão M, Jochim RC, Anderson JM, Lawyer PG, Pham VM, Kamhawi S, Valenzuela JG. Exploring the midgut transcriptome of Phlebotomus papatasi: comparative analysis of expression profiles of sugar-fed, blood-fed and Leishmania-major-infected sandflies. BMC Genomics. 2007;8:300. doi: 10.1186/1471-2164-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethmuller J, Riehle A, Grassme H, Gulbins E. Membrane rafts in host-pathogen interactions. Biochimica Et Biophysica Acta. 2006;1758:2139–2147. doi: 10.1016/j.bbamem.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Shimp RL, Jr, Rowe C, Reiter K, Chen B, Nguyen V, Aebig J, Rausch KM, Kumar K, Wu Y, Jin AJ, Jones DS, Narum DL. Development of a Pfs25-EPA malaria transmission blocking vaccine as a chemically conjugated nanoparticle. Vaccine. 2013;31(28):2954–2962. doi: 10.1016/j.vaccine.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–88. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The malERA Consultative Group on Drugs. A research agenda for malaria eradication: Drugs. PLOS Med. 2011;8:e1000402. doi: 10.1371/journal.pmed.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rodríguez J, Ghosh AK, Kanzok SM, Dinglasan RR, Wang S, Bongio NJ, Kalume DE, Miura K, Long CA, Pandey A, Jacobs-Lorena M. Multiple pathways for Plasmodium ookinete invasion of the mosquito midgut. Proc Natl Acad Sci USA. 2014;111(4):E492–500. doi: 10.1073/pnas.1315517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubaida Mohien C, Colquhoun DR, Mathias DK, Gibbons JG, Armistead JS, Rodriguez MC, Rodriguez MH, Edwards NJ, Hartler J, Thallinger GG, Graham DR, Martinez-Barnetche J, Rokas A, Dinglasan RR. A bioinformatics approach for integrated transcriptomic and proteomic comparative analyses of model and non-sequenced anopheline vectors of human malaria parasites. Mol Cell Proteomics. 2012;12(1):120–131. doi: 10.1074/mcp.M112.019596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, Andersson L, Cullis PM, Southgate CC. Biochemistry. 1981;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- World Health Organization Global Malaria Programme. World Malaria Report: 2013. WHO Press; Geneva, Switzerland: 2013. [Google Scholar]
- Wass MN, Stanway R, Blagborough AM, Lal K, Prieto JH, Raine D, Sternberg MJ, Talman AM, Tomley F, Yates J, 3rd, Sinden RE. Proteomic analysis of Plasmodium in the mosquito: progress and pitfalls. Parasitology. 2012;139:1131–1145. doi: 10.1017/S0031182012000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15(12):496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner GE, Ponsa N, Garman GW, Poudel S, Bell JA, Sattabongkot J, Coleman RE, Vaughan JA. Population dynamics of sporogony for Plasmodium vivax parasites from western Thailand developing within three species of colonized Anopheles mosquitoes. Malar J. 2006;5:68. doi: 10.1186/1475-2875-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The amino acid sequence of AgSGU was searched against the NCBI non-redundant database using BLASTp. Proteins are listed by BLAST E-value.
Immunofluorescence assay control using normal rabbit IgG (NRIgG) with midgut cryosections from sugar-fed An. gambiae counterstained with Evans blue (EB); (left panel) Brightfield, (middle panel) NRIgG, and (right panel) merged image of NRIgG (green), DAPI nuclei stain (blue) and EB protein counterstain (red). (B) Same as panel (A) but with midgut cryosections from An. gambiae 24 hours after blood feeding.
(A) Plates coated with recombinant AgSGU (1 μg/ml) and probed with AgSGU-specific whole IgG (10 μg/ml), or Fab (10 μg/ml). IgM was detected with either Mouse anti-Rabbit IgM (mu chain)-Biotin at 10 μg/ml (IgG1 Ab) followed by Goat anti-mouse IgG (H+L) HRP at 0.2 μg/ml, or Goat anti-Rabbit IgM HRP at 1:4000. IgG was detected with Goat anti-Rabbit IgG (gamma chain) HRP at 1:40,000. Absorbance readings were normalized to that of pre-immune rabbit serum, normal IgG, IgM, or Fab. (B) Standard membrane feeding assay using P. falciparum NF54 and An. gambiae mosquitoes.