Abstract
β-catenin is a multi-functional protein that plays an important role in the mature central nervous system; its dysfunction has been implicated in several neuropsychiatric disorders, including depression. Here we show that β-catenin mediates pro-resilient and anxiolytic effects in mice in the nucleus accumbens, a key brain reward region, an effect mediated by D2-type medium spiny neurons. Using genome-wide β-catenin enrichment mapping, we identify Dicer1—important in small RNA (e.g., microRNA) biogenesis—as a β-catenin target gene that mediates resilience. Small RNA profiling after excising β-catenin from nucleus accumbens in the context of chronic stress reveals β-catenin-dependent microRNA regulation associated with resilience. Together, these findings establish β-catenin as a critical regulator in the development of behavioral resilience, activating a network that includes Dicer1 and downstream microRNAs. We thus present a foundation for the development of novel therapeutic targets to promote stress resilience.
Despite decades of research, the molecular pathophysiology of depression remains elusive. One molecular player implicated in neuropsychiatric illnesses including depression is β-catenin1–5. In addition to playing a structural role at synapses, β-catenin mediates the transcriptional output of canonical Wnt signaling6–8. This multi-functionality has made it difficult to untangle the mechanism through which β-catenin might contribute to pathological states. We recently demonstrated the involvement of upstream Wnt signaling in the nucleus accumbens (NAc) in mouse depression models, with impaired signaling mediating susceptibility to social stress and increased signaling mediating resilience9. We thus began by studying the behavioral role of β-catenin in this brain region.
β-catenin in NAc mediates pro-resilient, antidepressant, and anxiolytic responses
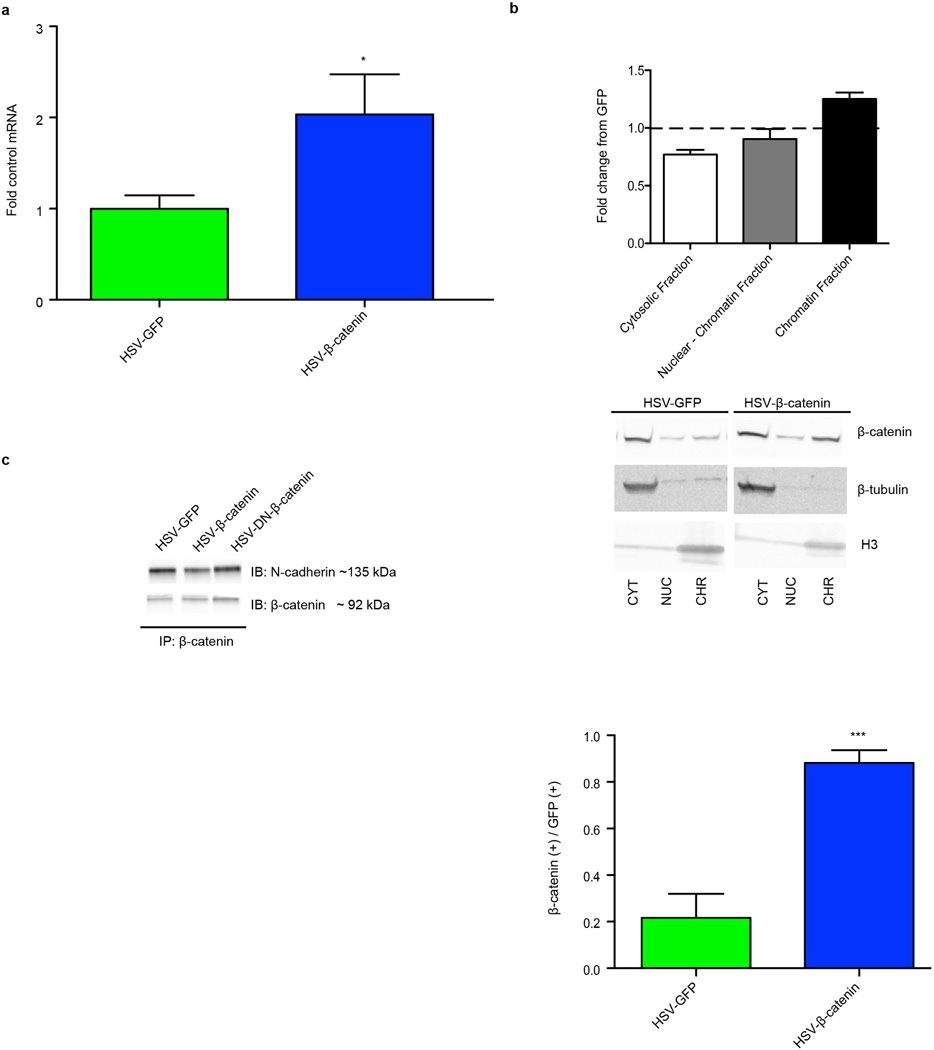
We overexpressed β-catenin in a Herpes simplex virus (HSV) vector in NAc (Fig. 1a; Extended Data Fig. 1a), which increases β-catenin solely in the nuclear compartment, as measured by subcellular fractionation and immunohistochemistry (IHC), whereas global N-cadherin/β-catenin complexes were unaffected (Extended Data Fig. 1b,c). This suggests that HSV-β-catenin selectively activates the transcriptional function of the protein, without having direct effects on N-cadherin at synapses, consistent with prior work in cultured cells10.
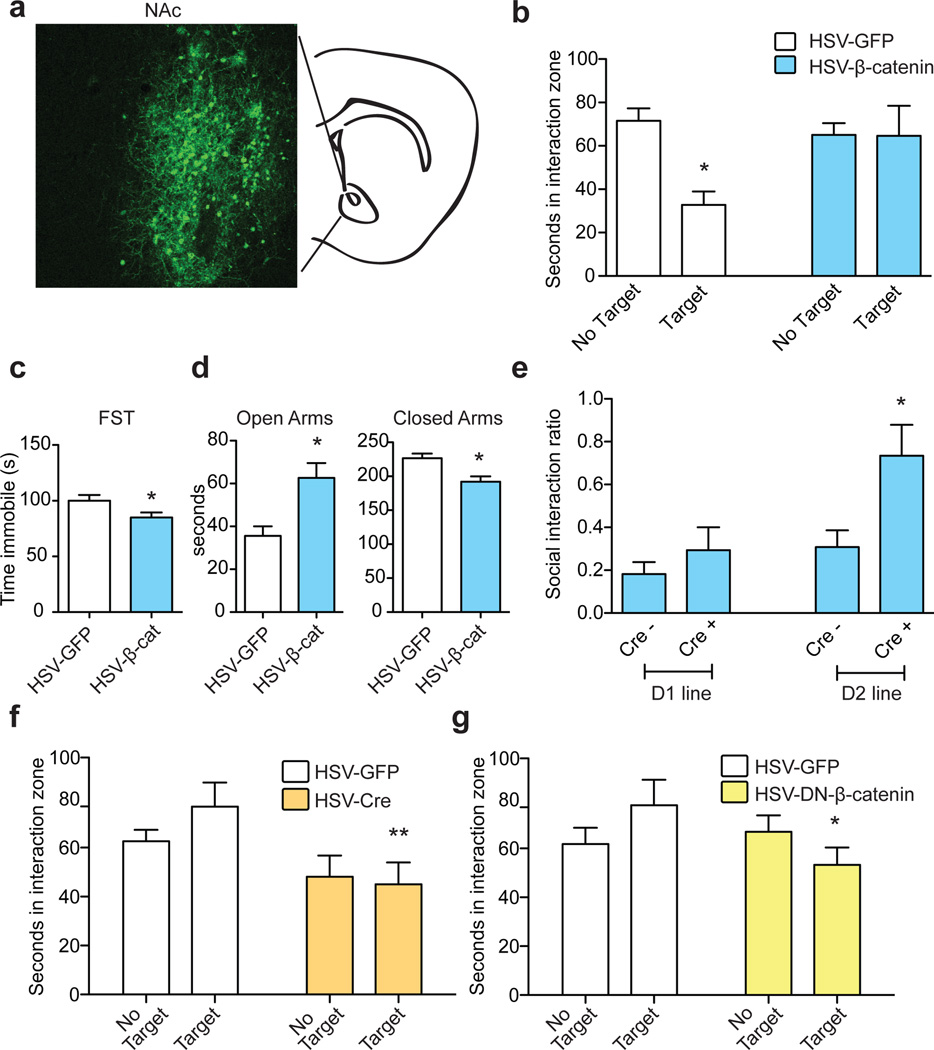
Figure 1. β-catenin in NAc mediates pro-resilient, antidepressant, and anxiolytic responses.
a, IHC illustrating viral transgene expression mediated by HSV-β-catenin with coronal cartoon of NAc highlighted. b, Pro-resilient effect of HSV-β-catenin on social interaction after ASD (*P<0.05, two way ANOVA, n = 8 GFP, n=10 β-catenin). c, Antidepressant-like effect of β-catenin in the FST (*P<0.05, two-tailed t-test, n=6 GFP, n=7 β-catenin). d, Anxiolytic-like effect of β-catenin in the EPM (Closed arms: * P<0.01, Open arms: * P<0.01, two-tailed t-test, n=6 GFP, n=7 β-catenin). e, Cell-type specific overexpression of β-catenin in ASD (D2 Cre– vs. Cre+: * P<0.05, two-tailed t-test, n = 13 D2Cre-, n=8 D2Cre+). f, Effect of knocking down β-catenin in a sub-threshold defeat procedure (**P<0.01, two way ANOVA, effect of virus only when target present, n=6 GFP, n=5 Cre). g, Effect of DN-β-catenin in sub-threshold defeat (*P<0.05, two way ANOVA, interaction effect, n=5 GFP, n=4 DN). Data presented as mean and SEM and is representative of at least two experiments. See Methods and Supplementary Table 9 for detailed statistics.
We next over-expressed β-catenin in NAc during accelerated social defeat stress (ASD)11, 12. We found that, while HSV-GFP injected control animals developed social avoidance, an indicator of depression-like behavior, overexpression of β-catenin prevented this phenotype (Fig. 1b). Furthermore, in baseline behavioral assays, β-catenin mediated an antidepressant-like response in the forced swim test (FST) (Fig. 1c), and anxiolytic effects in the elevated plus maze (EPM) (Fig. 1d). We saw no changes in sucrose preference or cocaine conditioned place preference (data not shown), suggesting that β-catenin does not cause hedonic changes. To confirm the pro-resilient effect of β-catenin, we utilized a stabilized β-catenin mutant (S33Y)13, and found identical results as for wildtype (WT)-β-catenin in the ASD and FST (Supplementary Notes), with no change in sucrose preference (data not shown). Finally, cell-type specific overexpression of β-catenin in D2- but not D1-type medium spiny neurons (MSNs) in NAc (Fig. 1e, Extended Data Fig. 2a) induced a pro-resilient phenotype.
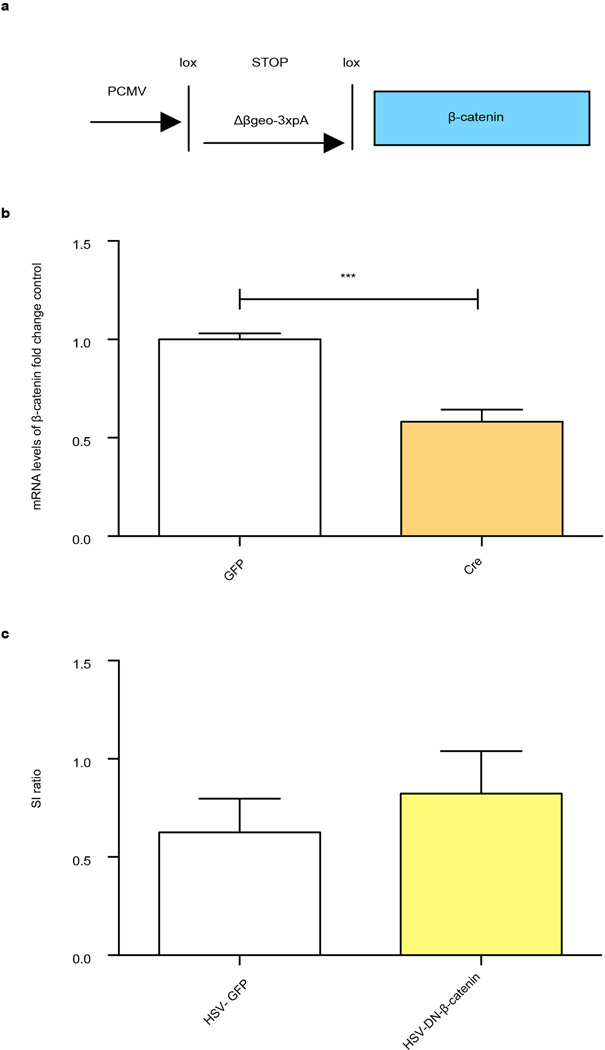
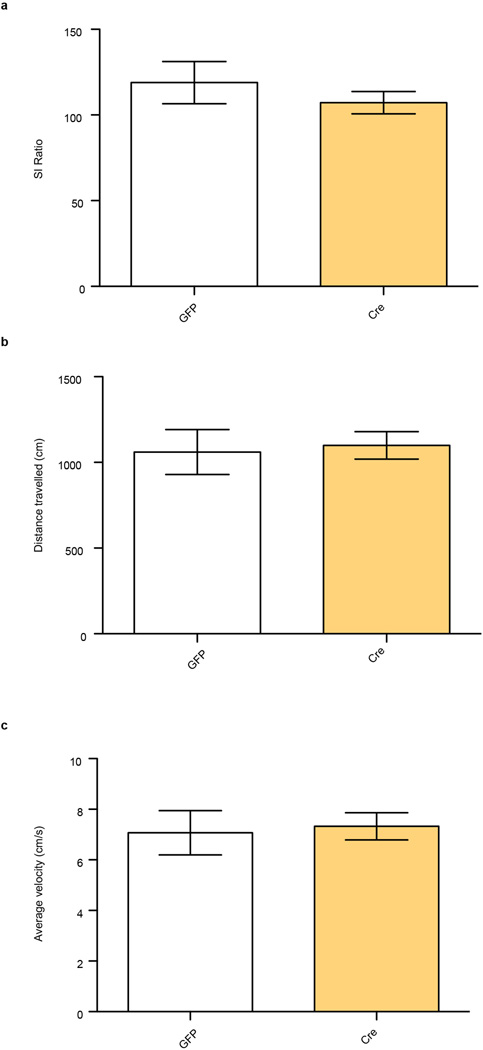
We also investigated the consequences of blocking β-catenin signaling in NAc with two approaches: excising β-catenin from NAc of conditional floxed mice (Extended Data Fig. 2b) and overexpressing a behaviorally-validated dominant negative (DN)-β-catenin mutant (Extended Data Fig. 2c)14. Both manipulations promoted susceptibility to stress in mice subjected to a sub-threshold defeat procedure (Figs. 1f, g). Excising β-catenin from NAc caused no change in social interaction (SI) or locomotion in control animals, demonstrating a specific association with stress (Extended Data Fig. 3a, b, c). These results establish a critical role for β-catenin signaling in NAc in behavioral resilience.
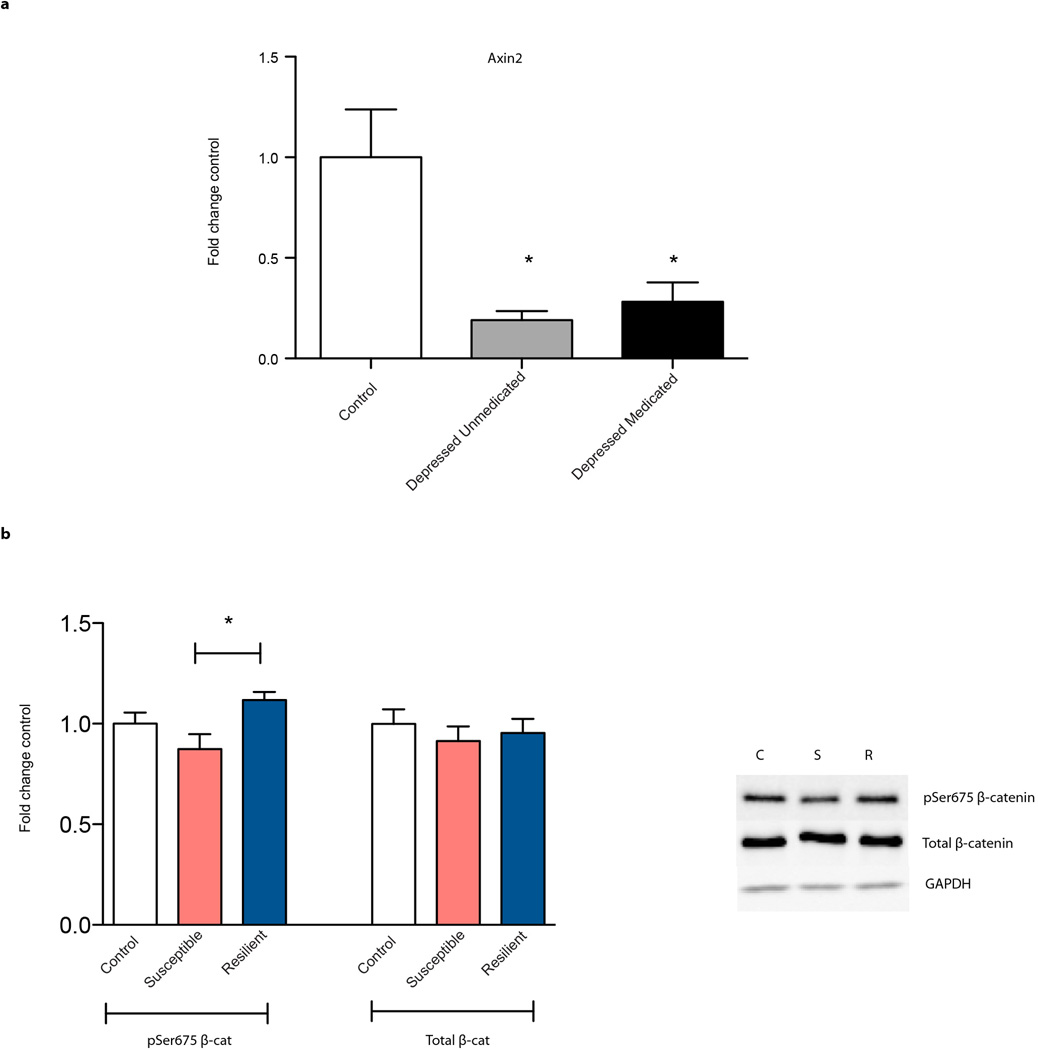
To explore the endogenous activity of β-catenin in depression, we examined its transcriptional activity in postmortem NAc of depressed humans. Axin2, a universal readout of activated canonical β-catenin signaling, was robustly suppressed in NAc of depressed humans (Fig. 2a, Supplementary Table 1, Extended Data Fig. 4a). In contrast, total N-cadherin and β-catenin mRNA levels were unchanged, pointing specifically to β-catenin nuclear function alterations in depression. There was also suppression of TCF3 and TCF4 (T cell transcription factors 3 and 4) levels in depressed patients; these are two of several transcription factors through which β-catenin acts. Together, these data demonstrate downregulation of the transcriptional output of β-catenin in NAc in human depression (Fig. 2a).
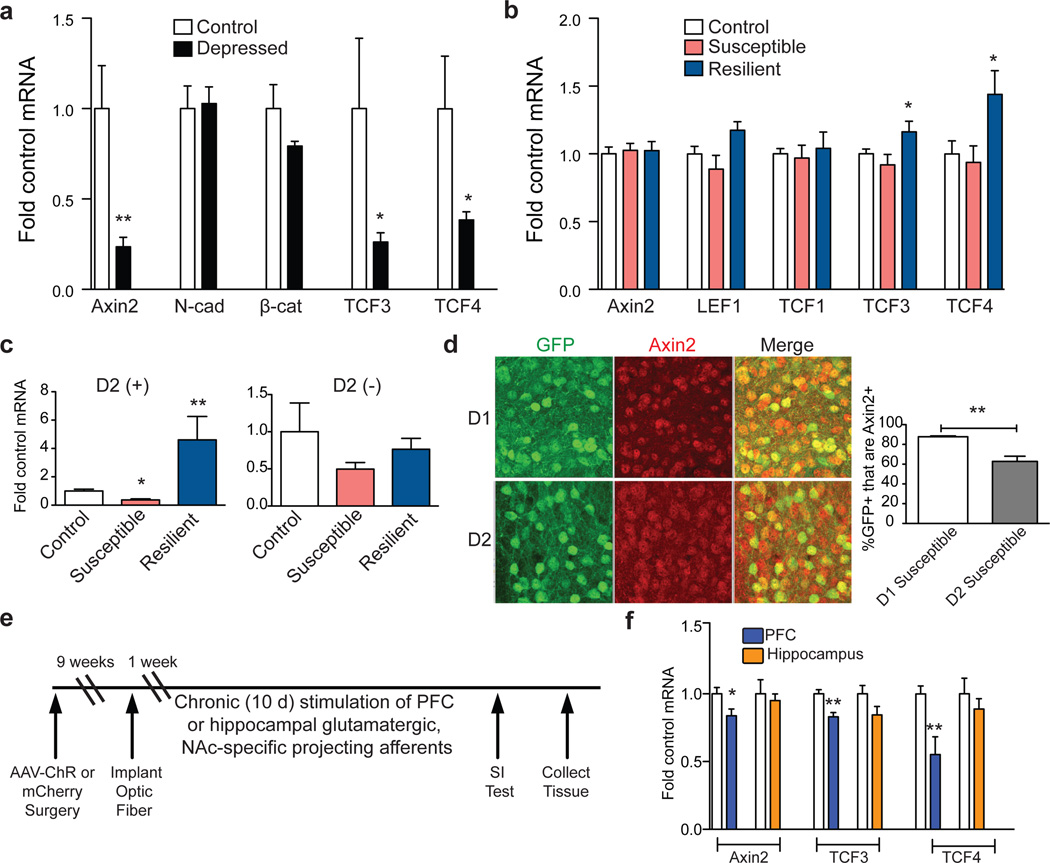
Figure 2. Regulation of β-catenin signaling in human depression and mouse CSDS.
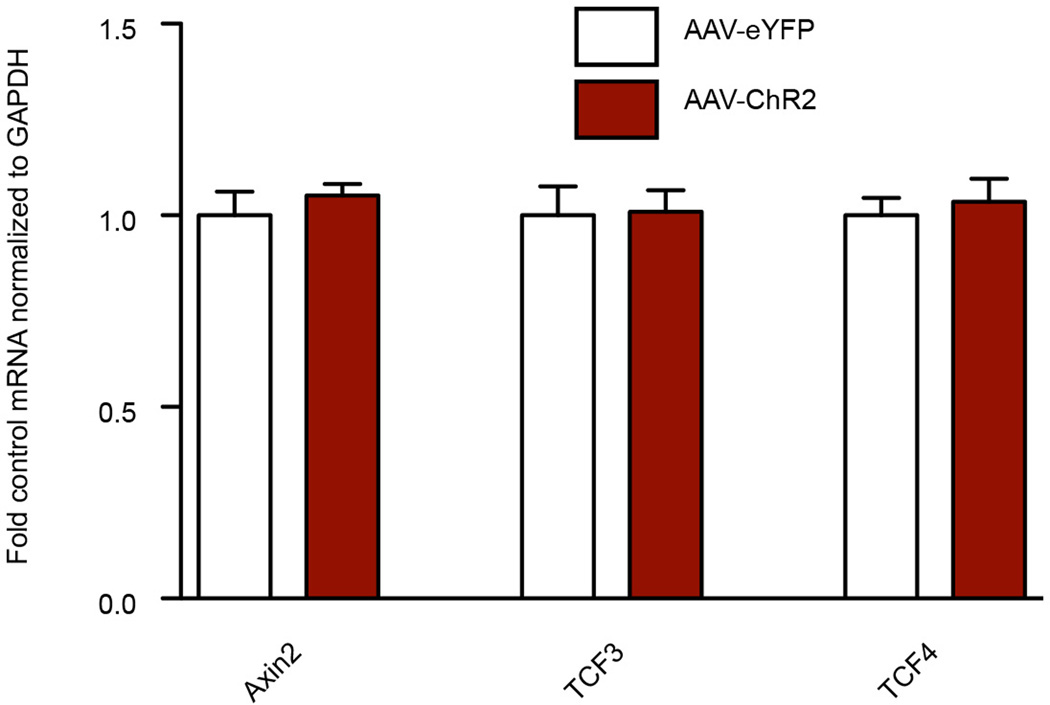
a, mRNA from human NAc (Axin2: **P<0.01; TCF3: *P<0.05; TCF4: *P<0.05, two-tailed t-test, n=6 control, n=10 depressed). b, mRNA from mouse control, susceptible, and resilient NAc 48 hours post CSDS (TCF3: *P<0.05; TCF4: *P<0.05, n= 8 control, susceptible, n=6 resilient). c, Axin2 is upregulated in D2(+) MSNs only in resilience (Axin2 D2(+):**p<0.01, control vs. resilient p<0.05, *p<0.01 susceptible vs. resilient, n=4 control, n=5 susceptible, n=3 resilient; Axin2 D2(−):NS, P>0.05, n=3 control, n=5 susceptible, n=3 resilient, one way ANOVA). d, Percentage of cells positive for Axin2 plus GFP in D1- or D2-GFP susceptible mice after CSDS (**P<0.01, two-tailed t-test, n=3/group). e, Optogenetic stimulation protocol. f, mRNA expression in NAc after repeated stimulation from PFC or hippocampus in ChR2 vs. mCherry (Axin2: *P<0.05, n=6 mCherry, n=5 ChR; TCF3: **P<0.01, n=4–6; TCF4: **P<0.01, n=6 mCherry, n=4 ChR, two-tailed t-test). Human data is from one experiment, all other data are representative of at least two experiments. All data presented as mean and SEM.
We next investigated Axin2 mRNA levels in mouse NAc 48 hours after chronic social defeat stress (CSDS). We found no difference between susceptible and resilient animals (Fig. 2b). However, resilient animals displayed increased TCF3 and TCF4, indicating that resilience may be associated with upregulation of β-catenin signaling (Fig. 2b). To probe this, we examined the levels of phospho-Ser675 β-catenin, a form with enhanced transcriptional activity, as well as total β-catenin at this time point. We found upregulation in resilient vs. susceptible animals of phospho-Ser675 β-catenin but not total β-catenin (Extended Data Fig. 4b). At 10 days after CSDS, we found elevated levels of Axin2 in resilient mice only (p <0.05, Supplementary Notes).
Cell-type specific β-catenin signaling regulates behavioral resilience
Given the small magnitude of change observed above, we questioned whether the cell-type specific behavioral effects in Fig. 1e corresponded to differential regulation of β-catenin signaling in D2 vs. D1 MSNs. Using FACS-isolated NAc neurons from D2-GFP mice, we found robust induction of Axin2 expression in D2(+) neurons of resilient mice, and significantly reduced Axin2 levels in susceptible vs. resilient mice, 48 hours post CSDS, effects not seen in D2(−) cells (Fig 2c). Furthermore, Axin2 IHC with D1- or D2-GFP transgenic mice subjected to CSDS revealed downregulation of β-catenin transcriptional activity in D2 vs. D1 MSNs in susceptible mice (Fig. 2d). In sum, upregulation of β-catenin signaling occurs in D2 MSNs in resilient mice, with downregulation seen in susceptible animals.
Because glutamatergic neurotransmission regulates β-catenin transcriptional activity and stress susceptibility15, 16, we tested whether medial prefrontal cortex (PFC) or hippocampus, two important glutamatergic inputs to NAc, control β-catenin signaling in NAc. Using previously validated constructs and stimulation protocols17, 18, we found that optogenetic stimulation of glutamatergic PFC terminals robustly suppressed β-catenin activity in NAc as indicated by decreased Axin2, TCF3, and TCF4, whereas stimulation of hippocampus terminals had no effect (Fig. 2 e, f). Repeated burst firing of dopamine afferents from the ventral tegmental area (VTA) also had no effect (Extended Data Fig. 5). Thus, PFC to NAc stimulation specifically elicited a molecular ‘signature’ of susceptibility, indicating that activation of this circuit could mediate the maladaptive suppression of β-catenin activity in NAc.
Genome-wide mapping of β-catenin after social defeat stress
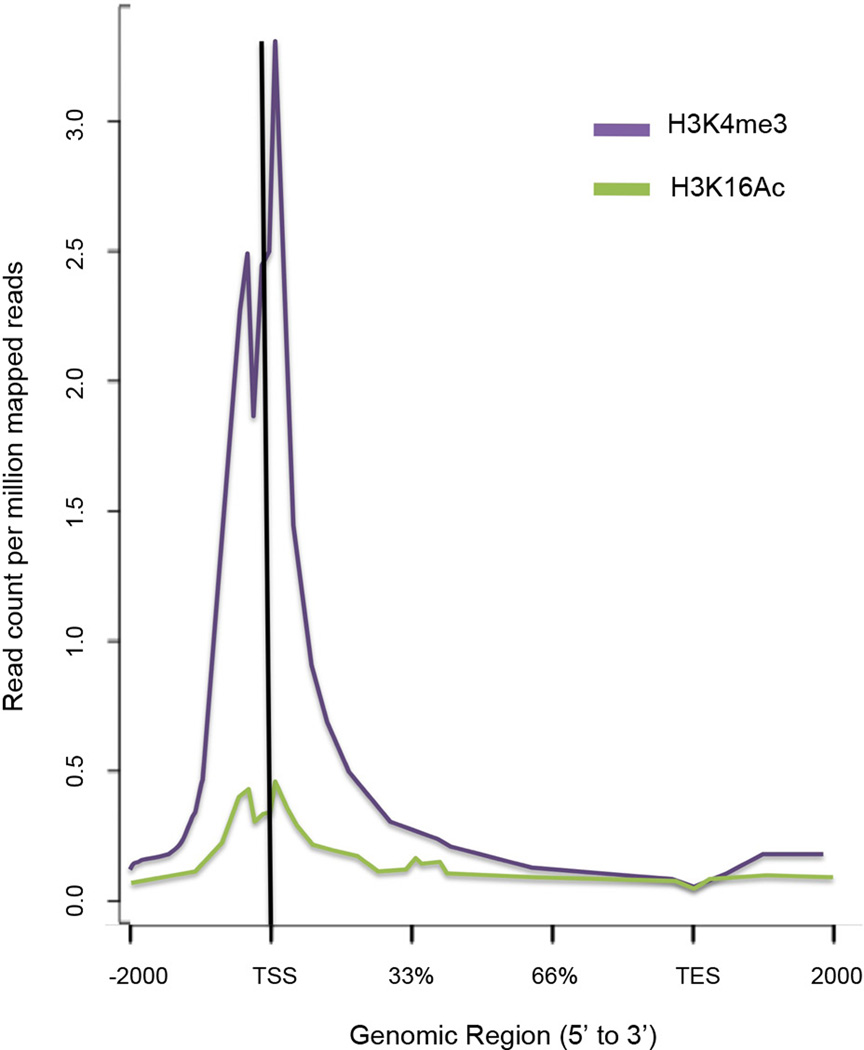
We next conducted β-catenin chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) on NAc of control, susceptible, and resilient mice after CSDS. We first validated our β-catenin ChIP protocol by examining an LEF/TCF consensus sequence in the promoter of a known β-catenin target gene, CaMKIV. We found enrichment of β-catenin at the LEF/TCF site, but not a distant site, in NAc of resilient mice only (Fig. 3a). Through ChIP-seq19, 20 we then examined global β-catenin enrichment after CSDS, and found major differences in peak numbers (Fig. 3b, Supplementary Data 1). Control and resilient conditions were associated with 10–15 fold higher absolute peak numbers compared to susceptible conditions, suggesting profound global alterations in β-catenin activity, consistent with our biochemical data (Fig. 2). Enrichment of β-catenin in resilient animals (Fig. 3b) only occurred at transcriptionally active sites as indicated by high basal binding of two transcriptional activation marks H3K4me3 and H4K16ac (Fig. 3c, Extended Data Fig. 6). However, we did not observe global changes in these two histone marks after CSDS (Extended Data Figs. 7, 8), suggesting that β-catenin may be recruited to active, open regions of chromatin through the presence of other, direct DNA-binding transcription factors.
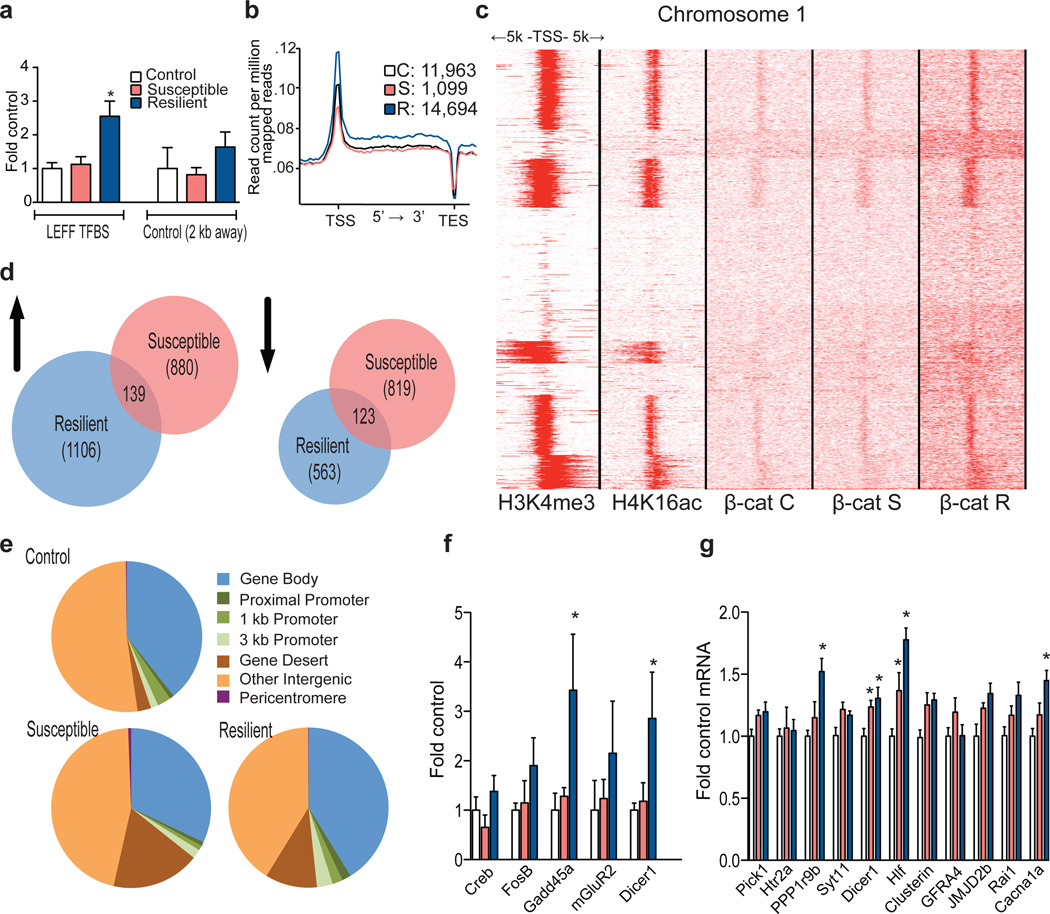
Figure 3. β-catenin ChIP-seq in NAc 48 hours post CSDS.
a, qChIP validation of β-catenin ChIP (* P<0.05, 1-way ANOVA, post-hoc test control vs. resilient and susceptible vs. resilient at LEFF transcription factor binding site (TFBS), n=4/group). b, Plot of β-catenin binding across gene regions. TSS: transcription start site, TES: transcription end site. Individual peak numbers per condition indicated in inset. c, Heatmap showing β-catenin binding 5kb up- and downstream of TSSs on chromosome 1 in control (c), susceptible (s), and resilient (r) NAc; binding profiles of H3K4me3 and H4K16ac under basal conditions are also shown. d, Number of increased (up arrow) vs. decreased (down arrow) β-catenin binding sites at promoters in resilient vs. control or susceptible vs. control conditions. e, Genome-wide distribution of β-catenin binding. f, qChIP validation of ChIP-seq (Gadd45a: *P<0.05, 1 way ANOVA; Dicer1: *P<0.05, 1 way ANOVA, n=4 control, susceptible, n=3 resilient). g, mRNA validation of β-catenin ChIP-seq (Dicer1: *P<0.01, 1-way ANOVA, n=13 control, n=11 susceptible, n=7 resilient). Data presented as mean and SEM and is representative of at least two experiments. Color-coding in f and g as in a.
Using Ingenuity Pathway Analysis, we demonstrated a predicted β-catenin network to be upregulated in NAc of resilient vs. susceptible mice (Extended Data Fig. 9), a prediction specific to β-catenin. Concomitantly, there were nearly twice as many increases as decreases in β-catenin binding in resilient vs. control mice at promoter regions. In contrast, susceptible vs. control animals displayed equivalent numbers of upregulated and downregulated β-catenin binding events (Fig. 3d). These results support our hypothesis that resilience is associated with genome-wide enrichment of β-catenin. Examining the distribution of β-catenin peaks across the genome (Fig. 3e) revealed similar results: redistribution of β-catenin binding towards promoters and gene bodies in resilience, and redistribution away from promoters/gene bodies and towards gene deserts in susceptibility.
To validate the β-catenin ChIP-seq data, we conducted quantitative ChIP (qChIP) on independent biological samples at genes that showed significant peaks in resilience or upregulation in resilient vs. susceptible animals, thus confirming significant β-catenin enrichment at several promoters (Fig 3f). As further validation, we examined mRNA levels of genes found in our ChIP-seq list that coincided either with in silico lists of predicted or known β-catenin targets21, 22 (Supplementary Table 2) or with the H3K4me3 and H4K16ac ChIP-seq datasets (Supplementary Data 2). We found robust upregulation of many of these genes in NAc of resilient mice (Fig. 3g).
Dicer1 and microRNA regulation by β-catenin after social defeat stress
One gene validated by qChIP and qPCR was Dicer1, a critical component of microRNA (miRNA) biogenesis23. Thus, selective enrichment of β-catenin binding at Dicer1 in resilient mice (Fig. 4a), and subsequent validation of this effect (Fig. 3f, g), indicated that Dicer1 represents a robust target of β-catenin in NAc. To study Dicer1’s behavioral effects, we knocked it down locally in NAc (Extended Data Fig. 10), and conducted sub-threshold defeat. Control animals injected with HSV-GFP displayed normal social interaction; however, animals with Dicer1 knockdown demonstrated social avoidance (Fig. 4b), which mimicked the effects of blocking β-catenin signaling (Fig. 1). Importantly, we can rule out confounding effects of long-term Dicer1 loss on neuronal viability24, since our experimental paradigm was limited to two weeks.
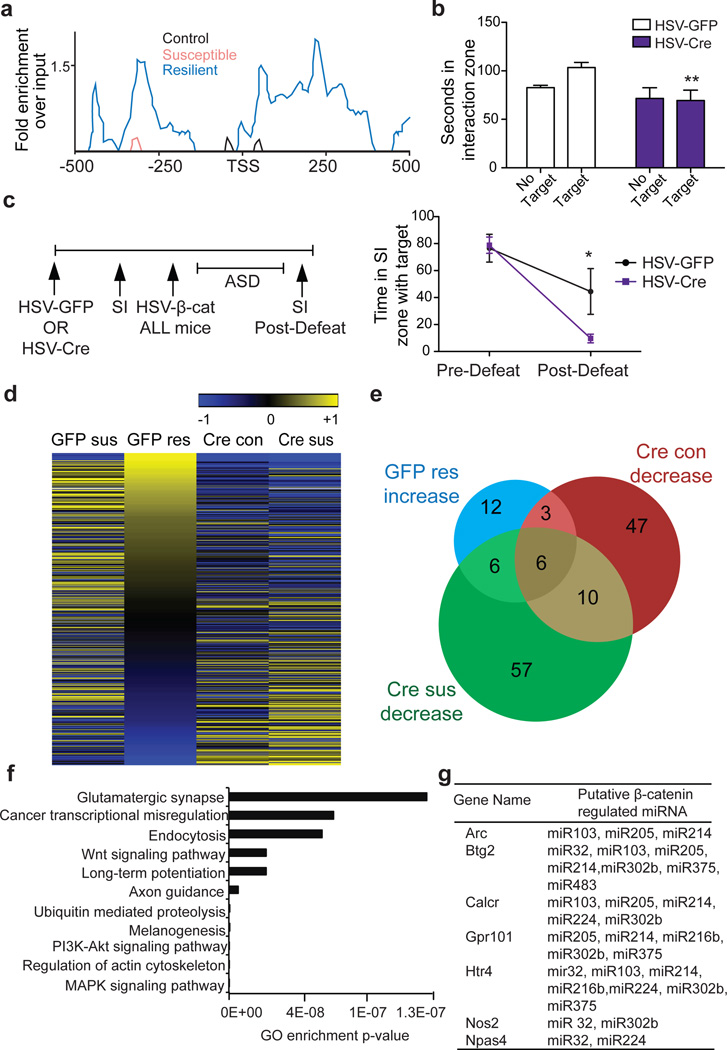
Figure 4. Dicer1 bridges β-catenin and miRNA regulation in CSDS.
a, β-catenin ChIP-seq enrichment around the Dicer1 TSS. b, Effect of NAc Dicer1 knockdown (HSV-Cre) in sub-threshold defeat with HSV-GFP as control (**P<0.01, effect of virus, two-way ANOVA, n=7 Cre, n=8 GFP). c, (left) Schematic of floxed Dicer1 deletion followed by β-catenin rescue; (right) social interaction before and after ASD with HSV-β-catenin (*P<0.05, interaction effect, matching two-way ANOVA, n=7/group). d, Heatmap of CSDS-regulated miRNA expression changes with (Cre) or without (GFP) β-catenin knockdown. Log 2 fold changes of all altered miRNAs among all groups are shown. e, Venn diagram showing increased miRNAs in GFP-resilient mice (GFP-res) overlap with decreased miRNAs in β-catenin knockout in non-stressed (Cre-con) or susceptible (Cre-sus) animals. f, Top 11 most enriched gene ontology terms of target genes of overlapping miRNAs in panel e. g, Predicted targets of β-catenin-dependent miRNAs display downregulation by mRNA-seq in resilient mice after CSDS. Data presented as mean and SEM and is representative of at least two experiments.
To assess whether the behavioral effect of Dicer1 was related to β-catenin signaling, we first expressed HSV-Cre or HSV-GFP in NAc of floxed Dicer1 mice and found no difference in SI under baseline, non-stressed conditions (Fig. 4c). We then injected all mice with HSV-β-catenin in NAc and subjected them to ASD. β-catenin overexpression blocked the development of social avoidance in mice expressing normal Dicer1 levels, but not in mice with NAc Dicer1 knockdown (Fig. 4c). This indicates that at least part of the pro-resilient effect of β-catenin is mediated through Dicer1.
Finally, these data prompted us to examine the global miRNA profile in NAc in response to CSDS and study its dependence on β-catenin. We injected AAV-GFP or AAV-Cre in NAc of floxed β-catenin mice, subjected them to CSDS or control conditions, and performed small RNA-seq (Supplementary Table 3). We first compared each group—GFP susceptible (GFP-sus), GFP resilient (GFP-res), Cre control (Cre-con), and Cre susceptible (Cre -sus) to the “GFP-con” condition. We could not study the Cre resilient condition, because virtually no mice are resilient upon β-catenin knockout from NAc. We found downregulation of numerous miRNAs, including many that were upregulated in resilience, when β-catenin was knocked out from control animals (Cre-con, Fig. 4d, Supplementary Table 4). Interestingly, a subset of miRNAs was upregulated following β-catenin knockout, which may represent miRNAs that are regulated by repressive factors under β-catenin control. We identified 66 miRNAs that were significantly downregulated in NAc after β-catenin deletion (Cre-con, Fig. 4e). We also identified downregulated miRNAs (n=79) in the Cre-sus condition, many of which were decreased in Cre-con, further substantiating our hypothesis that pro-adaptive miRNA responses are lost in the absence of β-catenin, enhancing susceptibility to stress (Fig. 4e). miRNAs that overlapped between any two groups (up in GFP-res, but down in Cre-con or Cre-sus), presumably represent the most biologically important β-catenin- and stress-regulated miRNAs (Fig. 4e, Supplementary Table 5). This subset controls several meaningful gene categories (Fig. 4f), including Wnt and glutamatergic signaling. Finally, to identify potential miRNA targets, we overlapped predicted targets of these β-catenin-regulated miRNAs (Supplementary Table 5) with mRNA-seq data from NAc after CSDS. We thus found several interesting, novel genes to be significantly repressed in resilience (Fig. 4g).
We also examined other small RNAs for regulation by CSDS. Piwi-interacting RNAs (piRNAs), small RNAs widely studied in germ line cells, were detected recently in brain and found to play a functional role in spine morphology and synaptic plasticity25, 26. 163 piRNAs were detectable in our dataset with read counts in at least one condition, supporting the notion of piRNA expression in brain (Supplementary Table 6). Although the majority of them were expressed at low levels, approximately 20 piRNAs appear to be regulated by CSDS (Supplementary Table 7). Examining additional small RNA categories that might be regulated by Dicer1 revealed several differentially expressed candidates (Supplementary Table 8), laying the groundwork for future investigation.
Discussion
The present study demonstrates that β-catenin in D2 MSNs activates a network in NAc that mediates behavioral resilience, whereas deficits in this pathway contribute to depression-related pathology. PFC inputs to NAc appear to be particularly important in controlling this β-catenin regulation. D2 MSNs, which comprise the indirect or ‘no-go’ pathway27–30, may be more important for mediating flexible behavioral choices in aversive contexts compared to reward-motivated behavior31–33. We thus posit that enhanced β-catenin signaling in NAc D2 MSNs of resilient mice permits increased behavioral flexibility, which allows them, despite having the same experience as susceptible mice, to overcome generalizing avoidance of all mice, a process independent of hedonic responses. This has parallels in humans: resilient individuals are more successful at managing stress and recovering from it34.
Our β-catenin ChIP-seq approach provides a valuable resource for mining the molecular targets that drive resilience. One validated target is Dicer1, which establishes a novel connection between β-catenin signaling and miRNAs in brain. Among the regulated miRNAs are those that feedback and regulate β-catenin signaling35. The cell type-specific role of β-catenin, and the inherent complexity of stress susceptibility vs. resilience, which involves many additional regulatory steps beyond Dicer1, presumably explains the relatively small number of β-catenin-dependent miRNAs observed in this study. miRNAs provide a crucial layer of post-transcriptional gene regulation in neural development, plasticity, and in an increasing number of brain disorders36–38. The present study, by identifying specific miRNAs associated with susceptibility or resilience, offers a template for future studies to induce resilience in inherently more susceptible individuals.
Full Methods (online)
Animals
For all experiments, 7–9 week old male mice were used. Unless otherwise noted for transgenic lines, c57bl/6 mice from Jackson Laboratories were used. All mice were housed on a 12-hour light/dark cycle with ad libitum access to food and water. CD1 retired breeder mice were obtained from Charles River Laboratories. The following transgenic mouse lines were used. From Jackson Laboratories: β-catenin conditional floxed mice (stock # 004152) and Dicer1 conditional floxed mice (stock # 006001). Additionally, D1-Cre, D2-Cre, D1-GFP, and D2-GFP male mice that were backcrossed to a c57bl/6 background were used for experiments as described in the text. For the D1-Cre/D2-Cre cell-type specific overexpression experiments, wild-type littermates were used as controls. The Mount Sinai Institutional Animal Care and Use Committee approved all animal protocols utilized in this study. For all experiments, extensive laboratory experience was used to estimate required sample sizes. Animals were randomly assigned to experimental groups and whenever possible, experimenters were blinded to the group. (For example, in behavioral experiments by assigning numbers to animals and in IHC experiments by hiding group designation until after quantification and analysis.)
Viral Mediated Gene Transfer
Stereotactic surgery was performed on mice under ketamine/xylazine anesthesia. Vectors were infused bilaterally into NAc at a rate of 0.1 µl/min with the following coordinates: +1.6 mm A/P, +1.5 mm M/L, ± 4.4 mm D/V from bregma. A total of 0.5 µl/side was infused except for the HSV-LS virus, in which case 0.7 µl was infused total. All vectors used were cloned into p1005 HSV or LS1L HSV. Mouse β-catenin constructs were provided by Dr. Steven Borkan (Boston University). WT and DN constructs were used, with the DN construct containing amino and carboxy-terminal truncations. Because this is a complicated mutant, we behaviorally validated it by demonstrating a failure to rescue β-catenin loss of function impairments in social interaction (Extended Data Fig. 2c). Human β-catenin S33Y construct (Addgene Plasmid#19286) was originally from Dr. Eric Fearon (Michigan). This mutant contains an S33Y mutation that prevents phosphorylation at Ser33 by GSK3β, thus preventing β-catenin degradation. For cell-type specific overexpression, an HSV carrying β-catenin in a lox-stop cassette was used (Supplementary Fig. 2a) in conjunction with D1- and D2-Cre transgenic mouse lines. Viral-Cre was used for local knockdown of β-catenin or Dicer1 in conditional floxed mice.
Behavior
10 day chronic social defeat stress (CSDS), an accelerated 4 day defeat procedure (ASD), and a sub-threshold defeat procedure have been described previously and represent an ethologically validated model of depression9, 11–12 We used ASD over 4 days (4 days of defeat, 2x/day) to coincide with periods of maximal HSV-mediated transgene expression in some experiments as described, which induces the same degree of behavioral deficits in normal mice as our standard 10-day CSDS procedure. For all defeats, social interaction (SI) was measured either 24 hours or 1 week following the last defeat. For all tissue analysis, including ChIP, mice were killed 48 hours after the last defeat (24 hours after SI) of a 10 day CSDS paradigm unless otherwise specified. Elevated plus maze and forced swim tests were performed as described previously12.
Postmortem human tissue
Human postmortem NAc cDNA was generated and analyzed as before9. Briefly, brain tissue was obtained from the Dallas Brain Collection, where tissue is collected from the Dallas Medical Examiner’s Office and UT Southwestern’s Tissue Transplant Program following consent of next-of-kin. Tissue was analyzed and matched for age, postmortem interval, RNA integrity number (RIN), and pH (see Supplementary Table 1) and this same tissue set was used in previously published work39. Samples were subjected to a standard dissection before snap freezing in 40°C isopentane and storage at 80°C; further dissection of NAc was performed on frozen tissue. The UT Southwestern Institutional Review Board reviewed and approved the collection of this tissue for research use. We should note that there was no difference in expression of Axin2 between medicated and unmedicated depressed patients, although all patients were clinically depressed at their time of death (Supplementary Fig. 4a). We thus combined the medicated and unmedicated groups into one overall depressed group as presented in Fig. 2.
RNA isolation and qPCR
RNA was extracted and purified using a protocol combining Trizol/choloroform extraction with the Qiagen RNeasy Micro kit, with a motorized mini-pestle vibrator to homogenize the tissue. After extraction, purity and concentration were measured on the Nanodrop spectrophotomoter. RNA was then reverse transcribed into cDNA with the iScript DNA synthesis kit (BioRad). GAPDH was utilized to normalize quantification. Primers were designed to flank exon/intron boundaries and were created using the open-source software Primer3. Real-time qPCR analysis was performed with the ΔΔCT method to obtain relative fold-change of expression as compared to control samples40. BLAST and dissociation curve analysis was also performed to ensure specificity of primer design.
Western blotting
NAc was dissected bilaterally using 14 gauge steel circular punches. The tissue was then sonicated in radioimmunoprecipitation assay (RIPA) buffer with a desktop sonicator (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, with protease and phosphatase inhibitors) and centrifuged. The supernatant was collected and the protein concentration was quantified using the Lowry method. Laemelli sample buffer was added to the protein lysate and equal amounts of protein were loaded onto precast SDS-PAGE gels with molecular weight ladders. Samples were transferred to activated PVDF membranes, blocked, and incubated in primary antibody overnight. Blots were washed, and then incubated with Licor secondary fluorescent antibodies. After further washing, the blots were scanned and images analyzed with Image-J software. The following antibodies were used: phospho-Ser675 β-catenin (Cell Signaling #4176; Ser675 is phosphorylated by PKA), total β-catenin (Cell Signaling #9562), GAPDH, β-tubulin, and total H3. All antibodies are commercially available and have been validated for use in the laboratory. Pre-incubating the tissue with calf intestinal phosphatase and demonstrating a decrease in signal was performed to validate the phospho-Ser675 β-catenin antibody.
Optogenetics
For glutamatergic nerve terminal stimulation, mice were injected unilaterally with AAV-CAMKIIa-ChR2-mCherry or AAV-CAMKIIa-mCherry with the coordinates of: (−3.6 AP, +3.05 ML, −4.85 DV) for ventral hippocampus and (+1.9 AP, +0.5 ML, −3.0 DV) for PFC unilaterally. After 9 weeks of recovery to allow for expression in terminals, a second sterotactic surgery was performed to implant an optic fiber targeting the NAc shell with coordinates of (+1.4 AP, +1.5 ML, −4 DV), again unilaterally, ipsilateral to virus expression. After allowing one week for recovery, the mice underwent 10 days of daily 5-minute stimulation sessions outside of their home cage as described41–42. Stimulation parameters were either 20 Hz, 30 pulses/burst, with 10 seconds between bursts (hippocampus); or 30 Hz, 90 pulses/burst, 10 seconds between bursts (PFC) to roughly balance the relative intensity of NAc innervation from these two afferent regions. Unilateral NAc tissue was then dissected 48 hours later for biochemical experiments. Constructs and stimulation parameters have been previously validated17, 18. AAV-ChR2 was used to stimulate VTA cell bodies with a phasic protocol (20 Hz, 5 spike/burst, 10 sec between bursts) given susceptible mice exhibit increased firing rate and bursting events following defeat12, 43.
Co-immunoprecipitation (Co-IP)
A co-IP kit (Roche) was used as follows. 4 punches of NAc were lysed in 300 µl of the provided lysis buffer. 10% total lysate was reserved and the rest was centrifuged and the supernatant transferred to a clean microcentrifuge tube. It was pre-cleared by incubation with protein G-agarose for 3 hours on a rotator at 4°C. The beads were centrifuged, and the supernatant was transferred to fresh tubes, where they were incubated with 5 µl of β-catenin antibody (Cell Signaling #9581) for one hour before 50 µl of a homogeneous protein G-agarose suspension was added and then incubated overnight at 4°C on a rotator. The complexes were centrifuged and the supernatant was removed, the beads were washed 2x with lysis buffer 1, 2x with buffer 2, and once with buffer 3. Protein sample buffer was added and the samples boiled for 3 minutes. Complexes were then analyzed as described under Western blotting.
Nuclear/cytoplasmic fractionation
NAc punches were homogenized with a glass Dounce tissue grinder and loose pestle in Buffer A (1 M Tris-HCl, 1 M sucrose, 1 M DTT, protease and phosphatase inhibitors). 10% of the lysate was reserved to assay total protein levels, and the rest was centrifuged at 3700 RPM for 10 minutes. The supernatants were then removed, centrifuged at 7500 RPM for 7 minutes, and the resulting supernatants were stocked as the cytoplasmic fraction. Buffer B (1 M Tris-HCl pH 7.5, 0.1 M EDTA, 0.1 M EGTA, 1 M sucrose, 1 M DTT, 10 % NP-40, protease and phosphatase inhibitors) was added to the pellets from the first centrifugation and the samples were kept on ice for 10 minutes before centrifuging again at 3700 RPM for 10 minutes. The supernatants were discarded and Buffer C (1 M Tris-HCl, 37.5% glycerol, 5 M NaCl, 0.1 M EDTA, 0.1 M EGTA, 1 M DTT, 10% NP-40, protease and phosphatase inhibitors) was used to re-suspend the nuclear fraction. The fractions were then processed for Western blotting as above or further separated into chromatin and non-chromatin nuclear fractions. Tubulin and total H3 were used as loading controls and to verify appropriate cytoplasmic and nuclear enrichment.
Immunohistochemistry (IHC)
Mice were anaesthetized with chloral hydrate followed by trans-cardial perfusion of 10 ml of filtered PBS, followed by 25 ml of filtered 4% paraformaldehyde (PFA) in PBS, pH 7.4. Brains were dissected out and post-fixed overnight in PFA. They were then rinsed in PBS and placed in 30% sucrose in PBS. For the IHC in Figure 2, once the brains sank, coronal 35 µm sections through the NAc were taken on a freezing microtome and kept in PBS with 0.01% sodium-azide. The slices were washed 3x in PBS for 10 minutes and then blocked for 3–4 hours (3% normal goat serum, 0.3% TritPBS) in net wells. They were incubated in primary antibody overnight diluted in block (rabbit Anti-Axin2, Abcam; Mouse Anti-GFP, Life Technologies) at 4 degrees. The slices were then washed 3x in PBS, followed by a 1-hour incubation in secondary antibody (Alexa-Fluor Anti-Rabbit & Anti-Mouse 680 & 800 diluted 1:1K in PBS). The slices were washed 4x in PBS and then mounted on charged slides and allowed to dry overnight. They were dehydrated, cover-slipped with Depex mounting medium, and sealed with clear nail polish. Z-stacks were taken on a Zeiss LSM 710 confocal microscope at 64x. Settings were kept identical for all images taken. The specificity of the Axin2 antibody was validated by competing the antibody with the immunizing protein. Average values of 3–5 images/mouse were used. For quantification purposes, the percent of Axin2+/GFP+ cells was counted per image, with Axin2+ being defined as >20% above background levels.
For the IHC in Supplementary Figure 1, coronal sections (50 µm thick) were made with a vibratome; sections were collected into antifreeze solution consisting of Ethylene glycol, glycerol, and PBS. Free-floating sections were blocked using 3% BSA in 0.1% PBST for 1 hour. The sections were stained for 48 hours at room temperature with primary antibody, and overnight with secondary antibody. The sections were mounted with Prolong Antifade reagent with DAPI (Life Technologies). Z-stacked images were acquired with a Zeiss LSM780 multi photon confocal system and processed using ImageJ. The number of GFP(+) cells containing β-catenin staining was quantified by requiring the presence of β-catenin in the nucleus. To quantify β-catenin protein expression, we used the rabbit-conjugated primary antibody for total β-catenin (9562; Cell Signaling). We also amplified GFP staining using a chicken-conjugated primary antibody for GFP (Aves Laboratory). Stains were visualized using Chicken-Cy2 and Rabbit-Cy3 secondary antibodies (Jackson Immunolabs).
FACS
D2(+) and D2(−) cells from the NAc of D2-GFP mice were isolated using a fluorescence-assisted cell sorting (FACS) protocol. Briefly, 48 hours after our standard CSDS protocol, bilateral 12 gauge punches were taken from the NAc and digested with an enzyme cocktail for 30 minutes at 37°C before being triturated to obtain a homogeneous cellular preparation. Cells were then processed through a gradient, washed, and labelled with DAPI (viability marker) before being processed through an Influx sorter (BD Bioscience). D2(+)MSNs were sorted based on the size, internal complexity, and intensity of fluorescence with D2 cells emitting in the green channel (GFP). RNA was isolated using the Direct-zol RNA miniprep (Zymo Research) kit and cDNA was synthesized using the Iscript kit (Biorad). We confirmed the enrichment of D2 MSN-enriched genes in D2(+) cells and D1 MSN-enriched genes in D2(−) cells.
Quantitative chromatin immunoprecipitation (qChIP)
Four 14 gauge NAc punches from each mouse were placed in 1% formaldehyde in 1x PBS to fix the DNA with the associated proteins. After 12 minutes on the rotator, 2 M glycine was added to stop the fixation for 5 minutes. The punches were then placed on ice and rinsed 5x with ice-cold PBS. Tissue from 5 animals were pooled at this point and homogenized in SDS lysis buffer (10% SDS, 1M Tris-HCl, 0.5 M EDTA) with a desktop sonicator. ChIP dilution buffer (10% Triton X-100, 5 M NaCl, 1 M Tris-HCL pH 8.1, 0.5M EDTA, 10% SDS and protease inhibitors) was added and the chromatin underwent high power sonication with the Bioruptor for 30 cycles of 30 seconds on/30 seconds off on high power. Conjugated magnetic beads were used to IP β-catenin with the ChIP-validated β-catenin antibody (Cell Signaling #4176) overnight in block solution (0.5% BSA in 1x PBS). The IP reaction was collected with a magnetic rack, washed, and both the input chromatin and the IPed DNA were reverse cross-linked at 65° overnight. The DNA was then purified with RNAse, Proteinase K, and the Qiagen PCR purification kit. The Qubit was used to quantify both the input and IP’d DNA and RT-PCR was used to quantify differential binding on the genomic DNA.
ChIP-seq
ChIP was performed for β-catenin as above. At the PCR purification step, however, 3 replicates were pooled onto one spin column so that each replicate became the pooled sample of 15 mice or 60 14 gauge NAc punches (4 NAc punches/animal), totaling ~100 mg wet weight tissue per library, following established protocols for brain. Animals were pooled so that social interaction times of different replicates across a group were approximately equal. ChIP-seq libraries were then prepared with the Illumina ChIP-seq kit as per their protocol. 2 replicates/condition were used for β-catenin while 3 replicates were used for histone mark experiments. Histone mark ChIP-seq was performed similarly except no further pooling was performed at the PCR purification step. Additionally, fragments from ~200–400 bp were size selected for sequencing for β-catenin to compensate for the decreased yield of DNA. Libraries were validated on the Bioanalyzer for appropriate size selection and amplification before being sent to the Mount Sinai Genomics Core for sequencing. Homer44 was used to identify peaks in individual conditions and NGS plot was used to create genome-wide overviews of binding at gene bodies45. We used hierarchical clustering based on the H3K4me3 dataset to generate the heatmap in Fig. 3c. To further validate our β-catenin datasets, we found that, in all 3 treatment conditions, IPA pathway analysis independently identified β-catenin as an upstream regulator due to the enrichment of known β-catenin target genes.
Small RNA-seq and analysis
Small RNA (<200 bp) was isolated and enriched with Qiagen RNeasy mini kit (Cat# 74104) following instructions. The small RNA was then used for library preparation following Epicentre Scriptminer small RNA library kit (Cat# SMSP10908) with optimization. In brief, a 3’ adaptor tag was ligated to the small RNA, then a 5’ adaptor oligonucleotide was attached following removal of excess 3’ adaptor oligonucleotide with degradase. The Di-tagged RNA was purified with Zymo RNA Clean & Concentrator Kits (Cat # R1015) and followed with reverse transcription into cDNA using the cDNA Synthesis Primer and MMLV Reverse Transcriptase. After removing RNA template by addition of RNase, the di-tagged cDNA was amplified and individually barcoded with nine PCR cycles using indices and PCR primers provided in the kit. The library was purified with Zymo DNA Clean & Concentrator kit (Cat# D4003) and size selected with Pippin (Sage Science). The library concentration was confirmed on Agilent Bioanalyzer prior to sequencing. Multiplexed libraries were then pooled and sequenced on an Illumina Hiseq sequencer. In total, 4–12 libraries/condition were included in this study. Raw sequencing reads were processed by cutadapt (https://code.google.com/p/cutadapt/) to remove adapter sequence at 3’ end, and sequences shorter than 16 nt after this were discarded. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was applied to inspect the sequencing quality. We ensured our small RNA sequencing was of good quality as the majority of reads aligned to mature miRNAs (Supplementary Table 3). miRanalyzer was used to align the short reads to genomic annotations and quantify the expression of the non-coding RNAs46. All miRNA annotations were downloaded from miRBase (v. 20)47. piRNA annotations were merged from piRNABank and NCBI48–49. tRNA and mRNA (RefSeq) annotations were downloaded from UCSC genome browser. The general ncRNA annotations were obtained from RFam (http://rfam.xfam.org/)50. The pipeline was organized by Ruffus (https://code.google.com/p/ruffus/), and the code is accessible from GitHub (https://github.com/shenlab-sinai/miRNA_pipeline_for_miRanalyzer)51. The differential expression detection was applied by DESeq2 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html, http://biorxiv.org/content/early/2014/02/19/002832) with cutoffs of fold change 1.3 and P-value<0.05.
Statistics
One- or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test, or two-way student’s t-test were used for statistical analyses. All experiments represent at least 2–3 biological replicates unless otherwise indicated.
Extended Data
Extended Data Figure 1. Validation of HSV-β-catenin.
a, β-catenin mRNA levels following HSV-β-catenin vs. HSV-GFP injection into NAc (t4=2.240, *P<0.05, two-tailed t-test, n=3/group). b, Top Panel: Subcellular fractionation of NAc lysates from HSV-GFP or HSV-β-catenin injected mice. Bottom Panel: IHC of nuclear β-catenin 5 days post-injection with HSV-β-catenin vs. HSV-GFP (t4=9.84, *** P<0.001, two tailed t-test, n=3/group). c, β-catenin IP on virus-injected NAc. IP results are representative of 5 replications. All other data shown are representative of at least two experiments. Data is presented as mean and SEM.
Extended Data Figure 2. Other β-catenin manipulations.
a, Schematic of Cre-dependent HSV-lox-stop (LS1L)-β-catenin cassette. b, Validation of β-catenin knock down in the NAc of floxed β-catenin mice (t7=5.620, ***P<0.001, two tailed t-test, n=4 GFP, n=5 Cre). c, Failure of DN-β-catenin to rescue SI as compared to GFP after previous excision of β-catenin from NAc in floxed β-catenin mice undergoing defeat (t12=0.7157, P>0.05, two tailed t-test, n=7/group). Data is presented as mean and SEM. All data shown are representative of at least two experiments.
Extended Data Figure 3. No effect of β-catenin deletion on baseline behaviors.
a, Social interaction (SI) in control, non-stressed animals (t8=0.840, P>0.05, two tailed t-test, n=5/group). b, total distance traveled in arena (t8=0.251, P>0.05, two tailed t-test, n=5/group). c, average velocity (t8=0.251, P>0.05, two tailed t-test, n=5/group). Data is presented as mean and SEM. All data shown are representative of at least two experiments.
Extended Data Figure 4. Regulation of β-catenin signaling in human depression and after CSDS in mice.
a, Axin2 expression is suppressed in both medicated and unmedicated depressed patients, both groups of which were clinically depressed at their time of death (F2,13=7.425, P<0.01 one-way ANOVA, post-hoc test P>0.05 between depressed unmedicated and medicated groups, *P<0.01 for either depressed group vs. control, n = 6 control, n=5 unmedicated depressed, medicated depressed). b, Phospho-Ser675 β-catenin and total β-catenin levels from mouse control, susceptible, and resilient NAc 48 hours post CSDS (phospho-Ser675: F2,16= 3.477 *P<0.05, post-hoc test susceptible vs. resilient, n=5 for control, susceptible, n=8 for resilient). Data is presented as mean and SEM. Human data is from one experiment. All other data shown are representative of two experiments.
Extended Data Figure 5. Repeated optogenetic burst stimulation of VTA cell bodies has no effect on canonical β-catenin signaling in NAc.
Experiment was performed as in Figure 2 with the exception of the optic fiber, which was placed above VTA for cell body stimulation (P > 0.05, two tailed t-test, n = 8/group). Data is presented as mean and SEM. Data is from one experiment.
Extended Data Figure 6. Genome-wide enrichment of H3K4me3 and H4K16ac binding in NAc at TSSs.
NGS plot was used to visualize binding patterns.
Extended Data Figure 7. Genome-wide pattern of H3K4me3 binding to genic regions in NAc under control, susceptible (defeat), and resilient mice.
Note the lack of difference across the three conditions. Data is from one experiment.
Extended Data Figure 8. Genome-wide pattern of H4K16ac binding to genic regions in NAc under control, susceptible (defeat), and resilient mice.
Note the lack of difference across the three conditions. Shading represents standard error. Data is from one experiment.
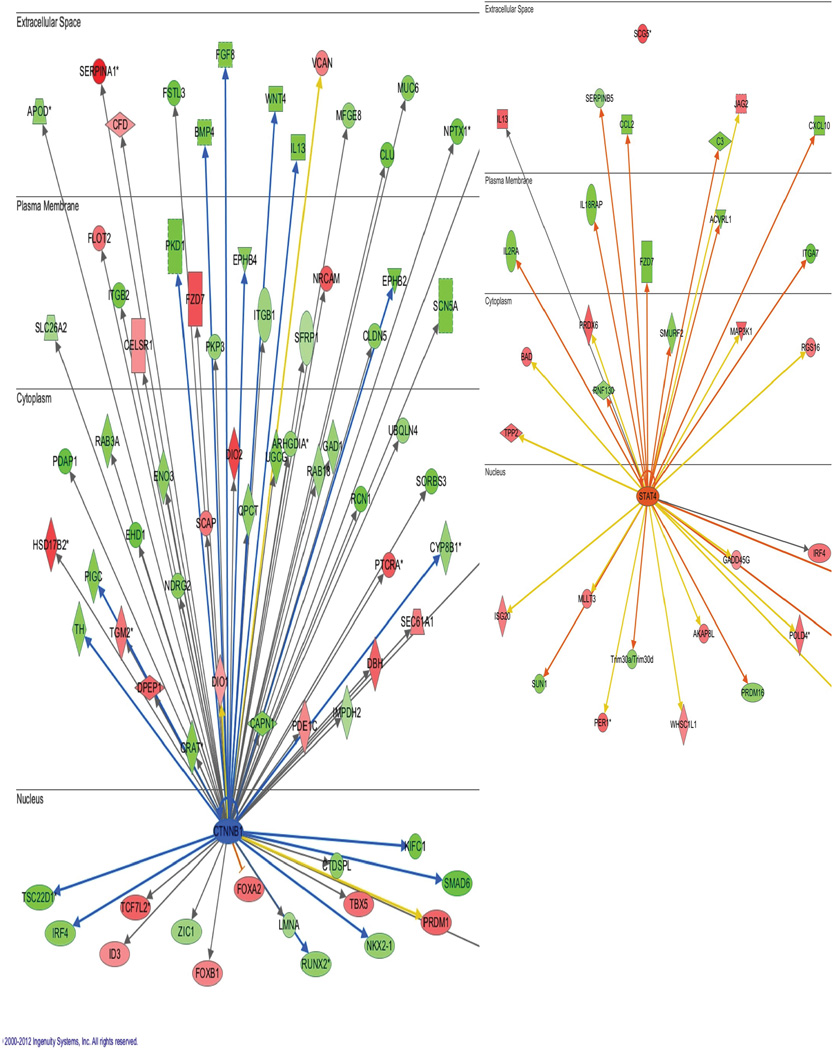
Extended Data Figure 9. Ingenuity pathway analysis (IPA) identifies a network of genes that show upregulated β-catenin binding at promoter regions in the NAc of resilient vs. susceptible mice.
Nodes represent differentially regulated genes, with green meaning up in resilient vs. susceptible and red meaning down in resilient vs. susceptible. The blue arrows indicate that the direction of regulation is consistent with IPA’s prediction of an upregulated β-catenin network in resilience; for example, a blue arrow means that a target gene that would be expected to be upregulated by β-catenin is in fact upregulated in this list. In contrast, yellow arrows indicate that the regulation observed is inconsistent with expectations, while gray arrows indicate lack of pre-existing data to formulate expectations of β-catenin action. Left panel shows mostly expected regulation of the β-catenin network (i.e., upregulation) in resilience; right panel shows non-specific changes occurring in a randomly chosen signal transducer and activator of transcription-4 (STAT4) network.
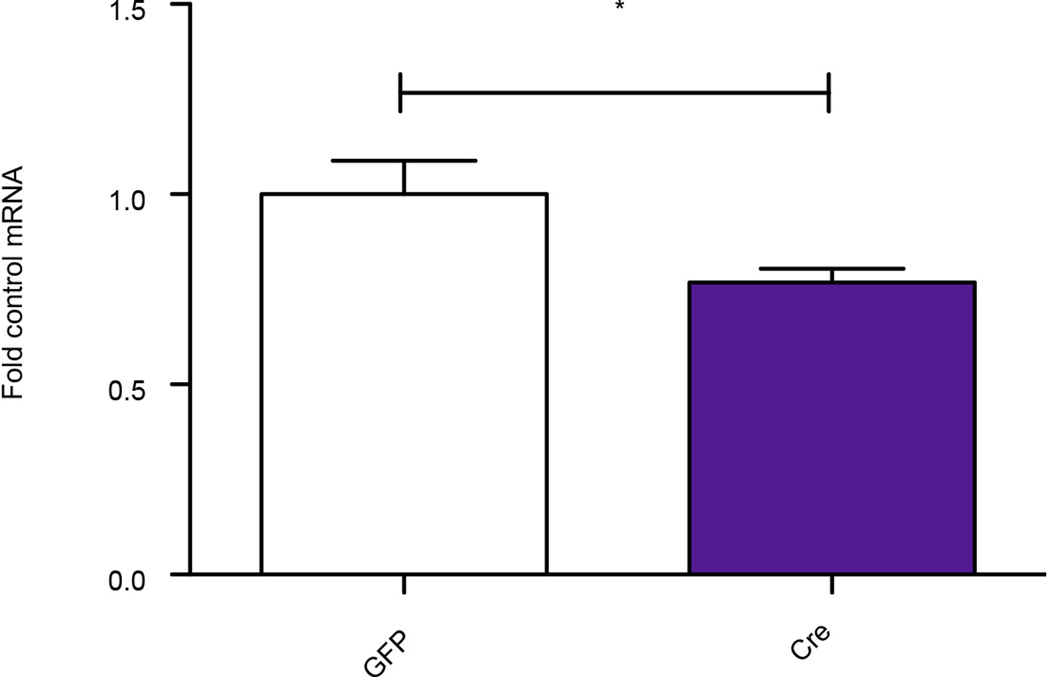
Extended Data Figure 10. Validation of local Dicer1 knockdown.
Note significant knockdown of Dicer expression in NAc after intra-NAc delivery of viral-Cre to floxed Dicer mice (t11=2.286, *P<0.05, two tailed t-test, n=7 GFP, n=6 Cre). Data is presented as mean and SEM and is representative of two experiments.
Supplementary Material
Acknowledgements
We thank Drs. O. Jabado and M. Mahajan from the Mount Sinai Genomics Core for support and Dr. Steven Borkan for providing β-catenin constructs.
This work was supported by grants from the National Institute of Mental Health.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions CD and JF conceived the project, designed research, conducted experiments, interpreted the results, and wrote the manuscript; HS, MR, DDW, KS, RB, BL, ER, PK, VV, DF, CP, EC,JK and EM conducted experiments; SG, CT provided reagents and tools; RN conducted experiments and provided reagents; NS, XL performed bioinformatic analysis; LS performed and supervised bioinformatic analysis; EJN conceived the project, designed and supervised research, interpreted the results, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information All sequencing data have been deposited into the Gene Expression Omnibus with accession number GSE61294 and GSE61295.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Madsen TM, Newton SS, Eaton ME, Russell DS, Duman RS. Chronic electroconvulsive seizure up-regulates β-catenin expression in rat hippocampus: role in adult neurogenesis. Biol. Psychiatry. 2003;54:1006–1014. doi: 10.1016/s0006-3223(03)00700-5. [DOI] [PubMed] [Google Scholar]
- 2.Beaulieu J-M, et al. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould TD, et al. Beta-catenin overexpression in the mouse brain phenocopies lithium-sensitive behaviors. Neuropsychopharmacology. 2007;32:2173–2183. doi: 10.1038/sj.npp.1301338. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Jope RS. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology. 2010;35:2143–2154. doi: 10.1038/npp.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens J, Kries J. von, Kühl M, Bruhn L. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 7.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 8.Van de Wetering M, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson MB, et al. A novel role of the WNT-dishevelled-GSK3β signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J. Neurosci. 2011;31:9084–9092. doi: 10.1523/JNEUROSCI.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadot E, et al. Regulation of S33/S37 phosphorylated beta-catenin in normal and transformed cells. J. Cell Sci. 2002;115:2771–2780. doi: 10.1242/jcs.115.13.2771. [DOI] [PubMed] [Google Scholar]
- 11.Berton O, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Kolligs FT, Hu G, Dang CV, Fearon ER. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Beta-catenin promotes survival of renal epithelial cells by inhibiting Bax. J. Am. Soc. Nephrol. 2009;20:1919–1928. doi: 10.1681/ASN.2009030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rada P, et al. Glutamate release in the nucleus accumbens is involved in behavioral depression during the Porsolt swim test. Neuroscience. 2003;119:557–565. doi: 10.1016/s0306-4522(03)00162-3. [DOI] [PubMed] [Google Scholar]
- 16.Abe K, Takeichi M. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53:387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Tye K, Prakash R, Kim S, Fenno L. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britt JP, et al. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron. 2012;76:790–803. doi: 10.1016/j.neuron.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, et al. diffReps: Detecting Differential Chromatin Modification Sites from ChIP-seq Data with Biological Replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hödar C, et al. Genome-wide identification of new Wnt/β-catenin target genes in the human genome using CART method. BMC Genomics. 2010;11:348. doi: 10.1186/1471-2164-11-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wexler EM, et al. Genome-wide analysis of a Wnt1-regulated transcriptional network implicates neurodegenerative pathways. Sci. Signal. 2011;4:ra65. doi: 10.1126/scisignal.2002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein E, Caudy aa, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 24.Cuellar TL, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee EJ, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajasethupathy P, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graybiel aM. The basal ganglia. Curr. Biol. 2000;10:R509–R511. doi: 10.1016/s0960-9822(00)00593-5. [DOI] [PubMed] [Google Scholar]
- 28.Gerfen C. THE NEOSTRIATAL MOSAIC: Multiple Levels of Compartmental Organization in the Basal Ganglia. Annu. Rev. Neurosci. 1992:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- 29.Kravitz A, Tye L, Kreitzer A. Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat. Neurosci. 2012;15:816–818. doi: 10.1038/nn.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobo MK, Nestler EJ. The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front. Neuroanat. 2011;5:41. doi: 10.3389/fnana.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S. Distinct roles of synaptic transmission in direct and indirect striatal pathways to reward and aversive behavior. Neuron. 2010;66:896–907. doi: 10.1016/j.neuron.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Darvas M, Palmiter R. Contributions of striatal dopamine signaling to the modulation of cognitive flexibility. Biol. Psychiatry. 2011;69:704–707. doi: 10.1016/j.biopsych.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yawata S, Yamaguchi T, Danjo T, Hikida T, Nakanishi S. Pathway-specific control of reward learning and its flexibility via selective dopamine receptors in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12764–12769. doi: 10.1073/pnas.1210797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Southwick SM, Charney DS. The science of resilience: implications for the prevention and treatment of depression. Science. 2012;338:79–82. doi: 10.1126/science.1222942. [DOI] [PubMed] [Google Scholar]
- 35.Veronese A, et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4840–4845. doi: 10.1073/pnas.1101734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosik KS. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 37.Im H-I, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Issler O, et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
References for Methods
- 39.Robison AJ, et al. Fluoxetine Epigenetically Alters the CaMKIIα Promoter in Nucleus Accumbens to Regulate ΔFosB Binding and Antidepressant Effects. Neuropsychopharmacology. 2014;39:1178–1186. doi: 10.1038/npp.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Lobo MK, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo JW, et al. BDNF is a negative modulator of morphine action. Science. 2012;338:124–128. doi: 10.1126/science.1222265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhury D, et al. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. MiRanalyzer: An update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 48.Sai lakshmi S, Agrawal S. piRNABank: A web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozomara A, Griffiths-Jones S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodstadt L. Ruffus: A lightweight python library for computational pipelines. Bioinformatics. 2010;26:2778–2779. doi: 10.1093/bioinformatics/btq524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.