Abstract
DNA assembly is one of the most important foundational technologies for synthetic biology and metabolic engineering. Since the development of the restriction digestion and ligation method in the early 1970s, a significant amount of effort has been devoted to developing better DNA assembly methods with higher efficiency, fidelity, and modularity, as well as simpler and faster protocols. This review will not only summarize the key DNA assembly methods and their recent applications, but also highlight the innovations in assembly schemes and the challenges in automating the DNA assembly methods.
Keywords: metabolic engineering, synthetic biology, pathway construction, pathway optimization, genome synthesis, automation
Key DNA assembly methods and their recent applications were summarized in this review.
INTRODUCTION
In the endeavors of metabolic engineering and synthetic biology, methods for putting genetic parts together into replicable and expressible DNA molecules are critical to rapid prototyping of the metabolic pathways or genetic circuits of interest. Although DNA can be chemically synthesized to a certain length, construction of larger fragments still relies on enzymatic assembly methods (Kosuri and Church, 2014). The first developed DNA assembly method is the restriction digestion and ligation method (Cohen et al., 1973), which led to the biotechnology revolution. Alhough popular, the limitations of this 40-year-old method restrict our ability of synthesizing complex DNA molecules. Increasingly, complicated DNA construct design involving multiple genes and intergenic components requires higher efficiency and fidelity in DNA assembly that is beyond the capability of the traditional cloning methods based on restriction digestion and ligation (Cobb et al., 2013). Moreover, the large size of these DNA constructs makes the selection of unique restriction sites extremely difficult. Even if restriction sites could be selected for a specific construct, they would most likely not be applicable for a different construct, which cripples the modularity of DNA assembly, a hallmark of synthetic biology. Actually, modularity is highly desired in various research projects. For example, combinatorial pathway library construction and screening have been demonstrated as an effective approach for pathway optimization, in which various genetic parts of similar functions are mixed and matched to search for particular combinations that improve the metabolic flux or other traits (Kim et al., 2013; Xu et al., 2013; Yuan et al., 2013). Similarly, in genetic circuit design, expedited prototyping requires assembly of many characterized prefabricated parts (Canton et al., 2008; Purnick and Weiss, 2009). Therefore, broadly applicable and highly efficient DNA assembly methods are desirable. Furthermore, to survey a greater range of combinations or designs, DNA assembly will most likely be performed in large scales via automation. High-throughput DNA assembly requires robust and standardized protocols, which necessitates improvements in assembly methods for even higher efficiency, fidelity, and modularity.
A number of new DNA assembly methods have been developed in the past decade. Some of them were built upon the traditional restriction digestion and ligation method, such as the Golden Gate method (Engler et al., 2008), while others harness different mechanisms, such as Gibson assembly (Gibson et al., 2009) and DNA Assembler (Shao and Zhao, 2009). By changing reaction protocols and linker regions between DNA parts to be assembled, these new methods have achieved improved efficiency, fidelity, and modularity, which have simplified both design and bench-side operation. Based on these methods, a number of innovations in assembly scheme and the resulting toolkits have been reported (Shetty et al., 2008; Xu et al., 2012; Casini et al., 2014a), which opened doors to a wide variety of applications. One major application with great industrial and scientific significance is the construction and engineering of metabolic pathways for production of chemicals and fuels. This review takes a glance at the state-of-the-art DNA assembly technologies and focuses on the applications of DNA assembly methods for pathway construction and engineering.
KEY DNA ASSEMBLY METHODS
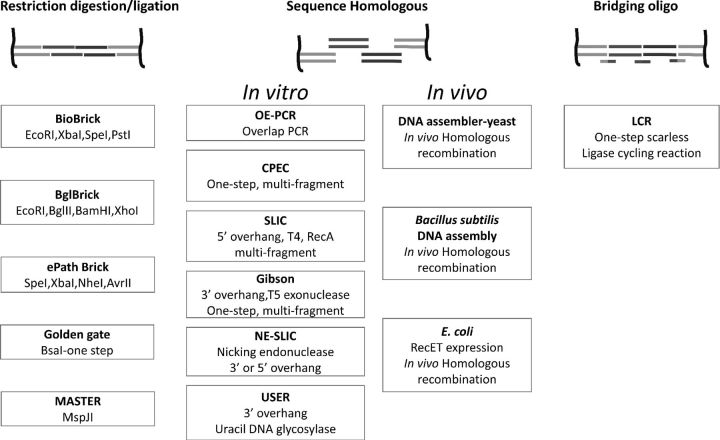
Based on the different assembly mechanisms employed, the various recently developed key DNA assembly methods can be divided into four groups, including restriction enzyme-based methods, in vivo and in vitro sequence homology-based methods, and bridging oligo-based methods (Fig. 1).
Figure 1.
Key DNA assembly methods.
Restriction enzymes-based methods
Restriction digestion and ligation using type II restriction enzymes and DNA ligase has been exploited as the standard cloning technique for about 40 years in molecular biology. A few improvements have been made based on the original method. The BioBrick™ standard was the first DNA assembly strategy that allows the sequential assembly of standard biological parts. It employs iterative cycles of restriction digestion and ligation reactions to assemble small DNA parts into a large DNA construct (Smolke, 2009; Sarrion-Perdigones et al., 2011). Each DNA part is flanked by EcoRI and XbaI restriction sites in the upstream end and by SpeI and PstI restriction sites in the downstream end. XbaI and SpeI are isocaudamers that generate two compatible sticky ends. Once ligated, the newly generated scar sequence (ACTAGA) between the two DNA parts is different from both original sites and therefore cannot be cut in subsequent digestions with either XbaI or SpeI. On the other end of the inserted fragment, EcoRI is restored, while a new XbaI site is introduced. Hence, the insertion can be repeated. The original BioBrick™ design had two extra nucleotides besides the natural 6-nucleotide scar which hampers its application in protein fusion as it creates frameshifts and premature stop codons. Improved revisions of this method have been developed to address this issue by starting with two standards specifically designed to assist assembly of fusion proteins (Phillips and Silver, 2006; Grünberg et al., 2009). A smaller 6-nucleotide scar sequence (ACTAGA) encoding threonine-arginine is generated in each conjunction, which eliminates open reading frameshifts and stop codons. More recently, a modified standard called BglBrick (Anderson et al., 2010) was reported. It addresses some key problems associated with the original BioBrick™ standards. The DNA parts are flanked by restriction sites of more efficient and methylation insensitive enzymes: BglII and BamHI. The scar sequence (GGATCT) encodes glycine-serine so that it is also suitable for protein fusion applications.
The Golden Gate method (Engler et al., 2008, 2009) relies on type IIs restriction enzymes, which are able to cleave DNA outside of their recognition site and produce an overhang of four arbitrary nucleotides (if the most popular BsaI is used). When designed properly, two digested fragments can be ligated to generate a product lacking the original restriction sites. This method also allows restriction digestion and ligation cycling in a one-pot reaction at 37 and 16 °C, which can greatly increase the efficiency, driving the reaction to completion. Notwithstanding, it is not suitable to assemble long DNA constructs due to the lack of unique restriction enzymes. However, an improvement has been made on BasI-based Golden Gate method, which uses SapI with a rarer cut site than BasI (Whitman et al., 2013). Another method termed methylation-assisted tailorable ends rational (Master) uses endonuclease MspJI which specifically recognizes methylated 4-bp sites, mCNNR (R = A or G), and generates a 4-bp arbitrary overhang like type IIs endonucleases (Chen et al., 2013). As it avoids cuts on corresponding type IIs sites within the fragments as in the Golden Gate method, the Master method is more suitable for assembling large DNA constructs. However, it requires expensive methylated primers and PCR amplification of parts which may introduce errors for long parts.
Although restriction enzyme-based methods are able to assemble multiple DNA parts into relatively large constructs, all DNA parts are required to be free of the restriction sites used in the assembly. Even though such sites can be removed by site-directed mutagenesis prior to assembly, it would require extra effort and cost to do so. Furthermore, restriction enzyme-based methods rely on annealing of short sticky ends that may have limited affinity and specificity when assembling multiple DNA parts in one pot.
Sequence homology-based methods: in vitro
Sequence homology-based methods usually utilize longer arbitrary overlapping regions between parts, which prevents the same issues as with restriction enzyme methods. Both in vitro and in vivo sequence homology-based methods have been developed.
Overlap extension polymerase chain reaction (OE-PCR) enables scarless assembly of DNA parts (Horton et al., 1989). Basic DNA parts are first amplified in separate PCRs with homologous ends between them. With the corresponding homologous regions, these DNA parts can anneal to each other and be extended by DNA polymerase in the second round of PCR to yield spliced DNA molecules. The resulting larger DNA fragments can then be inserted into plasmids using the restriction digestion and ligation method. To improve the efficiency of annealing, special GC-rich overlap sequences are designed to mediate reliable fusion PCR (Cha-aim et al., 2009). Recently, one method named circular polymerase extension cloning (CPEC) (Quan and Tian, 2009) was developed allowing assembly of multiple inserts with any vector in a one-step OE-PCR. With extended homologous regions, the fused DNA molecules circularize with a nick in each strand. After transformation, host Escherichia coli fixes the nicks to form intact vectors. Furthermore, it was shown that nonlinearized vectors can be used as templates for extension followed by DpnI digestion to eliminate the original plasmids (Bryksin and Matsumura, 2010).
In another sequence homology-based in vitro DNA assembly method called SLIC (sequence and ligation-independent cloning), recombination intermediates generated in vitro are transformed into cells. Endogenous DNA repair machinery was utilized to finish the repair and generate recombinant DNA molecules (Li and Elledge, 2007). The 3′ ends of the linearized vector and the overlap regions of the inserts are chewed back by T4 DNA polymerase in the absence of dNTPs and left as single stranded. Subsequently, the RecA protein and ATP are used to promote recombination before being used to transform E. coli. The gaps are also fixed by host E. coli in vivo. A follow-up method named SLiCE (Seamless Ligation Cloning Extract) (Zhang et al., 2012) uses inexpensive E. coli cell extracts to drive homology-mediated DNA assembly, which significantly reduces the cost. A disadvantage of this strategy is that the length of single-strand overlaps is not very controllable in the chew-back reaction. A modified method is called uracil-specific excision reagent cloning (USER) (Smith et al., 1993; Nour-Eldin et al., 2010). A single deoxyuridine residue is included in each primer in the PCR. The PCR products and the vector backbone are then excised by uracil DNA glycosylase and DNA glycosylase-lyase Endo VIII to produce 3′ overhangs. The deoxyuridine residues introduced in this method, however, significantly increase the cost of primers.
Similar to SLIC, the Gibson assembly method (Gibson et al., 2009) utilizes T5 exonuclease to chew back the 5′ ends to generate single-stranded complementary overhangs which are joined together covalently by fusion DNA polymerase and Taq DNA ligase. In a one-step isothermal in vitro reaction at 50 °C, the fragments can be assembled into a single circular DNA molecule. Similar to USER, Wang et al. (2013) developed a method termed nicking endonucleases for LIC (NE-LIC), in which nicking endonucleases (NEases) are exploited to generate overhangs of controlled lengths, although the NEase recognition sites are left as a scar. Notably, both Gibson assembly and NE-LIC feature an in vitro ligation reaction which was found to increase the efficiency of DNA assembly.
Colloms et al. (2013) reported a method named serine integrase recombinational assembly (SIRA). SIRA takes advantage of the recombination machinery of ϕC31 integrase from phage. ϕC31 cuts at attP and attB sites and rejoins the exchanged half sites to form new attL and attR sites in vitro. By the addition of phage recombination directionality protein, the reaction can be reversed which allows removal of recombined fragments. This unique feature of SIRA allows future editing of the DNA constructs.
In addition to these methods, two well-known commercial kits are available for plasmid construction. One kit is In-Fusion™ (Clontech), which is a proprietary version of the ‘chew back and anneal’ method to directionally clone one or more fragments into any vector. PCR-generated or vector-linearized DNA fragments with a 15-bp sequence overlap at their ends can be assembled in a fast and efficient way. Another kit is Gateway™ (Life Technologies), which utilizes a specific recombinase to mediate the recombination of specific overlap sequences. This method allows fast and efficient cloning of single genes into flexible vectors but is not suitable for assembly of large DNAs due to the remaining same sequences at each junction.
Sequence homology-based methods: in vivo
Homologous recombination occurs naturally in Saccharomyces cerevisiae with high efficiency and fidelity, which was exploited by Gibson et al. (2008a) to construct the Mycoplasma genitalium genome by assembling 25 DNA fragments and by Shao et al. (2009) to construct a large pathway by assembling multiple fragments nearly at the same time. In the method developed by Shao et al., or so-called DNA Assembler, all DNA parts to be assembled can be obtained either from PCR amplification or restriction digestion with homologous arms between neighboring parts in the pathway. All the linear DNA parts are directly transformed into S. cerevisiae. Circular plasmids are then constructed by its endogenous homologous recombination machinery.
With a similar mechanism, DNA assembly was also performed in other organisms such as Bacillus subtilis (Yonemura et al., 2007; Itaya et al., 2008) and certain plants (Zhu et al., 2008; Farre et al., 2012). Recently, a promising direct DNA cloning method in E. coli was reported by Fu et al. (2012; Zhang et al., 2000). Based on the discovery that the full-length Rac prophage proteins RecE and RecT facilitate highly efficient homologous recombination between two linear DNA molecules in E. coli, digested or sheared DNA was transformed together with PCR-amplified vector containing terminal homology arms into the host which over-expressed full-length RecET promoting formation of an intact vector. Using this strategy, ten megasynthetase gene clusters from 10 to 52 kb were cloned into expression vectors.
Bridging oligo-based methods
Different from sequence homology-based methods, De Kok et al. (2014) developed a DNA assembly method based on ligase cycling reaction (LCR). Single-stranded bridging oligos are designed to be complementary to two ends of neighboring DNA parts. Based on a one-step DNA assembly method named chain reaction cloning (Pachuk et al., 2000), the assembly conditions were systematically optimized. DNA parts to be assembled via LCR were amplified using 5′-phosphorylated primers. During the reaction, the double-stranded DNA parts are denatured at a high temperature, and then the upper (or lower) strands from the two parts to be adjoined anneal with the bridging oligo at a lower temperature and are subsequently joined scarlessly by thermostable ligase via a phosphodiester bond. In following cycles, the ligated strand serves as the template to assemble the complementary strands. By running multiple thermal cycles, linear DNA parts can be assembled into circular DNA constructs and then transformed into E. coli competent cells for amplification. By comparing this method with other available scarless and sequence-independent DNA assembly methods, the authors found that LCR method with optimized conditions had similar fidelity with yeast homologous recombination when assembling up to 12 DNA parts, whereas CPEC and Gibson isothermal assembly had lower fidelity under the same conditions.
INNOVATIONS IN DNA ASSEMBLY SCHEMES
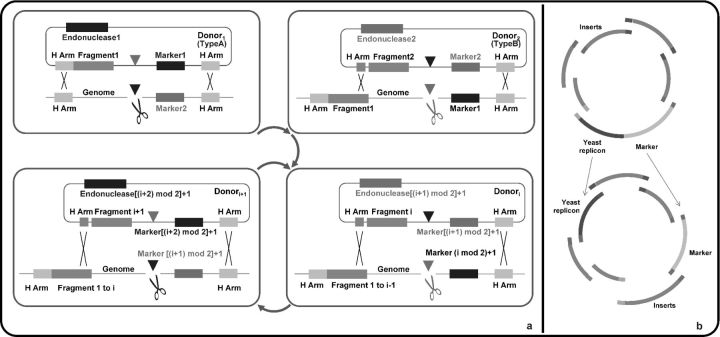
Besides the reaction mechanisms, the scheme by which DNA parts are put together greatly affects the quality and simplicity of assembly. Building multifragment pathways is usually problematic regardless of the method. In sequence homology-based methods, unexpected homology between fragments and nonhomologous end joining (NHEJ) will yield mis-assembled products. As usually the selection after assembly only determines whether the vector is present and replicable, there is no guarantee that each fragment is correctly assembled. It is common to find fragments omitted or swapped. Wingler and Cornish proposed an iterative integration scheme (Fig. 2) to ensure the incorporation of each fragment (Wingler and Cornish, 2011). The fragments are inserted into two types of serial donor plasmids: Type A with Homing Endonuclease 1, Recognition Site 2, and Marker 1; and Type B with Homing Endonuclease 2, Recognition Site 1, and Marker 2. Type A and B alternate in that series, in which the plasmids have homologous regions that can be linked head-to-tail. When the host is cured with the first donor (Type A), recognition site 1 on the host genome is cut, which facilitates integration of a part of the donor containing the first fragment, Marker 1, and Recognition Site 2 and replaces Marker 2 and Recognition Site 1 on the genome. Then the host is selected against Marker 1. Similarly, when the host is cured with the second donor (Type B), the host is changed back to Marker 2 and selected accordingly. The subsequent fragments are integrated in the same way. This work innovatively employed alternating markers to apply selective pressure for each fragment. However, the increased fidelity comes at a cost of time.
Figure 2.
Improved in vivo sequence homology-based DNA assembly methods in Saccharomyces cerevisiae. (a) In Wingler and co-workers' method (Wingler and Cornish, 2011), Type A and Type B are alternated in the series of donor plasmids harboring the fragments to be assembled. The homing endonuclease coded by the donor plasmid specifically cuts the integration site on the genome to promote integration of the next fragment. The markers are reused in turns to guarantee that the corresponding fragments are successfully appended. Mod represents modulo operation. ‘H. Arm’ represents homologous arms. (b) In Kuijpers and co-workers' method (Kuijpers et al. 2013), the yeast replicon and marker fragments on the vector backbone are separated to reduce the likelihood of generating false positives.
Sequential assembly is obviously less appealing over one-step assembly of multiple fragments if the required level of fidelity and efficiency could be reached. However, highly accurate and efficient one-step assembly is not easy to accomplish and requires additional engineering. In the in vivo homologous recombination-based assembly methods, NHEJ of the linearized vectors contributes to most of the false positives. One solution could be to introduce a counter-selective marker at the cloning site (Anderson and Haj-Ahmad, 2003). In order to survive, the host must have the counter-selective marker replaced by the designated inserts. To further minimize this problem, separating the essential elements on the vector, selection marker and episome has been proposed (Kuijpers et al., 2013). As the host organism needs at least 2 NHEJs to incorporate both elements, the probability of false positives was greatly reduced. With this new scheme, nine fragments were assembled by 60-bp overlapping regions with a correct yield of 95%.
Most of the DNA assembly methods rely on overlapping linker sequences, no matter short or long, shared between adjacent fragments. The linkers in some of the methods can be customized, which leaves freedom for further optimization. Liang et al. (2013) have optimized the 4-bp linkers for synthesizing customized recombinant transcription activator-like effectors (TALEs) with the Golden Gate method. TALEs are proteins that can target DNA sequences with specifically ordered central repeat domains (CRDs). Each CRD recognizes one nucleotide. Linkers with higher efficiency were selected by experiment. With these selected linkers, 13 fragments or DNA sequences coding for up to 31 CRDs were ligated together with a nuclease domain in one step. Without picking single colonies, 96% of the assembled TALE nucleases were found to be functional. Nonfunctional ones were sequenced and found to be correctly assembled too. With the improved efficiency and fidelity brought by the optimized linkers, colony picking could be avoided, which made this process much more automation-friendly compared with a closely related approach (Kim et al., 2013).
Building genetic circuits usually involves multiple design-built-test cycles, in which rapid prototyping is desired. Sequence homology-based methods such as the Gibson assembly method allow assembly of multiple standardized genetic parts in one step. However, as the DNA molecules coding for these circuits are likely to be highly repetitive, sequence homology-based methods will suffer from low efficiency and mis-assembly. Thus synthetic linkers that are unique for each fragment should be used instead of the endogenous sequences to ensure the complementary single-stranded ends of the fragments to anneal as designed. Moreover, the use of predefined sets of synthetic linkers modularizes the sequence homology-based assembly. As there is full freedom in designing the synthetic linkers, more parameters can be taken into consideration to further improve the efficiency and fidelity. Due to the large number of possible candidates, 4n, where is the number of base pairs in the linkers, computational approaches are usually preferred. A few algorithms have been implemented to generate the optimal orthogonal set of synthetic linker sequences. Most of these algorithms screens a large number of random oligo nucleotide sequences of a certain length. Although different thresholds are used, the screens commonly applied include: 1) orthogonality among the oligo sequences; 2) homology against host genomes; 3) melting temperature; 4) GC content and distribution; and 5) likelihood of forming hairpins and primer dimers. Using computationally designed linkers, it was possible to assemble over 30 parts with above 10% fidelity (Guye et al., 2013). Another work reached c. 85% fidelity when assembling five fragments with c. 80% identity (Torella et al., 2013). R2oDNA Designer is an online computational tool for designing the optimized linker set for Gibson assembly or similar methods (Casini et al.,2014). It was shown that designed linkers yielded an improved assembly efficiency compared to the conventional method (Casini et al., 2014).
On the other hand, the synthetic biology community keeps making great progress in standardization of assembly schemes with characterized genetic parts. Owing to the modularity of BioBrick™ standard and the contributions of the community, thousands of standard parts are available in the growing public registry (Partregistry, 2014), which have greatly facilitated rapid construction of gene circuits and beyond (Smolke, 2009; McLennan, 2012; Partregistry, 2014). Xu et al. (2012) developed ePathBrick, that is, a number of Biobrick™ vectors carrying characterized regulatory signal elements, which allows combinatorial assembly of promoters, ribosome binding sites (RBS), and terminators for fine tuning of gene expressions in pathways. ePathBrick vectors contain four isocaudomers that are compatible with the BioBrick™ standard, adding a fine-tuning toolkit to the growing BioBrick™ part collection. Toolkits based on Golden Gate method, such as MoClo (Weber et al., 2011) and GoldenBraid (Sarrion-Perdigones et al., 2011) have also been developed for modular assembly of large pathways. Due to the nature of Golden Gate assembly, these kits allow rapid construction of multiple parts in one pot. With these standardized parts, researchers will be able to test their designs by quickly assembling without ‘remachining’ them to fit each other.
SUCCESSFUL APPLICATIONS
Although most of the above-mentioned DNA assembly methods were developed in the past few years, there are already many successful applications. For example, one application of the DNA assembly methods is to discover novel natural products. Activation of cryptic or silent gene clusters encoding biosynthesis of natural products is usually difficult but is an important way to discover novel natural products. Recently, Shao et al. (2013) developed a novel plug-and-play strategy for natural product discovery in which a set of constitutive promoters, which were proven functional in the target expression host, were assembled upstream of each pathway gene to refactor silent biosynthetic pathways. With this strategy, a silent spectinabilin pathway from Streptomyces orinoci was successfully activated. Similarly, a cryptic polycyclic tetramate macrolactams (PTMs) biosynthetic gene cluster from Streptomyces griseus was successfully activated and three new PTMs were discovered (Luo et al., 2013).
Another application of the DNA assembly methods is to design and characterize gene circuits, which can not only help researchers gain better understanding of intracellular (Elowitz and Leibler, 2000; Atkinson et al., 2003) and intercellular (Bulter et al., 2004; Chen and Weiss, 2005; Wang et al., 2008) regulatory machineries but also have a wide range of potential applications (Wall et al., 2004; Purnick and Weiss, 2009). The rapid development of gene circuits should be attributed to the consistent effort in standardization of genetic parts and devices (Canton et al., 2008; Shetty et al., 2008). As discussed earlier, BioBrick™ parts can be readily assembled like Legos for prototyping of gene circuits, however, the sequential nature of the BioBrick standard would be time consuming if multiple parts are assembled. The Gibson assembly method allows one-pot assembly of multiple pieces of DNA regardless of restriction site availability. Using synthetic linkers, high-throughput isothermal assembly of complex gene circuits of up to 33 parts was made possible (Guye et al., 2013; Torella et al., 2013).
A third application of the DNA assembly methods is to synthesize genomes. The J. Craig Venter Institute synthesized a 583 kb M. genitalium genome by a combination of in vitro enzymatic and in vivo homologous recombination-based methods (Gibson et al., 2008). In the early stage, in vitro recombination method was utilized to assemble 25 DNA cassettes with an average length of 24 kb to eight 72 kb assemblies and subsequently assembled into four 144 kb assemblies. It was found that the efficiency of in vitro procedure declined as the assemblies became larger and the half-genome in size of 290 kb each was unable to be assembled. Therefore the in vivo S. cerevisiae recombination method was exploited to complete the final whole genome assembly. In the meantime, it was also found possible to directly assemble 25 DNA cassettes from the earliest stages into a complete genome in a single step by in vivo recombination in S. cerevisiae (Gibson et al., 2008). Two years later, a M. mycoides cell controlled by the chemically synthesized genome was created and exhibited the in silico designed phenotype (Gibson et al., 2010 ). The 16.3 kb mouse mitochondrial genome was also assembled via in vitro isothermal recombination method from 600 DNA pieces with 60 bp overlaps (Gibson et al., 2010). Very recently, a full designer S. cerevisiae chromosome was synthesized (Annaluru et al., 2014). These successes are excellent demonstrations of the power of modern DNA assembly methods.
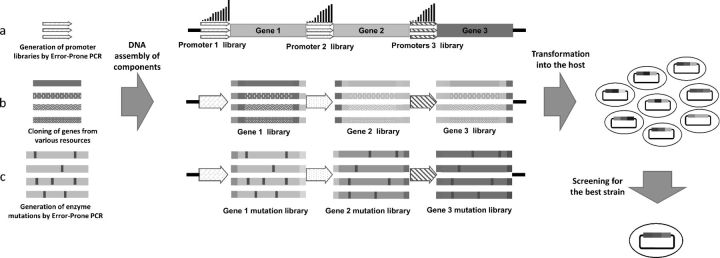
Another very important application of the DNA assembly methods is pathway optimization for metabolic engineering. DNA Assembler is an outstanding pathway optimization tool, especially in S. cerevisiae. Du et al. (2012) developed a simple and efficient strategy for multigene pathway optimization strategy termed customized optimization of metabolic pathways by combinatorial transcriptional engineering (COMPACTER) (Yuan et al., 2013). In this strategy, a library of promoters with varying strengths were used for each of the structural genes in either the xylose-utilizing pathway or the cellobiose-utilizing pathway and assembled together with the corresponding structural genes and terminators to generate a library of xylose- or cellobiose-utilizing pathways. Using a cell growth-based high-throughput screening strategy, a heterologous xylose-utilizing pathway with high efficiency and a heterologous cellobiose-utilizing pathway with the highest reported efficiency for both laboratory and industrial S. cerevisiae strains were isolated. The heterologous cellobiose-utilizing pathway was further optimized through directed evolution (Yuan and Zhao, 2013). Similar to COMPACTER, error-prone PCR-generated promoter mutants were assembled with corresponding genes by DNA Assembler to create the first round pathway library. Another pathway library was created based on the best pathway mutant from the first round and screened again. This iterative process was continued until no further improvement was observed. After three rounds of screening, the final industrial S. cerevisiae strain showed sixfold higher cellobiose consumption rate and ethanol productivity. Also using DNA Assembler, Eriksen et al. (2013) improved the cellobiose pathway by simultaneous directed evolution of cellodextrin transporter and β-glucosidase. Kim et al. (2013) developed a combinatorial pathway engineering approach to rapidly create/screen a highly efficient xylose-utilizing pathway in S. cerevisiae. A total of 20 xylose reductase homologues, 22 xylitol dehydrogenase homologues, and 19 xylulose homologues were cloned from different organisms. Using DNA assembler, the homologue libraries were first inserted into three different helper plasmids containing three pairs of promoters and terminators accordingly to generate expression cassettes. These cassettes were then PCR-amplified and assembled by DNA Assembler to create the xylose pathway library. The optimized pathway was then identified by rapid growth-based screening (Fig. 3).
Figure 3.
Schematic of three applications based on in vivo DNA assembler method. (a) Promoter libraries are generated by error-prone PCR and assembled in front of each gene in the pathway. (b) For each enzyme in the pathway, homologous genes from various microorganisms are cloned and are flanked by the arms homologous to their neighboring promoters and terminators. All expression cassettes are then assembled using the DNA assembler method. (c) All enzymes in the pathway are mutated via error-prone PCR to generate mutant libraries and are flanked by the arms homologous to their neighboring promoters and terminators. These expression cassettes are then assembled using the DNA assembler method.
As in gene circuit design, characterized prefabricated DNA parts can also accelerate the pathway optimization process. ePathBrick is a pathway fine-tuning toolkit compatible with BioBrick™ standard, which was recently used to optimize the fatty acid biosynthetic pathway in E. coli (Xu et al., 2013). The pathway was first engineered by gene knockout and over-expressions of a number of FAS related genes. Then, the entire pathway was divided to three modules: glycolysis (GLY), acetyl-CoA activation (ACA), and FAS. GLY, ACA, and FAS modules were assembled to five ePathBrick vectors with different copy numbers and promoter strengths. Eventually, the engineered strain achieved the highest fatty acid productivity ever reported. Also following BioBrick™ standard, a seven gene carotenoid biosynthetic pathway was fine-tuned by spanning the expression space with computationally designed RBS of various strengths (Salis et al., 2009; Zelcbuch et al., 2013). The RBS libraries were randomly assembled with the genes in the BioBrick™ iterative cloning process. After screening, an astaxanthin yield of fourfold higher than previously reported was achieved.
CONCLUSIONS AND FUTURE PROSPECTS
Over the past decade, DNA assembly technologies have made great advances. Most researchers of the synthetic biology and metabolic engineering community have adopted and benefited from these advances (Kahl and Endy, 2013). Nevertheless, there are still many challenges to overcome. The assembly quality still depends on the fragment sequences, while the efficiency and fidelity still need to be improved. High failure rate makes DNA assembly time and labor consuming. There is also room for the modularity to improve as well. Ideally, we should be able to ‘bolt on’ genetic parts with standard ‘fasteners’ and ‘adaptors’ without too much concern about compatibility.
Automated design will undoubtedly have broader applications (Ellis et al., 2011; MacDonald et al., 2011). Besides simple computer aided design tools for drawing the maps for one or several constructs, more high-throughput design tools will soon become available due to the needs in combinatorial assembly and multiplex genome engineering. Moreover, these high-throughput tools should be able to generate primer sequences according to the assembly methods and schemes as well as to interface with robotic liquid handlers in order to set up PCR and assembly reaction systems automatically. j5, a software package developed by the Joint BioEnergy Institute can help mass-process thousands of assemblies and generate the primer sequences for a number of commonly used assembly methods (Hillson et al., 2012). j5 can also generate the control files for automated liquid handlers to set up PCRs. By the same group, a cross-platform language has been proposed to simplify the programming of liquid handlers (Linshiz et al., 2013). As it is difficult for a generalized algorithm to fulfill all research purposes, specialized tools are also necessary. Liang et al. (2013) developed a high-throughput TALE synthesis platform. The computational tool in this work generated liquid handler control files based on the target binding sequences for setting up assembly reactions automatically.
With the development of laboratory automation devices, there is a potential to extend the level of automation in the assembly process. Most up-to-date robotic liquid handlers are able to carry out relatively accurate pipetting and transportation of labware (Kong et al., 2012). Some of the more complex platforms integrate temperature control units, shakers, and detection devices on board. Thus cherry-picking genetic parts, adding reagents, incubation, or even thermocycling could be readily automated, while steps after assembly reactions still have barriers to be fully automated. In this type of biofabrication line, any procedure that requires large amount of human intervention would be a throughput bottleneck. With the increasing needs, full automation of the assembly process may become the next game-changing technology. In the postassembly steps, colony picking especially, even if automated by colony picker and petri dish handlers, would still significantly lower the throughput of the whole process. The best scenario is when the assembly has enough fidelity that colony picking could be avoided. Although some DNA constructs can be verified by functional assays, lack of low-cost DNA analyzing devices still remains a technical hurdle. Application of microfluidics in molecular biology has made promising advances which may turn out to be the enabling technologies for high-throughput DNA assembly confirmation (Mueller et al., 2000; Dorfman et al., 2013). Taking advantage of high-throughput capillary electrophoresis, a cost-effective method for verification of DNA constructs has been reported (Dharmadi et al., 2014). With the decreasing cost of DNA sequencing (Shendure and Ji, 2008; Clarke et al., 2009), it may also become practical to directly sequence all assembled DNA molecules in the future. Further in the future, de novo synthesis of large DNA fragments may also be more affordable (Goldberg, 2013). De novo synthesis can help prefabricate DNA fragments with necessary linkers or mutations that make the downstream assembly easier. We envision that DNA assembly will soon become a cost-effective service provided by specialized corporations that handle orders on fully automated bio-assembly lines. By this way, synthetic biologists and metabolic engineers can focus more on designing and testing their ideas instead of the problematic assembly process of DNA molecules.
Acknowledgments
We thank the National Institutes of Health (GM077596), the National Academies Keck Futures Initiative on Synthetic Biology, Department of Defense Advanced Research Program Agency, Institute for Genomic Biology at the University of Illinois at Urbana-Champaign, and the Energy Biosciences Institute for financial support in our development and application of DNA assembly technologies. In addition, we state that there is no conflict of interest.
AUTHORS' CONTRIBUTION
R.C. and Y.Y. contributed equally to this publication.
Conflict of interest statement. None declared.
REFERENCES
- Anderson PR, Haj-Ahmad Y. Counter-selection facilitated plasmid construction by homologous recombination in Saccharomyces cerevisiae. Biotechniques. 2003;35:692–4. doi: 10.2144/03354bm05. 696, 698. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. BglBricks: a flexible standard for biological part assembly. J Biol Eng. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–8. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–5. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulter T, Lee SG, Woirl WWC, Fung E, Connor MR, Liao JC. Design of artificial cell-cell communication using gene and metabolic networks. P Natl Acad Sci USA. 2004;101:2299–304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–93. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- Casini A, Christodoulou G, Freemont PS, Baldwin GS, Ellis T, MacDonald JT. R2oDNA Designer: computational design of biologically neutral synthetic DNA sequences. ACS Synth Biol. 2014a doi: 10.1021/sb4001323. [DOI] [PubMed] [Google Scholar]
- Casini A, MacDonald JT, De Jonghe J, Christodoulou G, Freemont PS, Baldwin GS, Ellis T. One-pot DNA construction for synthetic biology: the Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 2014b;42:e7. doi: 10.1093/nar/gkt915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-aim K, Fukunaga T, Hoshida H, Akada R. Reliable fusion PCR mediated by GC-rich overlap sequences. Gene. 2009;434:43–9. doi: 10.1016/j.gene.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol. 2005;23:1551–5. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- Chen WH, Qin ZJ, Wang J, Zhao GP. The Master (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly. Nucleic Acids Res. 2013;41:e93. doi: 10.1093/nar/gkt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- Cobb RE, Ning JC, Zhao H. DNA assembly techniques for next-generation combinatorial biosynthesis of natural products. J Ind Microbiol Biotechnol. 2013;41:469–77. doi: 10.1007/s10295-013-1358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. P Natl Acad Sci USA. 1973;70:3240–4. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms SD, Merrick CA, Olorunniji FJ, Stark WM, Smith MCM, Osbourn A, Keasling KJ, Rosser SJ. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2013;42:e23. doi: 10.1093/nar/gkt1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kok S, Stanton LH, Slaby T, et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol. 2014;3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- Dharmadi Y, Patel K, Shapland E, Hollis D, Slaby T, Klinkner N, Dean J, Chandran SS. High-throughput, cost-effective verification of structural DNA assembly. Nucleic Acids Res. 2014;42:e22. doi: 10.1093/nar/gkt1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman KD, King SB, Olson DW, Thomas JD, Tree DR. Beyond gel electrophoresis: microfluidic separations, fluorescence burst analysis, and DNA stretching. Chem Rev. 2013;113:2584–667. doi: 10.1021/cr3002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yuan Y, Si T, Lian J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40:e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr Biol (Camb) 2011;3:109–18. doi: 10.1039/c0ib00070a. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen DT, Hsieh PC, Lynn P, Zhao H. Directed evolution of a cellobiose utilization pathway in Saccharomyces cerevisiae by simultaneously engineering multiple proteins. Microb Cell Fact. 2013;12:61. doi: 10.1186/1475-2859-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre G, Naqvi S, Sanahuja G, et al. Combinatorial genetic transformation of cereals and the creation of metabolic libraries for the carotenoid pathway. Methods Mol Biol. 2012;847:419–35. doi: 10.1007/978-1-61779-558-9_33. [DOI] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–6. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., 3rd One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. P Natl Acad Sci USA. 2008a;105:20404–9. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008b;319:1215–20. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Smith HO, Hutchison CA, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010a;7:901–5. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010b;329:52–6. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Goldberg M. BioFab: applying Moore's law to DNA synthesis. Ind Biotechnol. 2013;9:10–2. [Google Scholar]
- Grünberg R, Arndt K, Müller K. Fusion Protein (Freiburg) Biobrick assembly standard. 2009. http://hdl.handle.net/1721.1/45140.
- Guye P, Li Y, Wroblewska L, Duportet X, Weiss R. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res. 2013;41:e156. doi: 10.1093/nar/gkt605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillson NJ, Rosengarten RD, Keasling JD. j5 DNA assembly design automation software. ACS Synth Biol. 2012;1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–8. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat Methods. 2008;5:41–3. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- Kahl LJ, Endy D. A survey of enabling technologies in synthetic biology. J Biol Eng. 2013;7:13. doi: 10.1186/1754-1611-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Du J, Eriksen DT, Zhao HM. Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl Environ Microbiol. 2013a;79:931–41. doi: 10.1128/AEM.02736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013b;31:251–8. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- Kong F, Yuan L, Zheng YF, Chen W. Automatic liquid handling for life science: a critical review of the current state of the art. J Lab Autom. 2012;17:169–85. doi: 10.1177/2211068211435302. [DOI] [PubMed] [Google Scholar]
- Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers NG, Solis-Escalante D, Bosman L, van den Broek M, Pronk JT, Daran JM, Daran-Lapujade P. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013;12:47. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–6. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2013;3:67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- Linshiz G, Stawski N, Poust S, Bi C, Keasling JD, Hillson NJ. PaR-PaR laboratory automation platform. ACS Synth Biol. 2013;2:216–22. doi: 10.1021/sb300075t. [DOI] [PubMed] [Google Scholar]
- Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JT, Barnes C, Kitney RI, Freemont PS, Stan GB. Computational design approaches and tools for synthetic biology. Integr Biol (Camb) 2011;3:97–108. doi: 10.1039/c0ib00077a. [DOI] [PubMed] [Google Scholar]
- McLennan A. Building with BioBricks: Constructing a Commons for Synthetic Biology Research. Cheltenham: Edward Elgar; 2012. [Google Scholar]
- Mueller O, Hahnenberger K, Dittmann M, Yee H, Dubrow R, Nagle R, Ilsley D. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis. 2000;21:128–34. doi: 10.1002/(SICI)1522-2683(20000101)21:1<128::AID-ELPS128>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Geu-Flores F, Halkier BA. USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol Biol. 2010;643:185–200. doi: 10.1007/978-1-60761-723-5_13. [DOI] [PubMed] [Google Scholar]
- Pachuk CJ, Samuel M, Zurawski JA, Snyder L, Phillips P, Satishchandran C. Chain reaction cloning: a one-step method for directional ligation of multiple DNA fragments. Gene. 2000;243:19–25. doi: 10.1016/s0378-1119(99)00508-9. [DOI] [PubMed] [Google Scholar]
- Registry of Standard Biological Parts. 2014. Partregistry http://www.webcitation.org/6O8Ha2b2B.
- Phillips I, Silver P. A new biobrick assembly strategy designed for facile protein engineering. 2006. http://hdl.handle.net/1721.1/32535.
- Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–22. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–50. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juarez P, Fernandez-del-Carmen A, Granell A, Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One. 2011;6:e21622. doi: 10.1371/journal.pone.0021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZY, Zhao H, Zhao HM. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–9. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr Engineering bioBrick vectors from biobrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Day PJ, Walker MR. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and application for cloning into restriction digest-linearized vectors. PCR Methods Appl. 1993;2:328–32. doi: 10.1101/gr.2.4.328. [DOI] [PubMed] [Google Scholar]
- Smolke CD. Building outside of the box: iGEM and the BioBricks Foundation. Nat Biotechnol. 2009;27:1099–102. doi: 10.1038/nbt1209-1099. [DOI] [PubMed] [Google Scholar]
- Torella JP, Boehm CR, Lienert F, Chen JH, Way JC, Silver PA. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Res. 2013;42:681–9. doi: 10.1093/nar/gkt860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Hlavacek WS, Savageau MA. Design of gene circuits: lessons from bacteria. Nat Rev Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- Wang WD, Chen ZT, Kang BG, Li R. Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp Cell Res. 2008;314:699–706. doi: 10.1016/j.yexcr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Wang RY, Shi ZY, Guo YY, Chen JC, Chen GQ. DNA fragments assembly based on nicking enzyme system. PLoS One. 2013;8:e57943. doi: 10.1371/journal.pone.0057943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman L, Core M, Ness J, Theotdorous E, Gustafsson C, Minshull J. Advertorial: rapid, scarless cloning of gene fragments using the Electra Vector System™. Genet Eng Biotechnol. 2013;33 [Google Scholar]
- Wingler LM, Cornish VW. Reiterative recombination for the in vivo assembly of libraries of multigene pathways. P Natl Acad Sci USA. 2011;108:15135–40. doi: 10.1073/pnas.1100507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vansiri A, Bhan N, Koffas MA. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol. 2012;1:256–66. doi: 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- Yonemura I, Nakada K, Sato A, Hayashi J, Fujita K, Kaneko S, Itaya M. Direct cloning of full-length mouse mitochondrial DNA using a Bacillus subtilis genome vector. Gene. 2007;391:171–7. doi: 10.1016/j.gene.2006.12.029. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhao H. Directed evolution of a highly efficient cellobiose utilizing pathway in an industrial Saccharomyces cerevisiae strain. Biotechnol Bioeng. 2013;110:2874–81. doi: 10.1002/bit.24946. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Du J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Methods Mol Biol. 2013;985:177–209. doi: 10.1007/978-1-62703-299-5_10. [DOI] [PubMed] [Google Scholar]
- Zelcbuch L, Antonovsky N, Bar-Even A, et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013;41:e98. doi: 10.1093/nar/gkt151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–7. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. P Natl Acad Sci USA. 2008;105:18232–7. doi: 10.1073/pnas.0809737105. [DOI] [PMC free article] [PubMed] [Google Scholar]