Abstract
We used next-generation sequencing to identify IGH genetic variation in two closely related hypertensive rat lines that differ in susceptibility to end-organ disease (SHR-A3 and SHR-B2). The two SHR lines differ extensively at the IGH locus from the rat reference genome sequence (RRGS) and from each other, creating 306 sequence unique IGH genes. Compared to IGH genes mapped in the RRGS, 98 are null gene alleles (31 are null in both SHR lines, 45 are null in SHR-A3 only, and 23 are null in SHR-B2 only). Of the 306 divergent gene sequences, 126 result in amino acid substitution and, among these, SHR-A3 and SHR-B2 differ from one another at the amino acid level in 96 segments. Twelve pseudogenes in the RRGS had changes displacing the stop codon and creating probable functional genes in either or both SHR-A3 and SHR-B2. A further 5 alleles that encoded functional RRGS genes or open reading frames were converted to pseudogenes in either or both SHR-A3 and SHR-B2. These studies reveal that the pre-immune immunoglobulin repertoire is highly divergent among SHR lines differing in end organ injury susceptibility and this may modify immune mechanisms in hypertensive renal injury.
Introduction
Hypertension (high blood pressure) is a major risk factor for end organ disease such as stroke and renal failure. Risk of end organ disease shows strong heritability that appears to arise independently of genetic factors affecting blood pressure. The recent application of genome wide association studies (GWAS) to this problem has identified an unexplained gap of missing heritability, some of which may arise in regions of the genome in which extensive duplication has lead to concomitant emergence of both functional diversity and sequence complexity that is inadequately represented in GWAS marker sets 1, 2. One such region of the genome is the immunoglobulin heavy chain (IGH) locus, a region encompassing approximately 5Mb in the rat. A recent effort to assess the success of high throughput genotyping systems to capture the extent of human IGH locus genetic variation demonstrates the extreme degree of genetic diversity in this region and shows that genetic variation in the IGH locus has been under-represented in human GWAS 3.
Regions of the genome that are involved in host resistance to infection by simpler and more rapidly evolving pathogenic organisms may accumulate genetic variation that is subject to selection because it beneficially affects host-pathogen interaction. There are numerous examples of positive selection for alleles that provide important benefits to host-pathogen interaction, yet are potentially deleterious to overall health. For example, alleles at the major histocompatibility complex are associated with inflammatory diseases 4–6. The evolution of two distinct variants in human hemoglobin is associated with resistance to malaria infection but also creates susceptibility to sickle cell anemia 7. Variation in and around Apol1 is associated with resistance to trypanosome infection, but increases risk of hypertensive renal disease in individuals of African descent 8.
Resistance to norovirus infection is linked to a risk allele for Crohn’s disease 9. Thus, sometimes clearly deleterious health effects can arise in alleles subject to positive selection for host-pathogen resistance. A trigger mechanism may be needed to uncover the deleterious effects of a positively selected disease resistance allele. For example, type 1 diabetes in the rat can be induced by viral infection (or polynucleotide viral DNA mimics), but only in the presence of a susceptible genotype in the variable (V) genes of T-cell receptor beta gene locus 10, 11. Thus an initiating injury signal elicits disease only when the underlying immune response genotype is permissive.
The spontaneously hypertensive rat (SHR) is an inbred model of hypertension that exists in distinct lines in which susceptibility to hypertensive end organ damage (stroke and kidney disease) differs markedly across SHR lines. In spite of SHR lines arising from the same founder pair and sharing ~87% of the genome identical by descent, susceptibility to stroke and kidney disease are determined by underlying genetic variation 12. In our recent investigation of SHR lines that contrast in end organ injury susceptibility, we mapped the IGH locus as linked to large heritable variations in serum immunoglobulin levels 13. Re-sequencing of the IGH constant (C) gamma (IGHG) genes across SHR lines susceptible and resistant to hypertensive end-organ disease revealed high levels of amino acid substitution, including functional variation affecting interaction with immunoglobulin Fc receptors. In studies of the F2 progeny of an SHR-A3 x SHR-B2 intercross, variation at the IGH locus was associated with renal injury 13. There is growing evidence for the involvement of immune mechanisms in stroke and renal disease, end organ diseases to which SHR-A3 is susceptible, but which are resisted by SHR-B2 12, 14.
Our prior targeted resequencing effort at the IGH locus was restricted to the IGHG genes 13, and we were unable to fully define the potential of the V, diversity (D), joining (J) and IGHC genes other than IGHG to contribute to differences in disease susceptibility that may be attributable to functional variation in immunoglobulin repertoire across these two SHR lines. We have obtained whole genome sequence data for these two SHR lines that has allowed us to assemble a more complete view of genetic variation in the IGH locus that reveals very high divergence between the two SHR lines.
Results and Discussion
Illumina HiSeq genomic sequencing reads for each SHR line were aligned to the rat reference genome (Rn 3.4/rn4). We used this assembly in preference to the rn5 assembly because many of the known Brown-Norway IGH genes located in the rn4 assembly are not mapped in the rn5 assembly. Mean aligned sequence coverage depth was 43.6 (SHR-A3) and 48.5 aligned reads per base (SHR-B2). This is the highest density coverage yet reported for next generation sequencing of the rat genome 15–18. We have previously reported, using a 10K SNP panel, that 87% of the genomes of SHR-A3 and SHR-B2 are descended from a single common ancestor 19. Genetic divergence exists in the remaining 13% of the genome. This divergence is structured into haplotype blocks. Chromosome 6 contains only two such blocks of divergence, one in each in the telomeric regions. We previously used a high-density genome-wide SNP panel to define the distal telomeric block as approximately centered on chr6:143.7Mb with average genome-wide resolution of block boundaries of ~2Mb, indicating an approximate position of chr6:141.3Mb to chr6:146.1Mb 19. Detailed analysis of genetic variation between SHR-A3 and SHR-B2 using their assembled genome sequences allows this block to be refined with precision and we have determined that it extends between chr6:137,282,300–144,673,200. Table 1 indicates the rn4 annotated rat genes located in this block, whether they contain non-synonymous sequence variation across SHR-A3 and SHR-B2 and whether there is evidence of mRNA expression from these genes (presence of mRNA in NCBI gene database). Of the 19 genes in this block, only 4, including IGH met these criteria. The IGH locus was fully contained within this haplotype block and comprised 82% of the block.
Table 1.
Genetic variation in the divergent haplotype block in the distal region of chromosome 6. The genes present in this block are listed by symbol in positional order, defined by the position of the methionine initiation codon, except in the case of IGH where the entire extent of the locus is provided. The presence of Y (yes) or N (no) in coding variation column indicates whether sequence variation exists that yields amino acid changes between SHR-A3 and SHR-B2. In the mRNA column, these letters indicate if the NCBI nucleotide database supports transcription of these genes indicated as report of an mRNA sequence. Four gene symbols are presented in bold font to indicate genes for which there is evidence of both expression and coding sequence variation.
| Gene symbol | Coding variation | mRNA | initiation codon position |
|---|---|---|---|
| Siva1 | N | Y | chr6:137,631,420 |
| Akt1 | N | Y | chr6:137,657,515 |
| Zbtb42 | N | Y | chr6:137,664,936 |
| Pld4 | N | Y | chr6:137,744,724 |
| Cdca4 | N | Y | chr6:137,820,843 |
| Gpr132 | Y | Y | chr6:137,856,344 |
| Nudt14 | N | N | chr6:137,942,910 |
| Btbd6 | N | Y | chr6:137,984,061 |
| Brf1 | N | N | chr6:138,007,598 |
| Mta1 | N | Y | chr6:138,137,825 |
| Crip2 | N | Y | chr6:138,147,518 |
| Crip1 | N | Y | chr6:138,159,217 |
| IGH | Y | Y | chr6:138,247,564–143,302,037 |
| Adam6 | Y | Y | chr6:138,590,306 |
| Nucks1 | Y | Y | chr6:142,904,237 |
| Zfp386 | N | Y | chr6:143,311,806 |
| Vipr2 | N | Y | chr6:143,347,862 |
| Wdr60 | Y | N | chr6:143,534,961 |
| Ptprn2 | N | Y | chr6:143,787,995 |
We examined the genome-wide extent of polymorphism between each of the SHR rat lines and the reference genome. SHR-A3 had 1 base change every 386 bases (2,590 base changes per Mb) while SHR-B2 had 1 base change per 377 bases overall (2,652 base changes per Mb). Examination of the distribution of sequence divergence across the genome was performed by assessing the cumulative number of base changes per Mb in overlapping 5Mb blocks across the genome (i.e., block 1 = chr 1:0–5Mb, block 2 = chr 1:2–6Mb, block 3 = chr1:3–7Mb, etc). The most divergent region of the genome in SHR-B2 (compared with the reference genome) was identified as a 5Mb locus on the distal end of chromosome 6 (138–143Mb) in which there were 11,763 bases changes per Mb. This divergence was 37.3% greater than the next most divergent locus in the SHR-B2 genome and more divergent from the reference genome than any locus in SHR-A3. This most highly divergent locus aligns closely with the IGH locus and fully contains all IGH genes. In SHR-A3 the same 5Mb block has 1,954 base changes per Mb, making this locus slightly more similar to the rat reference genome than the genome-wide average for this rat line. This variation suggests that divergent evolution of this locus has occurred which may result in differences in the pre-immune immunoglobulin repertoire across these inbred rat lines.
All available Rattus norvegicus IGH genes from IGMT 20–22 (see Methods) were aligned to the rat reference sequence using the BLAT alignment tool. The position and orientation of the alignments was noted. We examined the location of each reference gene for the presence of SHR-A3 and SHR-B2 genome sequence alignments that span the entirety of the gene. The absence of alignment or the presence of gaps in the alignment resulted in the designation of a null allele, likely attributable to structural variation in this highly segmentally duplicated region of the genome. Supplemental Figure 1 provides examples of the sequence alignment at several IGHV genes that were called as null. We also sought the presence of simple sequence variations (single nucleotide polymorphisms and indels). When variations were observed, we reconstructed the specific allele present in each rat line and assembled a table of all allelic variation (Supplemental Table 1). Figure 1 provides a haplotype map indicating the distribution of alleles across the IGH locus. Allelic differences are summarized in Table 2, which indicates that 67.1% of IGH genes existed as two or more alleles across the three inbred rat strains investigated. SHR-A3 differed from the rat reference (Brown-Norway, BN) sequence across 144 genes, while SHR-B2 differed from BN in 213 genes.
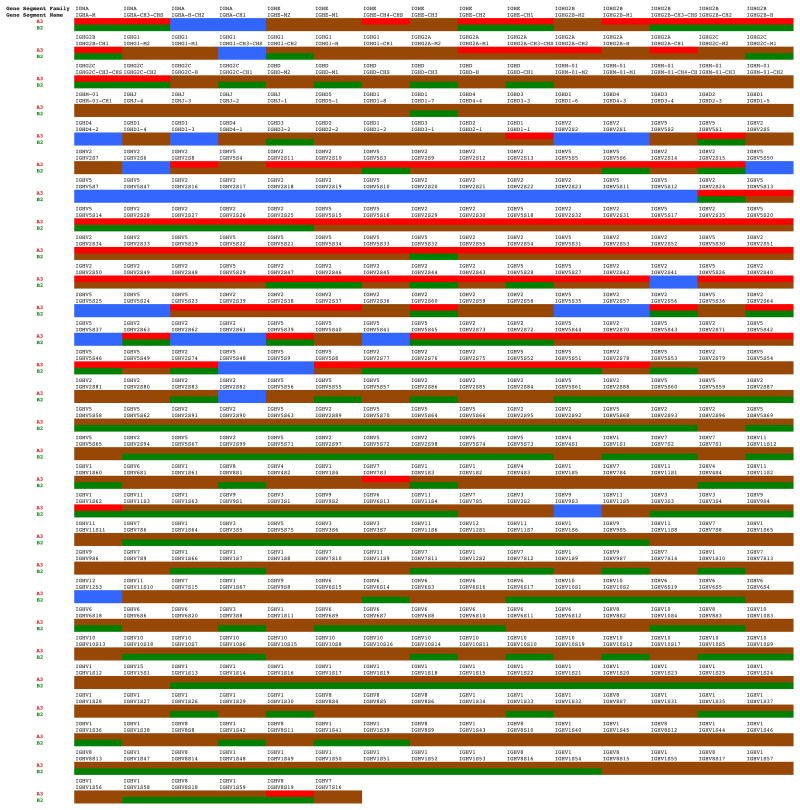
Figure 1.
Haplotype map of the IGH genes encoded from chr6:138,247,564–143,302,037. The map proceeds through the IGH locus from top left to bottom right. If a gene in SHR-A3 differs from the Brown-Norway reference sequence (BN) it is indicated in red. If a gene in SHR-B2 differs from BN it is indicated in green. If either SHR line has the same sequence in this gene, it is indicated in brown. If both SHR-A3 and SHR-B2 differ from BN, but not from one another, the gene is indicated in blue. Detected variation can range from a single nucleotide substitution to a null allele in which one or both SHR lines has no genome sequence alignment across the gene. The naming convention for IGH gene subgroups and genes follows that of the international ImMunoGeneTics information system 20, 22.
Table 2.
Overall genetic variation among 411 IGH genes in the Brown-Norway (BN) rat reference genome and the genomes of SHR-A3 and SHR-B2 (IBD = identical by descent).
| State | Identity | n | % |
|---|---|---|---|
| Mono-allelic | SHR-A3, SHR-B2, and BN are IBD | 136 | 33.1 |
| Di-allelic | SHR-A3 and BN are IBD, SHR-B2 differs | 133 | 32.4 |
| Di-allelic | SHR-B2 and BN are IBD, SHR-A3 differs | 61 | 14.8 |
| Di-allelic | SHR-A3, and SHR-B2 are IBD, BN differs | 35 | 8.0 |
| Tri-allelic | SHR-A3, SHR-B2 and BN all differ | 48 | 11.4 |
|
| |||
| Total | 411 | 100.0 | |
We identified numerous changes in gene sequence that altered amino acid composition of encoded protein. The most marked change was the presence of a large number of null alleles in both SHR-A3 and SHR-B2 (Table 3). In some instances this allelic loss was unique to either one or other of the two SHR lines, in other instances both lines were null. The existence of extensive structural variation in the human IGH locus has recently been described and includes haplotypes revealing complex rearrangements, segmental duplications and segmental losses 3.
Table 3.
Effect of coding sequence differences on gene functional potential in SHR-A3 and SHR-B2, compared with Brown-Norway reference sequence. (ORF = open reading frame).
| Functional effect | n |
|---|---|
| Functional in BN, Null in SHRA3 | 29 |
| Functional in BN, Pseudogene in SHR-A3 | 0 |
| ORF in BN, Null in SHRA3 | 5 |
| Pseudogene in BN, Null in SHRA3 | 47 |
| Pseudogene in BN, ORF in SHRA3 | 3 |
| Functional in BN, Null in SHRB2 | 17 |
| Functional in BN, Pseudogene in SHR-B2 | 4 |
| ORF in BN, Null in SHRB2 | 2 |
| Pseudogene in BN, Null in SHRB2 | 39 |
| Pseudogene in BN, ORF in SHRB2 | 9 |
A complete description of the functional state in SHR-A3 and SHR-B2, along with the amino acids affected by sequence change is provided in Supplemental Table 2. One of the objectives of our study was to extend our prior descriptions of functional IGHG variation across the two SHR rat lines to include information regarding changes in the large remaining portion of this locus that we have not previously investigated. The extent of potentially functional sequence differences observed included amino acid alterations in 96 IGH genes. This large degree of variation includes not only alleles at which one SHR line is null, while the other has a functional allele, but also includes twelve alleles in which one or both of the SHR lines possesses a potentially functional gene wherein the allele encoded in the reference BN sequence is a pseudogene. We have examined whether these twelve BN pseudogenes that lack a stop codon in SHR-A3 or SHR-B2 have retained functional recombination signal (RS) sequences necessary for their incorporation into an expressed immunoglobulin. In each case the V-RS was retained. Evidence of functionality will need to be extended to include expressed sequence before these genes can be considered with certainty to contribute to the pre-immune repertoire of SHR lines. However, the data we have developed supports the identification of several functional alleles in SHR-A3 and/or SHR-B2 that are pseudogenes in BN. A complete and detailed catalog of the IGH locus variation is provided in Supplemental Table 3.
In general, autoimmune diseases for which genetic susceptibility has been identified show association with three major gene groups: the major histocompatibility (MH) locus, T cell receptors (TR) and immunoglobulins (IG). This reflects the roles of these genes in presentation and recognition of antigens. There is growing evidence of the involvement of inflammation and immunity in risk of hypertensive end organ disease. Gene knockout studies 23 showed that mice deficient in Rag1 fail to demonstrate increased blood pressure response to deoxycorticosterone acetate or to angiotensin II (AII) infusion and that renal injury is reduced in AII-dependent hypertension in lymphocyte-deficient mice 24. Adoptive transfer of T regulatory lymphocytes reduces AII-mediated hypertension and renal damage 25. Similarly in stroke, the other major end organ disease of SHR-A3, there is evidence of immune involvement. Lymphocyte-deficient mice have much smaller infarcts in response to temporary middle cerebral artery occlusion (MCAO) and better neurological MCAO outcomes than intact controls 26, 27. Treatment with an immunosuppressant FTY720 (Fingolimod) reduced lymphocyte infiltration in MCAO and decreased brain injury 27.
The present study was motivated by our observation of remarkable sequence variation in IGHG subclasses between two closely-related hypertensive rat lines was associated with susceptibility to cardiovascular end-organ disease 13. This observation raises the possibility that end-organ injury risk involves an autoimmune mechanism. However, we recognized that the IGHG variation we sampled was likely to represent only a fraction of the potential genetic variation in the IGH locus and that disease association might be attributable to other variation in this locus. Because we lacked insight into the potential for variation across the rest of the IGH locus to contribute to disease susceptibility we used whole genome sequencing to uncover additional genetic variation in this locus.
One limitation to the use of next generation sequencing (NGS) is the reliance on a reference genome for alignment of the genome sequence reads. Our observation that both SHR-A3 and SHR-B2 have substantial regions in the IGH locus in which no alignment is observed indicates that structural variation may be an important source of variation in the genomic sequence at this locus. It also indicates that BN, nor any other single inbred rat strain, is likely to possess a complete repertoire of all rat IGH coding genes. Furthermore, SHR-A3 and/or SHR-B2 may contain additional IGH genes that are absent in BN, and consequently have not emerged in the reference sequence alignment approach employed in these studies. Though sequence complexity is an obstacle, it may be possible to identify additional IGH genes by de novo sequence assembly from our next generation sequence reads 28. This possibility is enhanced by the very high density sequence coverage we obtained, though it may be challenged by the structural complexity of this locus 29.
In regions of segmental duplication reliance solely on NGS alignment can reduce the accuracy with which the sequence can be assembled because both the subject and reference sequences may have regions that have been differently affected by segmental duplication and loss events. In some regions of the IGH locus we noted that one or both of our rat lines showed assemblies that lacked aligned sequence reads while in other regions read coverage was much greater than the genome-wide average. Supplemental Figure 2 shows the distribution of sequence read coverage for SHR-A3 and SHR-B2 across the IGH locus. The troughs and peaks in this distribution may reflect deletion and duplication events respectively. In some of the suspected duplicated regions in the IGH locus we observed what appeared to be lack of homozygosity. This might be attributable to duplicated segments that have undergone subsequent divergence. If such duplication is absent in the reference sequence, reads from duplicated regions in the subject sequence may align across the sequence from which the duplicated region arose and apparent heterozygosity may reflect differential mutation across alignments that arise from duplicated segments that have subsequently diverged by mutation. The recent emergence of NGS methods producing long sequence reads may provide greater clarity regarding different duplication events across individual inbred strains. The distribution of coverage depth across the IGH locus shares some similarity with the distribution of sequence variation (Supplemental Figure 3), notably in the proximal region of the locus. However, in more telomeric regions SHR-B2 frequently shows very high levels of sequence divergence in regions while coverage depth does not suggest duplication as the predominant factor leading to divergence.
The germline IGH genes from the variable (IGHV), diversity (IGHD) and joining (IGHJ) genes assemble by recombination during B cell genesis 30. This assembly furnishes individuals with immunoglobulin diversity that embodies the initial capacity to recognize antigens. Initiation of adaptive immunity in the B cell lineage begins with this genetically determined pre-immune immunoglobulin repertoire. Our studies show that the pre-immune repertoire is highly divergent among BN, SHR-A3 ad SHR-B2. These differences may influence the initiation and development of antibody production directed at both external and self-antigens. A recent report in an inbred-mouse model of rheumatoid arthritis that involves auto-antibody targeting of collagen epitopes reveals that a haplotype involving two amino acid substitutions in a single IGHV gene determines auto-reactive antibody production to antigen exposure, confirming the capacity of germline variation to be determinative of susceptibility in autoimmune disease 31.
Advances in high throughput sequencing are now being applied as a tool to characterize individual differences in antibody diversity both in the pre-immune repertoire and as immunity develops 32. This is accomplished by sequencing of the expressed IGH mRNA’s present in lymphocytes. Although genetic risk of renal disease in SHR is clearly polygenic and any single locus is unlikely to account for more than a part of risk, the association we have shown between hypertensive renal disease and germ-line variation in the IGH locus 13 provides a rationale to investigate immune mechanisms of hypertensive end organ disease. The pre-immune immunoglobulin repertoire provides the template for antibody selection that continues through a further process of immunoglobulin somatic mutation during B cell selection and is amplified by proliferation of the most reactive B cell clones. Knowledge of the germ-line encoded pre-immune antibody repertoire provides the classification system needed to structure the enormously diverse range of immunoglobulin sequences that are developed in response to antigenic stimulation. Examination of the expressed IGH repertoire prior to and during the development of end organ disease may also provide a pathway to identify the antigens that provoke immune responses that drive disease. The present report provides a vital foundation for such studies.
Methods
Rats
Studies were performed on genomic DNA isolated from the livers of inbred rats of the injury-prone spontaneously hypertensive line (SHR-A3, SHRSP/BbbUtx) that have been maintained in our facility for 15 years. We also used inbred rats from the injury-resistant SHR-B2 line that were bred in our facility from stocks originating from colonies held at Kinki University School of Medicine, Japan (SHR/Utx). These two lines are derived from a single pair of founders common to both lines. These two inbred lines are ~87% genetically identical by descent (IBD) and this identity is partitioned into discrete haplotype blocks in these strains 33. Animals were housed in an AAALAC-approved animal facility and provided a standard rodent chow diet and drinking water ad libitum. All animal use was prospectively reviewed and approved by the University’s Animal Welfare Committee.
Genome sequencing
Genomic DNA was extracted from samples of liver tissue obtained from male representatives of each rat line and quantified by UV spectroscopy. DNA samples were submitted to a commercial genome sequencing company (Axeq Technologies, Rockville, MD, 20850, United States) for sequencing. Sequencing was performed using the Illumina HiSeq 2500 platform.
Assembly and annotation
An average of 1.37 billion reads of 100 bp were obtained from each sample providing an average genome coverage of approximately 45X. We mapped the paired-end reads to the rat assembly [RGSC3.4 (Ensembl release 69)] using Novoalign software (Novocraft.com, version V2.08.01 using default settings 34. Novoalign performs local alignment using the Smith-Waterman algorithm. The typical default threshold command −t A,B is 20,3 where the threshold is calculated using: t = (L−A) * B where L is the read length (sum of pairs), A = log4(reference genome length), B <= gap extended penalty (default = 3), and the default gap opening penalty is 40 and the gap extend penalty is 6. An average of 95.4% of the total reads were aligned to the rat assembly. After generating BAM files, the mapped reads were sorted by coordinate location using SamTools 35 and Picard tools 36. We then labeled the PCR duplicates with Picard tools 36. Finally, we used the Genome Analysis Tool Kit (GATK version v2.3-9-ge5ebf34) 37 to perform local realignment, recalibration and variant calling. We removed false positive calls by using a post-calling filter that enforces that each variant has a mapping quality >30, a base quality >20, and a coverage ≥10, and the presence of the variant in reads from both orientations. The resulting variant call format files (vcf version 4.1) were annotated using snpEff software 38 together with the rat genome annotations (RGSC3.4.69). Finally we visually inspected the paired-end read data in the integrative genomics viewer (IGV, v 2.3.25 from Broad Institute) 39.
IGH locus analytical approach
We obtained genomic sequences for verified rat (Rattus norvegicus) IGH genes from the international ImMunoGeneTics information system 21, 22. Sequences were obtained for a total of 411 genes including IGH V, D and J genes as well as C genes (IGHA, IGHD, IGHE, IGHG AND IGHM) encoding the constant region of alpha, delta, epsilon, gamma and mu heavy chains of the IgA, IgD, IgE and IgM, respectively. The genomic positions and orientation of these genes was identified using the Blat tool at the UCSC genome web site to align the gene sequences with the Brown Norway (BN) rat reference (Rn 3.4, rn4) genome 40. Where sequences of IGH genes were insufficient in length to obtain Blat alignment, we examined, via IMGT, the gene-encoding clone sequence via the accession number provided and located the gene sequence and sufficient adjacent sequence to definitively identify the genome location by Blat. IGV was used to align the rat reference genome sequence with variant call files comparing the SHR-A3 and SHR-B2 genomes with the rat reference genome. Specific base changes were identified from the genome sequence of SHR-A3 and SHR-B2, assembled in BAM files and loaded into IGV. Effect of base changes on encoded amino acids was determined by translation using the ExPASy Translate tool 41. In numerous instances alignments of high throughput sequencing reads to the rat reference assembly resulted in gaps or the complete absence of alignment between SHR-A3 or SHR-B2 (or both) sequences and the BN reference genome. Such alignment gaps were considered to reflect likely areas of genomic deletion or the absence of genome duplication events and were considered to be null alleles.
Supplementary Material
Acknowledgments
The work reported has been supported by grants from NIH (R01DK069632-05 and R01DK081866) to PAD. PAD is grateful for additional endowed support from the Cullen Chair in Molecular Medicine at the University of Texas HSC Houston, which provided support for rat genomic DNA sequencing.
Footnotes
Conflict of Interest Disclosures.
None of the authors reports any conflict of interest or relationships that are relevant to the topic of the manuscript that may create or appear to create conflicts of interest.
References
- 1.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013;92(4):530–46. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reveille JD. Genetics of spondyloarthritis--beyond the MHC. Nature reviews Rheumatology. 2012;8(5):296–304. doi: 10.1038/nrrheum.2012.41. [DOI] [PubMed] [Google Scholar]
- 5.Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169(1):345–50. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandiedonck C, Knight JC. The human Major Histocompatibility Complex as a paradigm in genomics research. Briefings in functional genomics & proteomics. 2009;8(5):379–94. doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunn HF. The triumph of good over evil: protection by the sickle gene against malaria. Blood. 2013;121(1):20–5. doi: 10.1182/blood-2012-08-449397. [DOI] [PubMed] [Google Scholar]
- 8.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlsson B, Kindberg E, Buesa J, Rydell GE, Lidon MF, Montava R, et al. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII. 4 Norovirus infection. PLoS One. 2009;4(5):e5593. doi: 10.1371/journal.pone.0005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mordes JP, Cort L, Norowski E, Leif J, Fuller JM, Lernmark A, et al. Analysis of the rat Iddm14 diabetes susceptibility locus in multiple rat strains: identification of a susceptibility haplotype in the Tcrb-V locus. Mamm Genome. 2009;20(3):162–9. doi: 10.1007/s00335-009-9172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirabassi RS, Guberski DL, Blankenhorn EP, Leif JH, Woda BA, Liu Z, et al. Infection with viruses from several families triggers autoimmune diabetes in LEW*1WR1 rats: prevention of diabetes by maternal immunization. Diabetes. 2010;59(1):110–8. doi: 10.2337/db09-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto K, Yamori Y, Nagaoka A. Establishment of the Stroke-prone Spontaneously Hypertensive Rat (SHR) Circ Res. 1974;14(Suppl I):I143–I153. [Google Scholar]
- 13.Herring SM, Gokul N, Monita M, Bell R, Boerwinkle E, Wenderfer SE, et al. Immunoglobulin locus associates with serum IgG levels and albuminuria. J Am Soc Nephrol. 2011;22(5):881–9. doi: 10.1681/ASN.2010111148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Hicks MJ, et al. Hypertensive Renal Disease: Susceptibility And Resistance In Inbred Hypertensive Rat Lines. J Hypertension. 2013;31:2050–9. doi: 10.1097/HJH.0b013e328362f9a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, et al. The genome sequence of the spontaneously hypertensive rat: Analysis and functional significance. Genome Res. 2010 doi: 10.1101/gr.103499.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atanur SS, Diaz AG, Maratou K, Sarkis A, Rotival M, Game L, et al. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell. 2013;154(3):691–703. doi: 10.1016/j.cell.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X, Brenner M, Zhang X, Laragione T, Tai S, Li Y, et al. Whole-Genome Sequences of DA and F344 Rats with Different Susceptibilities to Arthritis, Autoimmunity, Inflammation and Cancer. Genetics. 2013 doi: 10.1534/genetics.113.153049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baud A, Hermsen R, Guryev V, Stridh P, Graham D, McBride MW, et al. Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat Genet. 2013 doi: 10.1038/ng.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell R, Herring SM, Gokul N, Monita M, Grove ML, Boerwinkle E, et al. High-resolution identity by descent mapping uncovers the genetic basis for blood pressure differences between spontaneously hypertensive rat lines. Circ Cardiovasc Genet. 2011;4(3):223–31. doi: 10.1161/CIRCGENETICS.110.958934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. [Accessed: 2nd February, 2014];IMGT®, the international ImMunoGeneTics information system. http://www.imgt.org.
- 21.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33(Database issue):D256–61. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzik TJ, Hoch NE, Brown KA, McCann LA, Rahman A, Dikalov S, et al. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J Exp Med. 2007;204(10):2449–60. doi: 10.1084/jem.20070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowley SD, Song YS, Lin EE, Griffiths R, Kim HS, Ruiz P. Lymphocyte responses exacerbate angiotensin II-dependent hypertension. American journal of physiology Regulatory, integrative and comparative physiology. 2010;298(4):R1089–97. doi: 10.1152/ajpregu.00373.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barhoumi T, Kasal DA, Li MW, Shbat L, Laurant P, Neves MF, et al. T regulatory lymphocytes prevent angiotensin II-induced hypertension and vascular injury. Hypertension. 2011;57(3):469–76. doi: 10.1161/HYPERTENSIONAHA.110.162941. [DOI] [PubMed] [Google Scholar]
- 26.Hurn PD, Subramanian S, Parker SM, Afentoulis ME, Kaler LJ, Vandenbark AA, et al. T- and B-cell-deficient mice with experimental stroke have reduced lesion size and inflammation. J Cereb Blood Flow Metab. 2007;27(11):1798–805. doi: 10.1038/sj.jcbfm.9600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shichita T, Sugiyama Y, Ooboshi H, Sugimori H, Nakagawa R, Takada I, et al. Pivotal role of cerebral interleukin-17-producing gammadelta T cells in the delayed phase of ischemic brain injury. Nat Med. 2009;15(8):946–50. doi: 10.1038/nm.1999. [DOI] [PubMed] [Google Scholar]
- 28.Jackman SD, Birol I. Assembling genomes using short-read sequencing technology. Genome Biol. 2010;11(1):202. doi: 10.1186/gb-2010-11-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nature methods. 2011;8(1):61–5. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefranc M-P, Lefranc G. The Immunoglobulin Facts Book. Academic Press; London, UK: 2001. [Google Scholar]
- 31.Raposo B, Dobritzsch D, Ge C, Ekman D, Xu B, Lindh I, et al. Epitope-specific antibody response is controlled by immunoglobulin VH polymorphisms. J Exp Med. 2014 doi: 10.1084/jem.20130968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Science translational medicine. 2013;5(171):171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell R, Herring SM, Gokul N, Monita M, Grove ML, Boerwinkle E, et al. High Resolution Identity by Descent Mapping Uncovers the Genetic Basis for Blood Pressure Differences Between SHR Lines. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.110.958934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novoalign software. [Accessed: September 19, 2013]; http://www.novocraft.com.
- 35.SAMtools software. [Accessed: September 19th, 2013]; http://samtools.sourceforge.net/
- 36.Picard software. [Accessed: September 19, 2013]; http://picard.sourceforge.net/
- 37.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14(2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.