Abstract
Brown fat activates uncoupled respiration to defend against cold and contributes to systemic metabolic homeostasis. To date, the metabolic action of brown fat has been primarily attributed to its role in fuel oxidation and uncoupling protein 1 (UCP1)-mediated thermogenesis. Whether brown fat engages other tissues through secreted factors remains largely unexplored. Here we show that Neuregulin 4 (Nrg4), a member of the EGF family of extracellular ligands, is highly expressed in adipose tissues, enriched in brown fat, and markedly increased during brown adipocyte differentiation. Adipose tissue Nrg4 expression was reduced in rodent and human obesity. Gain- and loss-of-function studies in mice demonstrated that Nrg4 protects against diet-induced insulin resistance and hepatic steatosis through attenuating hepatic lipogenic signaling. Mechanistically, Nrg4 activates ErbB3/ErbB4 signaling in hepatocytes and negatively regulates de novo lipogenesis mediated by LXR/SREBP1c in a cell-autonomous manner. These results establish Nrg4 as a brown fat-enriched endocrine factor with therapeutic potential for the treatment of obesity-associated disorders, including type 2 diabetes and non-alcoholic fatty liver disease.
Brown fat defends against cold through mitochondrial uncoupling protein UCP1, which links fuel oxidation to heat generation 1,2. Brown adipose tissue (BAT) also plays an important role in whole body energy balance and fuel metabolism. Genetic ablation of brown fat via transgenic expression of diphtheria toxin A renders mice prone to the development of obesity3, whereas activation of BAT thermogenesis by cold has been linked to increased energy expenditure, reduced adiposity, and lower plasma lipids4-6. Recent work has demonstrated that metabolically active BAT is present in adult humans 7-10, raising the prospect that augmenting brown fat abundance and/or function may provide an effective avenue for the treatment of obesity and its associated metabolic disorders 11,12. While sharing key molecular and metabolic characteristics with the classical rodent BAT, brown fat in humans appears to contain both classical and brown-like, beige/brite 13,14, adipocytes with distinct molecular signature and developmental origin 15-17.
To date, the metabolic action of brown fat has been primarily attributed to its role in fuel oxidation and UCP1-mediated thermogenesis. Brown adipocytes actively take up glucose and fatty acids for oxidation by mitochondria, the rate of which is increased under thermogenic conditions. Surprisingly, while mice lacking UCP1 had heightened sensitivity to cold 18, they were resistant to diet-induced obesity at ambient temperature and became more prone to weight gain only at thermoneutrality19,20. The latter is in striking contrast to the obesity-prone phenotype in mice lacking brown fat as a result of genetic ablation 3. These paradoxical findings strongly suggest that brown fat exerts effects on systemic nutrient and energy metabolism beyond its intrinsic ability to carry out uncoupled respiration. We postulated that brown fat may serve as a source of endocrine factors that modulate systemic energy metabolism.
Secreted factors are important in mediating metabolic crosstalk among tissues to maintain systemic nutrient and energy homeostasis. Adipose tissue hormones 21-23, gut-derived fibroblast growth factors 24,25, myokines 26, and bone-derived factors 27 participate in nutrient sensing and coordinate key aspects of metabolic homeostasis. The epidermal growth factor (EGF) family of extracellular ligands has been implicated in the regulation of tissue development and tumorigenesis via the ErbB family of receptor tyrosine kinases28-30. EGFR and ErbB2 are uniquely important in engaging a subset of EGF growth factors in cancer cell growth and survival. Interestingly, a distinct subgroup of growth factors called Neuregulins (Nrg1-4) also contains EGF-like domain and primarily signals through ErbB3 and ErbB4 to regulate diverse biological processes 28. Here we identified Neuregulin 4 (Nrg4) as a novel BAT-enriched secreted factor that attenuates hepatic lipogenic signaling and preserves glucose and lipid homeostasis in obesity. Impaired Nrg4 signaling may contribute to the development of type 2 diabetes and non-alcoholic fatty liver disease (NAFLD).
RESULTS
Identification of Nrg4 as a brown fat-enriched secreted protein
To identify BAT-enriched secreted factors, we analyzed the expression profile of the mouse secretome gene set across twelve tissues (GeneAltas MOE430) and during brown adipocyte (BAC) differentiation. The secretome database contains a set of 2,169 mouse genes predicted to encode secreted proteins 31, 1,378 of which have probe sets on Affymetrix mouse genome MOE430 chips. Our analysis identified a cluster of 26 genes whose expression is enriched in mouse brown fat and induced during differentiation of immortalized brown preadipocytes (Fig. 1a). Among these, Neuregulin 4 (Nrg4) exhibited a highly restricted pattern of expression in adipose tissues. Quantitative PCR (qPCR) analysis indicated that Nrg4 mRNA expression was enriched in BAT, albeit present at a lower level in WAT (Fig. 1b). In contrast, its expression in other tissues, including skeletal muscle, liver, brain, and heart, was relatively low. Consistent with microarray data, Nrg4 expression was markedly induced during adipocyte differentiation (Fig. 1c), reaching a higher level in differentiated immortalized brown adipocytes than 3T3-L1 white adipocytes and primary hepatocytes (Fig. 1b). Nrg4 expression in BAT, but not inguinal and epididymal WAT, was elevated by acute cold exposure, whereas its expression in both BAT and inguinal WAT was augmented by cold acclimation (Fig. 1d). Nrg4 was induced by norepinephrine in differentiated brown adipocytes, but not 3T3-L1 adipocytes (Fig. 1e).
Figure 1. Identification of Nrg4 as a brown fat-enriched secreted protein.
(a) Clustering analysis of genes predicted to encode secreted proteins that are induced during brown adipocyte (BAC) differentiation and enriched in BAT. (b) qPCR analysis of Nrg4 expression in mouse tissues (left) and cultured adipocytes and hepatocytes (right). Data represent the means of triplicates ± s.d. *p<0.05, BAC vs. 3T3-L1 or Hep. (c) qPCR analysis of Nrg4 mRNA expression during BAC differentiation. Data represent the means of triplicates ± s.d. (d) qPCR analysis of Nrg4 mRNA expression in adipose tissues from mice kept at room temperature (RT) or following acute cold exposure (Cold) or cold acclimation (Accl), n=5 in each group, data represent mean ± s.e.m. *p<0.05, RT vs. Cold or Accl. (e) qPCR analysis of Nrg4 mRNA expression in differentiated BAC and 3T3-L1 adipocytes treated with vehicle (Veh) or norepinephrine (NE, 1 μM) for 4 hrs. Data represent the means of triplicates ± s.d. *p<0.05, NE vs. Veh. (f) Immunoblots of media and lysates from HEK293 cells transiently transfected with indicated constructs using anti-SEAP, data are representative of three independent experiments. (g) Secretion of Nrg4 into culture media. Shown are immunoblots of total lysates from ErbB4-Min6 cells treated with DMEM or concentrated conditioned media (CM) from HEK293 (top) or BAC (bottom) expressing vector (Vec) or Nrg4, data are representative of three independent experiments.
Nrg4 belongs to a small family of epidermal growth factor-like (EGFL) domain-containing proteins that are synthesized as transmembrane precursors and undergo proteolytic cleavage 32. The released extracellular fragments act on target cells through autocrine, paracrine and endocrine mechanisms. The proteolytic cleavage of Nrg4 near the transmembrane domain is predicted to release a highly conserved N-terminal EGFL peptide (Supplementary Fig. S1)33,34. To map the residues required for Nrg4 cleavage, we generated a panel of Alanine mutants (a.a. 51-62) between the EGFL and transmembrane domains. To facilitate detection, full-length wild type and mutant Nrg4 proteins were fused to the C-terminus of secreted alkaline phosphatase (SEAP). Immunoblotting analysis of conditioned media (CM) from transfected HEK293 cells showed that SEAP-Nrg4 fusion protein was readily detectable in media (Fig. 1f), indicating that the extracellular fragment of Nrg4 undergoes proteolytic cleavage. Mutants with a.a.53-54 or a.a.53 replaced with Alanine had markedly decreased release of SEAP Nrg4 fusion proteins into media, despite having similar expression levels in total cell lysates. As such, Ser53 appears to be critical for Nrg4 cleavage and release.
Neuregulins transduce signals via activation of the ErbB receptors, particularly ErbB4. To examine whether native Nrg4 can be released into the extracellular space to activate ErbB4, we collected CM from HEK293 cells transiently transfected with a vector expressing full-length wild type Nrg4 and treated Min6 cells stably overexpressing ErbB4 (ErbB4-Min6). Compared to control, CM from Nrg4-transfected cells elicited strong ErbB4 tyrosine phosphorylation (Fig. 1g). Similarly, biologically active Nrg4 could be detected in culture media from differentiated immortalized brown adipocytes transduced with Nrg4 retrovirus. Despite significant efforts, an immunological assay with sufficient sensitivity and specificity for the detection of Nrg4 in plasma remains unavailable at present. Using HEK293 cells with reconstituted expression of ErbB receptor combinations, we found that recombinant NRG4 augments tyrosine phosphorylation of ErbB3 and ErbB4, but not ErbB2 (Supplementary Fig. S2a).
Nrg4 binding is restricted to the liver
A major function of brown fat is non-shivering thermogenesis. We next examined whether Nrg4 is required for defense against cold in Nrg4 null mice generated using a gene trap method. Unexpectedly, despite its abundant expression in BAT, Nrg4 appears to be dispensable for defense against hypothermia following cold exposure. Rectal body temperature was nearly indistinguishable between two groups (Fig. 2a). Expression of Ucp1 and deiodinase 2 (Dio2), the latter being an enzyme involved in thermogenic regulation, was induced to similar extent by cold exposure in WT and KO mice (Fig. 2b). In addition, BAT histology, hepatic gene expression, and plasma levels of insulin, adiponectin, and leptin also appeared similar (Fig. 2c and Supplementary Fig. S3). These observations raised the possibility that Nrg4 may not directly engage in BAT thermogenesis. Instead, this factor may act on other tissues following its release from adipose tissues.
Figure 2. Nrg4 binds to hepatocytes and is dispensable for the defense against cold.
(a) Rectal temperature in WT (filled, n=5) and Nrg4 KO (open, n=7) mice following cold exposure at 4°C. (b) qPCR analysis of BAT gene expression in WT (filled, n=5) and KO (open, n=6) mice kept at ambient room temperature (RT) or following cold exposure (Cold) for 4 hrs. Data in a-b represent mean ± s.e.m., and are representative of three independent experiments, *p<0.05, RT vs. Cold. (c) H&E staining of brown fat sections from mice following cold exposure. (d) Immunoblots of total lysates from ErbB4-Min6 cells treated with CM containing SEAP or SEAP-Nrg4Ex. (e) Binding assay on frozen tissue sections. Schematic diagrams of SEAP or SEAP-Nrg4Ex fusion proteins were shown on the left. (f) SEAP-Nrg4Ex binding to liver sections in the presence of excess recombinant GST or GST-Nrg4Ex. (g) SEAP-Nrg4Ex binding to liver sections in the presence of control CM or CM containing the extracellular domains of ErbB3 or ErbB4. Data in c-g are representative of three independent experiments. Scale bar=100 μm.
To identify the target tissue(s) of Nrg4, we generated a fusion protein between SEAP and the extracellular fragment of Nrg4 (SEAP-Nrg4Ex) and performed binding assays on frozen tissue sections35. The presence of Nrg4 binding sites in tissues can be readily detected by histochemical staining for alkaline phosphatase activity. SEAP-Nrg4Ex fusion protein was bioactive in receptor binding, as indicated by robust activation of ErbB4 phosphorylation (Fig. 2d). While SEAP-Nrg4Ex binding was near background levels in BAT, heart, skeletal muscle, and spleen, strong binding as indicated by the presence of SEAP enzymatic staining was detected on the liver section (Fig. 2e). In contrast, Nrg1 exhibited a broader profile of tissue binding (Supplementary Fig. S2b). Binding of Nrg4Ex to hepatocytes was specific as recombinant GST-Nrg4Ex, but not GST, nearly completely abolished the signal (Fig. 2f). To determine whether ErbB3 and ErbB4 may mediate Nrg4 binding to hepatocytes, we performed competitive binding assays in the presence of excess amounts of the extracellular fragments of ErbB3 or ErbB4. As shown in Fig. 2g, extracellular ligand-binding domains of ErbB3 and ErbB4 markedly reduced SEAP-Nrg4Ex binding on liver sections. These results illustrate that liver is a target tissue of Nrg4, likely through its direct binding to the ErbB receptors.
Nrg4 deficiency exacerbates diet-induced insulin resistance and hepatic steatosis through augmenting lipogenic signaling
Secreted factors released by adipose tissues exert diverse effects on metabolic homeostasis 21-23. To explore the role of Nrg4 in systemic metabolism, we analyzed metabolic parameters of WT and Nrg4-deficient mice fed standard chow or high-fat diet (HFD). Nrg4 KO mice were born at expected Mendelian ratio and gained similar body weight as control when fed standard chow (Fig. 3a). Upon HFD feeding, Nrg4 null mice gained slightly more weight, accompanied by significantly increased adiposity and reduced percent lean body mass. Plasma triglyceride (TAG) concentrations were higher in KO mice under fed condition (Fig. 3b). Fasting blood glucose and plasma insulin levels were higher in HFD-fed Nrg4 deficient mice (Fig. 3c). Glucose tolerance test (GTT) and insulin tolerance test (ITT) revealed that Nrg4 deficiency exacerbated glucose intolerance and insulin resistance following diet-induced obesity (Fig. 3d). While WAT and BAT histology appeared similar, Nrg4 null mouse livers developed more pronounced fat accumulation following HFD feeding, accompanied by increased liver TAG content and plasma ALT levels (Fig. 3e and Supplementary Fig. S4a). To assess whether hepatic Nrg4 may be responsible for these metabolic changes, we performed in vivo RNAi knockdown of Nrg4 in HFD-fed mice through tail vein injection of adenoviral vectors. Liver-specific Nrg4 knockdown did not alter plasma and liver TAG levels orhepatic gene expression (Supplementary Fig. S4c-d).
Figure 3. Nrg4 deficiency exacerbates diet-induced hepatic steatosis.
(a) Body weight, percent of fat and lean body mass in WT (filled, n=8) and Nrg4 KO (open, n=9) mice fed chow or HFD. (b) Plasma TAG levels, (c) Blood glucose and plasma insulin levels in HFD fed mice as in a. (d) GTT (left) and ITT (right) in HFD-fed WT (filled, n=7) and Nrg4 KO (open, n=7) mice. (e) Liver TAG content (left) and H&E staining of tissue sections (right, scale bar=100μm), n=7-9 for each genotype. (f) A cluster of differentially expressed genes involved in lipid metabolism. (g) qPCR analysis of hepatic gene expression in HFD-fed WT (filled, n=7) and Nrg4 KO (open, n=9) mice. Data represent mean ± s.e.m. *p<0.05, KO vs. WT. (h) Immunoblots of total liver lysates using indicated antibodies (top); pSREBP1 denotes precursor SREBP1 protein. Immunoblots of nuclear SREBP1 (nSREBP1) using liver nuclear extracts (bottom). Lamin A/C immunoblot was included as loading control.
Because Nrg4 binding was restricted to the liver, we next performed transcriptional profiling on total liver RNA from HFD-fed WT and KO mice to identify metabolic pathways targeted by Nrg4. Among the genes altered by Nrg4 deficiency was a cluster of genes involved in lipid metabolism. Notably, the expression of several genes involved in de novo lipogenesis, including glucose kinase (Gck), malic enzyme (Me1), fatty acid synthase (Fasn), stearoyl-CoA desaturase 1 (Scd1), and ELOVL fatty acid elongase 5 (Elovl5), was significantly elevated in Nrg4 deficient mouse livers (Fig. 3f-g). The expression of Fsp27, a lipid droplet protein associated with hepatic steatosis, and several proinflammatory cytokines was also increased (Fig. 3g and Supplementary Fig. S4b). In contrast, mRNA levels of genes involved in fatty acid β-oxidation (Ehhadh, Acox1, and Cpt1a), gluconeogenesis, and mitochondrial OXPHOS was comparable between two groups. The expression of several transcriptional regulators of hepatic metabolism, including PGC-1α, PGC-1β, ChREBP, and SREBP2, were also similar between control and KO livers. In contrast, mRNA levels of SREBP1c, a key regulator of de novo lipogenesis and triglyceride synthesis 36, were significantly increased in Nrg4 KO mouse livers. Importantly, protein levels of precursor SREBP1 (pSREBP1) and the cleaved and transcriptionally active isoform of SREBP1 in the nucleus (nSREBP1) were also elevated (Fig. 3h). Activation status of the AMPK and AKT pathways appeared similar between two groups. Together, we concluded that Nrg4 deficiency leads to aberrant induction of hepatic lipogenesis and predisposes mice to diet-induced hepatic steatosis.
Nrg4 cell-autonomously attenuates de novo lipogenesis in hepatocytes
To determine whether Nrg4 regulated de novo lipogenesis in a cell-autonomous manner, we performed studies in cultured primary mouse hepatocytes. Previous studies have demonstrated that liver expresses all ErbB receptors except ErbB237. However, ErbB4 expression in hepatocytes is relatively low in culture. To reconstitute ErbB signaling in hepatocytes, we transduced primary hepatocytes with control (GFP) or ErbB4 adenoviral vectors and examined the effects of Nrg4 on receptor activation and downstream signaling. As expected, ErbB4 expression was below detectable range in hepatocytes transduced with GFP adenovirus (Fig. 4a). Adenoviral-mediated expression of ErbB4 resulted in auto-phosphorylation that was moderately increased by GST-Nrg4Ex treatments. Interestingly, GST-Nrg4Ex treatment elicited a dose-dependent increase of tyrosine phosphorylation of endogenous ErbB3 and STAT5 proteins without affecting total ErbB3 and STAT5 levels. Because ErbB3 does not have intrinsic kinase activity, increased phosphorylation is likely due to ligand-dependent heterodimerization with ErbB4 in response to Nrg4. Previous studies have demonstrated that ErbB receptor activation stimulates the STAT5 signaling pathway in cultured cells and in vivo 38,39. Our results indicate that this signaling pathway is also operational in hepatocytes. Importantly, the stimulation of ErbB3 and STAT5 phosphorylation was absent when a kinase-dead mutant of ErbB4 (ErbB4 KD) was used (Fig. 4b). Treatments of hepatocytes with JNJ28871063, a pan-ErbB inhibitor, also abolished the response to GST-Nrg4Ex (Fig. 4c), suggesting that kinase activity of ErbB4 is required for mediating the stimulation of ErbB3 and STAT5 in response to Nrg4.
Figure 4. Nrg4 cell-autonomously attenuates de novo lipogenesis in hepatocytes.
(a) Immunoblots of hepatocytes transduced with GFP or ErbB4 adenovirus and treated for 15 min with GST or different doses (2, 10, 20 μg/ml) of GST-Nrg4Ex. (b) Immunoblots of hepatocytes transduced with adenovirus expressing GFP, WT ErbB4, or kinase dead ErbB4 (ErbB4-KD) and treated with GST or GST-Nrg4Ex. (c) Immunoblots of hepatocytes transduced with GFP or ErbB4 adenovirus and treated with GST, GST-Nrg4Ex (10 μg/ml) or GST-Nrg4Ex in the presence of vehicle or 10 μM pan-ErbB inhibitor JNJ28871063 (JNJ). Data in a-c are representative of three independent experiments. (d) qPCR analysis of gene expression in transduced primary hepatocytes treated with vehicle (DMSO) or T0901317 in the presence of GST or GST-Nrg4Ex. (e) Incorporation of 14C-acetate into lipids in primary hepatocytes treated as in d. (f) qPCR analysis of ErbB3 flox/flox (E3f/f) and null (E3KO) hepatocytes treated as in d. (g) Report gene assay in Hepa1 cells transiently transfected with 4xLXRE-luc in combination with indicated plasmids. Data in d-g represent the means of triplicates ± s.d. *p<0.05, GST vs. GST-Nrg4Ex, #p<0.05, vehicle vs. T0901317.
Hepatic lipogenesis is highly responsive to nutritional and hormonal signals. The induction of lipogenic gene program in the fed state requires transcriptional activation of SREBP1c, which is a direct target of nuclear hormone receptor liver-X receptor (LXR) 40,41. We next examined whether Nrg4/ErbB4 activation directly impacts the SREBP1c/lipogenesis axis in hepatocytes in response to LXR activation. Hepatocytes transduced with GFP or ErbB4 adenoviruses were treated with vehicle (DMSO) or T0901317, a potent agonist for liver-X receptor, in the presence of GST or GST-Nrg4Ex. As expected, T0901317 treatment strongly induced mRNA expression of Srebp1c and lipogenic genes, such as Fasn and Scd1, in hepatocytes transduced with GFP (Fig. 4d). Treatment of ErbB4-transduced hepatocytes with GST-Nrg4Ex significantly lowered the stimulatory effects of T0901317 on these genes. The expression of Abca1, another LXR target gene, was also reduced in response to ErbB4/Nrg4 activation. Consistently, incorporation of 14C-acetate into lipids in hepatocytes was significantly inhibited by activation of Nrg4/ErbB signaling (Fig. 4e). Using hepatocytes isolated from ErbB3 flox/flox and ErbB3 flox/flox:Alb-Cre mice, we found that ErbB3 deletion impaired the attenuation of Srebp1c and Fasn expression by Nrg4 (Fig. 4f). STAT5 has been demonstrated to exert inhibitory effects on nuclear hormone receptor signaling 42. To examine whether STAT5 activation directly modulates the transcriptional activity of LXR, we performed reporter gene assay using a luciferase construct containing LXR responsive element (LXRE). As expected, LXR/RXR increased the LXRE reporter luciferase activity in HEK293 cells (Fig. 4g). Cotransfection of constitutively active STAT5 significantly inhibited the increase of LXRE reporter gene expression, suggesting that STAT5 activation may transrepress LXR and attenuate its transcriptional activity.
Adipose tissue Nrg4 expression is reduced in murine and human obesity
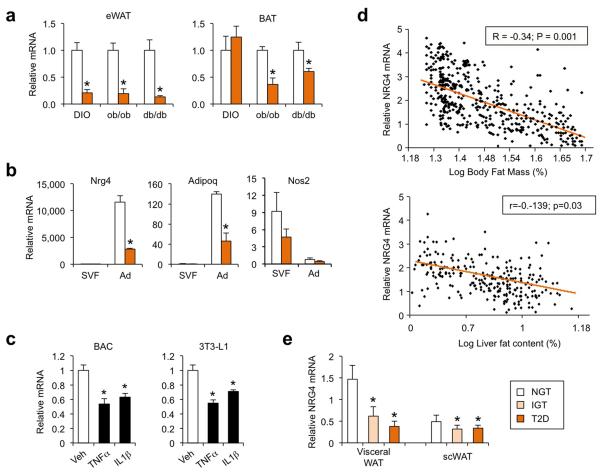
We next examined whether adipose Nrg4 expression is altered in mouse and human obesity. Compared to lean control, mRNA levels of Nrg4 in epididymal WAT were markedly lower in mice with high-fat diet (HFD)-induced obesity (DIO) or genetic obesity caused by leptin (ob/ob) or leptin receptor (db/db) deficiency (Fig. 5a). While Nrg4 mRNA expression in brown fat was similar between lean and DIO mice, its levels were significantly lower in BAT from ob/ob and db/db mice, more severe models of obesity. Fractionation studies indicated that Nrg4 mRNA expression was detected nearly exclusively in adipocytes, but not in the stromal vascular fraction, and was significantly lower in the obese group than lean control (Fig. 5b). Adiponectin (Adipoq) and nitric oxide synthase 2 (Nos2) were included as control for adipocytes and stromal vascular cells, respectively. Adipose tissue inflammation impairs insulin sensitivity and metabolic homeostasis in part through augmented proinflammatory cytokine signaling 43-45. To determine whether proinflammatory cytokines contribute to obesity-induced downregulation of Nrg4 in adipocytes, we treated differentiated brown adipocytes and 3T3-L1 adipocytes with TNFα, a prototypical obesity-induced cytokine 46. We found that Nrg4 mRNA expression was decreased by TNFα treatments in both brown and white adipocytes (Fig. 5c). Similar inhibition of Nrg4 expression was also observed following interleukin 1β (IL1β) treatments. Thus, the reduction of adipocyte Nrg4 expression is likely a consequence of augmented proinflammatory cytokine signaling in obesity.
Figure 5. Adipose tissue Nrg4 expression is reduced in murine and human obesity.
(a) qPCR analysis of Nrg4 mRNA expression in epididymal white fat (eWAT) and BAT from lean (open) or obese (orange) mice. For DIO, WT male mice were fed standard chow (n=5) or HFD (n=6) for three months. For genetic obesity, a group of WT (n=3) and ob/ob (n=4) and a separate group of WT (n=5) and db/db (n=6) mice were analyzed. (b) qPCR analysis of Nrg4 mRNA expression in stromal vascular fraction (SVF) and adipocyte fraction (Ad) isolated from eWAT from lean (n=3) or DIO (n=3) mice. Data in a-b represent mean ± s.e.m. and are representative of three independent experiments, *p<0.05, lean vs. obese. (c) qPCR analysis of Nrg4 mRNA expression in differentiated brown or 3T3-L1 adipocytes following treatments with vehicle (Veh), TNFα (10 ng/ml), or IL1β (40 ng/ml) for 6 hrs. Data represent the means of triplicates ± s.d., representative of three independent experiments. *p<0.05 vs. Veh. (d) Association between relative human scWAT NRG4 mRNA level and log percent body fat mass (top) or log liver fat content (bottom). (e) qPCR analysis of NRG4 mRNA expression in scWAT and visceral WAT from individuals with normal glucose tolerance (NGT), impaired glucose tolerance (IGT), and type 2 diabetes (T2D). Data represent mean ± s.e.m. *p<0.05 vs. NGT.
To investigate whether Nrg4 expression is also reduced in human obesity, we examined its mRNA levels in subcutaneous WAT (scWAT) of a cohort of individuals with a wide range of body fat mass, and found that scWAT NRG4 mRNA levels inversely correlated with log percent body fat mass and log liver fat content (Fig. 5d). When assessing NRG4 expression in BMI-matched individuals, we found that NRG4 mRNA levels in both subcutaneous and visceral adipose tissues were significantly lower in individuals with impaired glucose tolerance (IGT) or type 2 diabetes (T2D) compared to those with normal glucose tolerance (NGT) (Fig. 5e). Together with the observations that Nrg4 deficient mice developed more severe diet-induced hepatic steatosis, our results strongly suggest that inadequate Nrg4 expression may be causally linked to the pathogenesis of NAFLD.
Transgenic expression of Nrg4 improves diet-induced metabolic disorders
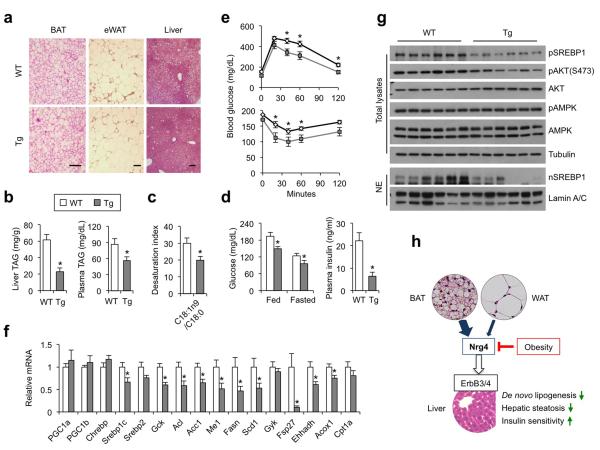
A key prediction from these observations is that elevating Nrg4 levels will protect mice from obesity-associated metabolic disorders. To test this, we generated Nrg4 transgenic mice using aP2 promoter/enhancer and chose line #111 that exhibited adipose-selective overexpression for metabolic studies. Nrg4 transgenic mice were indistinguishable from WT littermates when fed standard chow. Further, body temperature, BAT gene expression, and plasma parameters were similar between control and transgenic mice before and after cold exposure (Supplementary Fig. S5c). Upon HFD feeding, transgenic mice gained slightly but significantly less body weight and adiposity than control (Supplementary Fig. S6a-b). Histological analyses and measurements of liver lipid content indicated that transgenic mice had significantly improved hepatic steatosis (Fig. 6a-b). Plasma TAG levels were also lower in the TG group (Fig. 6b). Fatty acid profile analysis of liver TAG demonstrated that transgenic mice have reduced levels of major fatty acid species, including palmitic acid (C16:0), oleic acid (C18:1), and linolenic acid (C18:2) (Supplementary Fig. S6c). Importantly, the desaturation index (C18:1n9/C18:0), which reflects cellular SCD1 activity, was significantly lower in the transgenic group (Fig. 6c). Nrg4 transgenic mice had lower blood glucose and insulin concentrations (Fig. 6d), and improved glucose tolerance and insulin sensitivity (Fig. 6e). Accordingly, we found that the levels of SREBP1c mRNA and protein were significantly decreased in transgenic mouse livers (Fig. 6f-g). The expression of lipogenic genes, including Gck, Acl, Acc1, Me1, Fasn, and Scd1, and proinflammatory cytokines, including Tnfα, Il1β, and Ccl2 (Supplementary Fig. S6d), was lower in the transgenic mouse livers. Plasma ALT levels also trended lower in the transgenic group (Supplementary Fig. S6e). These results demonstrate that transgenic expression of Nrg4 in adipose tissues is sufficient to attenuate hepatic lipogenesis and ameliorate diet-induced fatty liver disease and insulin resistance.
Figure 6. Transgenic expression of Nrg4 alleviates diet-induced fatty liver through attenuating lipogenesis.
(a) H&E staining of tissue sections (scale bar=100 μm), data are representative of three independent experiments. (b) Liver TAG content (left) and plasma TAG levels (right) in ad lib HFD-fed WT (open, n=8) or Tg (gray, n=9) mice. (c) Desaturation index of liver C16 and C18 fatty acids. (d) Blood glucose and plasma insulin levels. (e) GTT (top) and ITT (bottom) in HFD-fed WT (black line, n=9) and Tg (gray line, n=8) mice. (f) qPCR analysis of hepatic gene expression in mice as in e. Data in b-f represent mean ± s.e.m., *p<0.05, WT vs. Tg. (g) Immunoblots of total liver lysates (top) and nuclear extracts (bottom), data are representative of two independent experiments. (h) Model of Nrg4 as a brown fat-enriched secreted protein that preserves metabolic homeostasis in diet-induced obesity. Nrg4 binds to the liver and attenuates hepatic lipogenic signaling via activating the ErbB3 and ErbB4 receptors.
DISCUSSION
Brown and beige adipocytes are capable of activating uncoupled respiration through UCP1-mediated proton leak and heat generation. This intrinsic thermogenic function is critical for the defense against cold and contributes to whole body energy balance. However, whether brown fat engages other metabolic tissues via its release of endocrine factors remain poorly understood. Here we define a novel mechanism through which brown adipocytes may regulate systemic metabolism independent of UCP1-mediated thermogenesis. While Nrg4 expression is enriched in BAT, both brown and white adipocytes likely contribute to the release of Nrg4 into circulation (Fig. 6h). The significance of white fat as a source of Nrg4 may be even more relevant in humans given the paucity of brown/beige fats in adults. Importantly, transgenic rescue of Nrg4 expression in adipose tissues is sufficient to ameliorate the severity of fatty liver and insulin resistance, raising the prospect of developing Nrg4 as an effective therapeutic biologic for the treatment of NAFLD and type 2 diabetes.
Excess stimulation of SREBP1-mediated lipogenesis has been observed in mouse models of fatty liver disease as well as in human NAFLD 36,47,48. Importantly, liver-specific transgenic expression of the transcriptionally active form of SREBP1c increases hepatic lipogenesis, leading to excess adiposity, hepatic steatosis, and hypertriglyceridemia 36,49,50, whereas inhibition of the SREBP pathway had the opposite effects on hepatic fat accumulation and whole body energy metabolism 51,52. Several lines of evidence support the notion that Nrg4 directly acts on the liver and regulates hepatic lipogenesis. First, the extracellular EGF-like domain of Nrg4 exhibited strong binding to hepatocytes when fused to SEAP. Nrg4 binding in the liver was nearly completely abolished when excess GST-Nrg4Ex or CM containing the extracellular domains of ErbB3 and ErbB4 were included as competitors in the binding assays. As such, Nrg4 likely directly binds to ErbB3 and/or ErbB4 on hepatocyte cell surface. Using cultured hepatocytes with reconstituted ErbB4 expression, we found that Nrg4 was capable of triggering dose-dependent activation of endogenous ErbB3 and downstream signaling pathway, most notably transcription factor STAT5. ErbB3 does not have an active kinase domain but can heterodimerize with ErbB4 in response to ligand binding, resulting in receptor phosphorylation and the activation of downstream effectors. Given that hepatocytes express very low levels of ErbB2 37, and that EGFR does not bind to Nrg4 34, our data are consistent with ErbB3 and ErbB4 serving as major Nrg4 receptors in the liver. Finally, the activation of Nrg4 signaling in hepatocytes resulted in cell-autonomous inhibition of the SREBP1c/lipogenic pathway through transrepression of LXR by STAT5. While the liver appeared to be a major target tissue of Nrg4, it cannot be ruled out that Nrg4 actions in other tissues, including the central nervous system, may contribute to its regulation of systemic metabolism.
Nrg4 mRNA expression is enriched in, but not exclusively restricted to, brown fat. Similar to our work, a recent study identified Nrg4 as a transcript exhibiting strong enrichment in brown adipocytes 53. Given the abundance of white fat in the body, it is likely that Nrg4 released from both BAT and WAT sources may contribute to systemic metabolic regulation. Further, the paucity of brown and brown-like fats in humans raises the possibility that Nrg4 released from white fat may assume an even greater functional role in mediating the adipose-hepatic crosstalk. The levels of Nrg4 mRNA are significantly reduced in adipose tissues in murine and human obesity. Our studies demonstrate that Nrg4 undergoes proteolytic cleavage to release the biologically active extracellular ligand. However, due to a lack of robust immunological assays, important questions regarding Nrg4 levels in circulation and its nutritional regulation remain to be addressed. Despite this, our in vivo gain- and loss-of-function studies illustrate the significance of the Nrg4/ErbB signaling pathway in balancing hepatic lipogenesis in obesity. No specific and effective therapies are currently available to treat NAFLD. Future studies are needed to explore the potential of Nrg4 as a therapeutic biologic for treating obesity-associated disorders, such as NAFLD and type 2 diabetes.
ONLINE METHODS
Identification of brown fat-enriched secreted factors
The identification of gene cluster encoding putative brown fat-enriched secreted proteins was based on mouse secretome dataset available at Secreted Protein Database (spd.cbi.pku.edu.cn) 31, mouse tissue microarray dataset (GSE9954, GEO database), and brown adipocyte differentiation time course microarray dataset. Among 2,169 high-confident genes predicted to encode secreted proteins, 1,378 of which have annotated Affymetrix probe sets and were included in the analysis. Genes that were induced more than 2-fold in day 7 differentiated brown adipocytes compared to preadipocytes and enriched by more than 2-fold in brown fat were considered as putative brown-fat enriched secreted factors.
Animal studies
All animal studies were performed following the guideline established by the University Committee on Use and Care of Animals at the University of Michigan. Mice were housed under 12/12 hr light/dark cycles with free access to food and water. For chow feeding, mice were fed with Teklad 5001 laboratory diet. For high-fat diet (HFD) feeding, mice were fed with a diet containing 60% of calories from fat (D12492, Research Diets Inc.) starting at 10 to 12 weeks of age. Nrg4 knockout mice were purchased from the Mutant Mice Regional Resource Center (MMRRC) at the University of California, Davis, and backcrossed for more than ten generations to the C57BL/6J background.
The transgenic vector was constructed by placing mouse Nrg4 cDNA that contains part of 5′ and 3′ UTR between the aP2 enhancer/promoter sequence and human growth hormone polyadenylation signal. Mouse Nrg4 cDNA (including 5′UTR and partial 3′UTR) were TA-cloned into pCR2.1 TOPO vector. The transgenic vector was linearized by using HindIII and ApaI to release the transgenic cassette for microinjecting into fertilized eggs from C57BL/6J mice. Transgenic mice were generated at the Transgenic Animal Model Core at the University of Michigan. For acute cold exposure, mice were transferred to prechilled cages with no bedding and single-housed in a 4°C cold room. Mice has free access to food and water during cold exposure. Rectal temperature was measured every hour using Microtherma 2T probe (ThermoWorks).
Human studies
In a cross-sectional study, we investigated NRG4 mRNA expression in paired omental and subcutaneous (SC) adipose tissue samples (n=642). Individuals fulfilled the following inclusion criteria: 1) Absence of any acute or chronic inflammatory disease as determined by a leucocyte count > 7000 Gpt/l, C-reactive protein (CrP) > 10.0 mg/dl or clinical signs of infection, 2) Undetectable antibodies against glutamic acid decarboxylase (GAD), 3) No thyroid dysfunction, 4) No alcohol or drug abuse, 5) No pregnancy. All study protocols have been approved by the ethics committee of the University of Leipzig. All participants gave written informed consent before taking part in the study. All subjects had a stable weight, defined as the absence of fluctuations of >2% of body weight for at least 3 months before surgery. Adipose tissue was immediately frozen in liquid nitrogen after explantation. BMI was calculated as weight divided by squared height. Insulin sensitivity was assessed using the HOMA-IR index or with the euglycemic-hyperinsulinemic clamp method. Human NRG4 mRNA expression was measured by TaqMan-based qPCR assay. Human NRG4 mRNA expression was calculated relative to the mRNA expression of HPRT1, determined by a premixed assay developed for NRG4 and HPRT1 (Applied Biosystems).
Adipocyte differentiation and treatments
Brown preadipocyte isolation and differentiation were performed as previously described 54. SV40 T-large antigen immortalized brown preadipocytes were cultured in DMEM with 10% fetal bovine serum (FBS) for 2 days after reaching confluence (denoted as day 0 of differentiation). BAC differentiation was induced by adding a cocktail containing 0.5mM IBMX, 125μM Indomethacin, 1μM dexamethasone to maintenance media containing 10% FBS, 20nM insulin and 1nM T3. Three days after induction, cells are cultured in the maintenance media alone. Total RNA was isolated at different days during brown adipocyte differentiation for gene expression analysis.
3T3-L1 preadipocytes were cultured in DMEM with 10% bovine growth serum (BGS) until 2 days post confluent (count as d0). Differentiation was induced by adding a cocktail containing 0.5mM IBMX, 1μM dexamethasone and 1μg/mL insulin to DMEM supplemented with 10% FBS. Three days after induction, cells were cultured in DMEM containing 10% FBS plus 1μg/mL of insulin for another 2 days followed by maintenance in DMEM supplemented with 10% FBS. TNFα (10ng/mL) and IL1β (40ng/mL) treatment were carried out in mature adipocytes cultured in the maintenance media.
Adipose tissue fractionation
Epididymal WAT was dissected from lean and HFD-fed obese C57BL/6J mice. The tissues were minced into 2~3mm pieces, digested in Krebs Ringer Bicarbonate HEPES buffer (KRBH, containing 10 mM bicarbonate and 30 mM HEPES, pH 7.4) supplemented with 3% fatty acid free bovine albumin, 500nM adenosineand 3mg/mL collagenase II, and shaked gently at 37°C for 40min. Digested tissues were filtered through 200μm nylon filters into 50 ml conical tubes followed by centrifugation for at 800rpm for 30sec. The infranatant containing stromal vascular fraction was transferred into another tube using a needle syringe. The top layer containing mature adipocytes was washed again for a total of 3 times with KRBH buffer supplemented with 3% bovine albumin, and used for total RNA isolation. After the final removal of infranatant, the adipocyte fraction was lysed for RNAs. The combined infranatants were centrifuged at 4000rpm for 5min to obtain cell pellets containing stromal vascular fraction.
Min6 cell culture and treatments
Min6 cells stably expressing ErbB4 were a gift from Dr. Peter Dempsey (University of Michigan), and were cultured in DMEM supplemented with 15% FBS, 1.7g/500mL sodium bicarbonate, 2.5uL/500mL β-mercaptoethanol and 1% Pen/Strep. Before conditioned media treatment (for 15min), the cells were starved in serum-free DMEM for 4 hrs.
Transient transfection and Nrg4 binding assay
Hormone binding assay was performed as previously described 35,55. 293T cells were transfected with vectors expressing SEAP or SEAP-Nrg4Ex. 24 hours after transfection, cells were switched to serum-free media for additional 2 days before the media were collected and concentrated using Centricon. Briefly, frozen tissue slices were incubated with SEAP or SEAP-Nrg4Ex conditioned media for 45 min at room temperature before they were washed four times in 0.1% tween-20 containing PBS and fixed in a solution containing 20 mM HEPES (pH 7.4), 60% acetone, and 3% formaldehyde. After inactivating endogenous alkaline phosphatase at 65 °C for 30 min, the enzymatic activity derived from the fusion protein was detected using NBT/BCIP substrate. For competition binding, frozen tissue slices were pre-incubated for 30 min with 4μg/μL GST or GST-Nrg4Ex or 40 fold concentrated conditioned media from HEK293 cells expressing extracellular domain of ErbB3 or ErbB4, followed by one hour of SEAP-Nrg4Ex conditioned media co-incubation with those competitors.
To pinpoint the exact cleavage site of Nrg4, serial alanine mutations encompassing Nrg4’s predicted metalloprotease cleavage site were generated through site-directed mutagenesis PCR on top of the WT SEAP-Nrg4 expressing vector. HEK293 cells were transfected with equal amount of plasmids expressing either WT or mutant Nrg4 in the presence of polyethylenimine. The original 10% BGS DMEM were changed to 0.5% FBS DMEM 12 hrs after transfection. 48 more hours later, both media and total cell lysates were collected for western blotting against SEAP. For generating extracellular domain (ECD) of ErbB3 and ErbB4, we obtained ErbB3 and ErbB4 ECD expressing plasmids from Dr. Leahy 56. Conditioned media were collected as above and were concentrated 40-fold using Amicon ultra-centrifugal filter units from Millipore.
Hepatocyte isolation and treatment
Primary hepatocytes were isolated as previously described 57 by using collagenase type II from C57BL/6J mice. Hepatocytes were maintained in DMEM medium containing 10% BGS at 37°C and 5% CO2. Adenovirus infection was performed in the same day of isolation. After 24 hrs, cells were treated with GST and GST-Nrg4Ex (10 μg/mL) with vehicle (DMSO) or T0901317 (5 μM) for 24hrs. For signaling, cells were switched to DMEM supplemented with 0.1% BSA for 12hrs before GST and GST-Nrg4Ex treatment.
Luciferase reporter assay
Hepa 1 cells in 24-well plate were transiently transfected with 4xLXRE-Luc (30 ng/well) in the presence or absence of expression vectors for LXR (30 ng/well), RXR (30 ng/well), and constitutive active STAT5 (caSTAT5, 100 or 200 ng/well). Twenty four hrs after transfection, the cells were treated with vehicle (DMSO) or T0901317 (10 μM) for an additional 24 hrs before harvesting for luciferase assay. All the reporter assays were repeated at least three times in triplicates.
Gene expression analysis
Total RNA from white adipose tissue was extracted using a commercial kit from Invitrogen. RNAs from other tissues and cultured cells was extracted using TRIzol method. For quantitative real-time PCR (qPCR) analysis, equal amount of RNA was reverse-transcribed using MMLV-RT followed by quantitative PCR reactions using SYBR Green (Life Technologies). Relative abundance of mRNA was normalized to ribosomal protein 36B4. For detecting the coding isoform of Nrg4 using Taqman PCR, we designed sense primer encompassing the junction of exon3 and exon6 (5′ CCCAGCCCATTCTGTAGGTG3′), anti-sense primer in exon6 (5′ACCACGAAAGCTG-CCGACAG 3′), and a taqman probe in exon6 but between sense and anti-sense primers (5′ 6-FAM-CGGAGCACGCTGCGAAGAGGTT-BHQ 3′). Taqman PCR was carried out using the Taqman Universal PCR Master Mix system (Applied Biosystems). Relative abundance of the Nrg4 coding isoform was normalized to ribosomal protein 36B4.
For microarray study of liver gene expression, total liver RNA isolated from HFD-fed WT and Nrg4 KO mice was used to generate probes for Affymetrix Mouse MG-430 PM array strips. Sample processing and data analyses were performed according to the manufacturer’s instruction. The dataset has been deposited into the NCBI Gene Expression Omnibus (GEO) database with accession number GSE53877.
Immunoblotting analyses
For total lysates livers were homogenized in a lysis buffer containing 50 mM Tris (pH 7.5), 150mM NaCl, 5mM NaF, 25mM β-glycerolphosphate, 1mM sodium orthovanadate, 10% glycerol, 1% tritonX-100, 1 mM dithiothreitol (DTT), and freshly added protease inhibitors. For liver nuclear extracts, frozen livers were homogenized using a Dounce homogenizer in ice-cold homogenization buffer containing 0.6% NP40, 150mM NaCl, 10mM HEPES (pH=7.9), 1mM EDTA, and protease inhibitor cocktail. The homogenates were briefly centrifuged at 450 rpm at 4°C to remove tissue debris. The suspension was transferred to a new tube and centrifuged at 3,000 rpm for 5 min at 4°C. The nuclei pellet was washed with homogenization buffer and resuspended in a low-salt buffer containing 20mM Tris (pH=7.5), 25% glycerol, 1.5mM MgCl2, 200μM EDTA, 20mM KCl, and protease inhibitors. Nuclear proteins were extracted following the addition of a high-salt buffer (½ volume) containing 20mM Tris (pH=7.5), 1.5mM MgCl2, 200μM EDTA, 1.2M KCl, and protease inhibitors at 4°C for 2hrs. Immunoblotting experiments were performed using specific antibodies against SREBP1 (sc-13551), tubulin (T6199), ErbB3 (sc-285). The following antibodies were from cell signaling technology: Lamin A/C (#2032), phospho-ErbB4 (Y1284) (#4757), ErbB4 (#4795), phospho-ErbB3 (Y1289) (#4791), phospho-ErbB2 (Y1221/1222) (#2243), phospho-STAT5 (Y694) (#9359) and total STAT5, phospho-AKT (S473)(#4060) and total AKT (#4691), phosphor-AMPK (Thr172) (#2535) and total AMPK (#2532).
Metabolic measurements and liver lipid analysis
Plasma concentrations of triglycerides (Sigma) and non-esterified fatty acid (Wako Diagnostics) were measured using commercial assay kits. Liver triglyceride was extracted and measured as previously described 58. Body fat and lean mass were measured using an NMR analyzer (Minispec LF90II, Bruker Optics). For liver lipid analysis, lipids were extracted by the method of Bligh-Dyer in the presence of an internal standard (T21:0 TAG, 10 nmol/mg protein) and separated on silica gel 60-Å plates that were developed with a nonpolar acidic mobile phase (70:30:1, v/v/v, hexane/ethyl ether/acetic acid). Briefly, spots corresponding to TAG were visualized with 0.01% rhodamine 6G and identified with TAG standard. The bands were scraped, extracted, and treated with acidic methanol. Quantitative GC analysis of resulting FA methyl esters was conducted (Hewlett-Packard 5890 GC; Hewlett-Packard, Palo Alto, CA, USA) with a 30-m×0.32 mm Omegawax 250 column (Sigma) coupled with a flame ionization detector.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Dempsey for providing ErbB4-Min6 cells, Y. Yarden for providing ErbB expression plasmids, D. Leahy for expression constructs for ErbB3 and ErbB4 extracellular domains, and A. Saltiel for discussions. We are grateful to Q. Yu and L. Wang for technical support, the lab members for discussion, and the staff at the University of Michigan Transgenic Animal Model Core and the metabolic phenotyping core supported by Michigan Diabetes Research and Training Center (DK020572) and Nutrition Obesity Research Center (DK089503). This work was supported by National Institutes of Health (DK077086 and DK095151, J.D.L.; DK097608, X.S.) and a grant from Novo Nordisk. S.L. and G.X.W. were supported by Scientist Development Grant and Predoctoral Fellowship, respectively, from the American Heart Association.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS J.D.L., G.X.W., and X.Y.Z. conceived the project and designed research. G.X.W., X.Y.Z., Z.C., S.L., and Z.X.M. performed metabolic and molecular studies; M.K., A.D., and M.B. performed human studies. D.Z., A.L.O., and X.S. performed lipid profile analysis. G.X.W. and J.D.L. wrote the manuscript.
COMPETING FINANCIAL INTERESTS This work was partially supported by a research agreement with Novo Nordisk.
REFERENCES
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Kozak LP, Harper ME. Mitochondrial uncoupling proteins in energy expenditure. Annual review of nutrition. 2000;20:339–363. doi: 10.1146/annurev.nutr.20.1.339. [DOI] [PubMed] [Google Scholar]
- 3.Lowell BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 4.Yoneshiro T, et al. Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of clinical investigation. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartelt A, et al. Brown adipose tissue activity controls triglyceride clearance. Nature medicine. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 6.van der Lans AA, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of clinical investigation. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American journal of physiology. Endocrinology and metabolism. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 8.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 10.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 11.Enerback S. Human brown adipose tissue. Cell metabolism. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. The Journal of biological chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature medicine. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell metabolism. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enerback S, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 19.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, et al. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. The Journal of clinical investigation. 2003;111:399–407. doi: 10.1172/JCI15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadowaki T, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. The Journal of clinical investigation. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocrine reviews. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 23.Waki H, Tontonoz P. Endocrine functions of adipose tissue. Annual review of pathology. 2007;2:31–56. doi: 10.1146/annurev.pathol.2.010506.091859. [DOI] [PubMed] [Google Scholar]
- 24.Angelin B, Larsson TE, Rudling M. Circulating fibroblast growth factors as metabolic regulators--a critical appraisal. Cell metabolism. 2012;16:693–705. doi: 10.1016/j.cmet.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes & development. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nature reviews. Endocrinology. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 27.Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481:314–320. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. Journal of cellular physiology. 2009;218:460–466. doi: 10.1002/jcp.21635. [DOI] [PubMed] [Google Scholar]
- 29.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annual review of pharmacology and toxicology. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 30.Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Current opinion in cell biology. 2007;19:124–134. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, et al. SPD--a web-based secreted protein database. Nucleic acids research. 2005;33:D169–173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odiete O, Hill MF, Sawyer DB. Neuregulin in cardiovascular development and disease. Circulation research. 2012;111:1376–1385. doi: 10.1161/CIRCRESAHA.112.267286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes NV, Newsam RJ, Baines AJ, Gullick WJ. Characterization of the cell membrane-associated products of the Neuregulin 4 gene. Oncogene. 2008;27:715–720. doi: 10.1038/sj.onc.1210689. [DOI] [PubMed] [Google Scholar]
- 34.Harari D, et al. Neuregulin-4: a novel growth factor that acts through the ErbB-4 receptor tyrosine kinase. Oncogene. 1999;18:2681–2689. doi: 10.1038/sj.onc.1202631. [DOI] [PubMed] [Google Scholar]
- 35.Muller H, Dai G, Soares MJ. Placental lactogen-I (PL-I) target tissues identified with an alkaline phosphatase-PL-I fusion protein. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1998;46:737–743. doi: 10.1177/002215549804600606. [DOI] [PubMed] [Google Scholar]
- 36.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of clinical investigation. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carver RS, Stevenson MC, Scheving LA, Russell WE. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. 2002;123:2017–2027. doi: 10.1053/gast.2002.37060. [DOI] [PubMed] [Google Scholar]
- 38.Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE. ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. The Journal of biological chemistry. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 39.Jones FE, Welte T, Fu XY, Stern DF. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. The Journal of cell biology. 1999;147:77–88. doi: 10.1083/jcb.147.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes & development. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz JR, et al. Role of LXRs in control of lipogenesis. Genes & development. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 43.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annual review of immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 44.Odegaard JI, Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339:172–177. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nature medicine. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 46.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 47.Kohjima M, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. International journal of molecular medicine. 2008;21:507–511. [PubMed] [Google Scholar]
- 48.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. The Journal of biological chemistry. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 49.Knebel B, et al. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PloS one. 2012;7:e31812. doi: 10.1371/journal.pone.0031812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimano H, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. The Journal of clinical investigation. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang JJ, et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell metabolism. 2011;13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Moon YA, et al. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell metabolism. 2012;15:240–246. doi: 10.1016/j.cmet.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosell M, et al. Brown and White Adipose Tissues. Intrinsic differences in gene expression and response to cold exposure in mice. American journal of physiology. Endocrinology and metabolism. 2014 doi: 10.1152/ajpendo.00473.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein J, Fasshauer M, Klein HH, Benito M, Kahn CR. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- 55.Lin J, Linzer DI. Induction of megakaryocyte differentiation by a novel pregnancy-specific hormone. The Journal of biological chemistry. 1999;274:21485–21489. doi: 10.1074/jbc.274.30.21485. [DOI] [PubMed] [Google Scholar]
- 56.Leahy DJ, Dann CE, 3rd, Longo P, Perman B, Ramyar KX. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr Purif. 2000;20:500–506. doi: 10.1006/prep.2000.1331. [DOI] [PubMed] [Google Scholar]
- 57.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Li S, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell metabolism. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.