Abstract
The living cells are arranged in a complex natural environment wherein they interact with extracellular matrix and other neighboring cells. Cell-cell interactions, especially those between distinct phenotypes, have attracted particular interest due to the significant physiological relevance they can reveal for both fundamental and applied biomedical research. To study cell-cell interactions, it is necessary to develop co-culture systems, where different cell types can be cultured within the same confined space. Although the current advancement in lab-on-a-chip technology has allowed the creation of in vitro models to mimic the complexity of in vivo environment, it is still rather challenging to create such co-culture systems for easy control of different colonies of cells. In this paper, we have demonstrated a straightforward method for the development of an on-chip co-culture system. It involves a series of steps to selectively change the surface property for discriminative cell seeding and to induce cellular interaction in a co-culture region. Bone marrow stromal cells (HS5) and a liver tumor cell line (HuH7) have been used to demonstrate this co-culture model. The cell migration and cellular interaction have been analyzed using microscopy and biochemical assays. This co-culture system could be used as a disease model to obtain biological insight of pathological progression, as well as a tool to evaluate the efficacy of different drugs for pharmaceutical studies.
I. INTRODUCTION
Cell culture in an ex-vivo environment has been in practice for decades and has long been used to develop in vitro models for studying various mechanisms underlying the cellular physiology. With the advancement in modern biotechnologies, it has become possible to improve the complexity of these models and thus making it more physiologically relevant. In order to achieve better insight into cell functioning, the in vitro models need to be able to mimic natural phenomena that are occurring in vivo. Cell interaction is an important phenomenon, which is vital for physiological functioning at tissue levels1,2 and also playing key roles in cancer biology3,4 and wound healing.5 These physiological relevances have led to the development of several innovative and effective assays, such as wound healing assays and migration assays.6 Cell interaction may occur among cells of the same phenotype, such as between endothelial cells during wound healing.7 It may also happen between cells of different phenotypes, such as between endothelial cells and tumor cells during tumor-induced angiogenesis.8,9 It is therefore desirable to develop co-culture systems, where different types of cells grow on the same substrate and the cellular interactions can be induced and monitored in real time. For this purpose, cell patterning is necessary to control their spatial distribution. In many prior studies, cell patterning was achieved by active methods using forces or fields, such as standing surface acoustic waves,10 magnetic field,11 optical force,12 dielectrophoresis,13 or optoelectronic methods.14 The other important or probably more natural strategy of cell patterning was by passive methods utilizing the biochemical or biophysical affinity between cells and functionalized microarrays or micropatterns, which were created by various microfabrication techniques, such as photolithography, soft lithography, microcontact printing,15 inkjet printing,16 and dip-pen lithography.17
In the most recent decade, microfluidics has proved to be an effective tool for many cell-based studies.18 Compared to conventional methods, the miniature size of microfluidic devices helps to create a microenvironment that better mimics the natural cellular environment.19 In addition, the ability to precisely control flows20 and generation of biochemical gradient21,22 also facilitates to adjust and maintain favourable conditions for cell growth and introduce biochemical cues to influence the cell fate.23 Researchers have taken advantage of these microfluidic platforms and successfully developed in vitro models for studying various important physiological phenomena, such as wound healing,24,25 cancer metastasis,26,27 and vascular angiogenesis.28,29
Among these innovative microfluidic devices, some are dedicated for co-culture system, which is a rather challenging task compared to handling a single type of cells on a chip. Prior reports have shown various designs, including valves,30 laminar flow between immiscible liquids,31,32 separate compartments for dynamic co-culture,33 and detachable substrates, where the cells are individually cultured on different substrates.34 Some researchers have also used scaffold materials such as collagen to create barriers between two cell types in a 3-dimensional co-culture system.35 Many microfluidic co-culture systems are targeted to specific applications, such as macrophage-osteoblast interaction36 or bacterial cancer targeting.37 It is very common that an actuating component, such as a syringe pump, is needed to control the barriers between distinct compartments for different cell types, which, however, makes the overall system bulky and the operation complicated. Moreover, if two different substrates, such as polydimethylsiloxane (PDMS) and polystyrene, are used together for cell culture, the cell behaviour may vary due to different substrate properties. Therefore, it is necessary to develop a model that is easy to manipulate, compact in size, and has uniform and controllable material property.
In this work, we developed a simple method to create a planar co-culture system using a PDMS chip without any actuating component. Scotch tape was used for selective surface treatment to create distinct hydrophilic and hydrophobic PDMS compartments, and hence fluid flow could be confined locally. Two types of cell models (HS5 and HuH7) were cultured in distinct hydrophilic compartments, while the hydrophobic compartment serves as a barrier in between. Once both cell types reached confluence, the barrier was removed easily by introducing liquid into it thereby creating a co-culture region for cell migration and interaction. These two particular cell types were chosen for a proof-of-concept study on tumor-stromal interactions, which could be of great interest and significance in cancer metastasis and subsequent survival of metastasized tumor cells.38 The substrate hydrophobicity was characterized and various biochemical assays, such as micro bicinchoninic acid (BCA) assay for adsorbed protein quantification, Prestoblue cell viability assay, and Cyquant cell proliferation assay for cell adhesion quantification, were carried out to evaluate the effect of this co-culture system.
II. EXPERIMENTAL METHODS
A. Chip fabrication
The microfluidic chip was fabricated by soft lithography,39 including UV lithography, replica molding, and embossing. Negative photoresist SU-8 2050 (MicroChem, MA, USA) was spin-coated onto a silicon wafer followed by soft baking at 95 °C for 45 min, exposed with UV irradiation (365 nm) through a photomask, baked at 95 °C for 15 min, and finally developed using SU-8 developer solution (MicroChem, MA, USA). The thickness of the SU-8 mold was approximately 150 μm. PDMS (Sylgard 184, Dow Corning, MI, USA) was cast on the SU-8 mold, degassed, and cured for 2 h at 70 °C for complete crosslinking. The chip design (Figure 1) comprises three compartments separated from each other by arrays of rectangular pillars (cross-section 200 μm × 100 μm), which extends over 1 cm along the microchannels. The width of the side channels is 800 μm, and the width of the central channel is 500 μm. A PDMS-coated cover slip was used as chip substrate. Both the PDMS chip and the substrate were treated with a plasma-cleaner (Harrick Plasma, NY, USA) at input power of 100 W and RF of 8–12 MHz and bonded together.
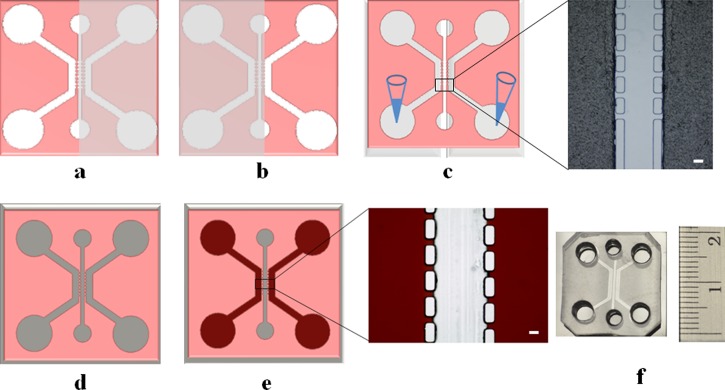
FIG. 1.
Microfluidic chip preparation. (a) Step 1: Scotch tape is used as a stencil to cover the central channel and a side channel on the right; only the side channel on the left is plasma treated. (b) Step 2: Scotch tape covers the central channel and the side channel on the left; only the side channel on the right is plasma treated. (c) Step 3: temporary channels are formed by covering both side channels with Scotch tape. Inset shows a microscopic image of the tape covering the plasma-treated side channels. Gelatin is introduced to coat both side channels to enhance surface wettability. Scale bar: 100 μm. (d) Step 4: a completely sealed chip is assembled by plasma treating and bonding: the surface-treated (in steps 1–3) PDMS replica and a PDMS-coated glass cover slip. (e) Step 5: the entire chip is dried in the air and the central channel recovers its hydrophobicity after 24 h, while the side channels remain hydrophilic because of gelatin coating. Inset shows a microscopic image of side channels filled with red dye solution, while the liquid does not overflow into the central channel owing to surface tension [Figure S1 in the supplementary material (multimedia view)52]. Scale bar: 100 μm. (f) The final microfluidic co-culture chip ready for use.
B. Selective plasma treatment
Although PDMS is a convenient and cheap material for microfabrication, the native PDMS surface shows poor affinity as a substrate for living cell culture due to its strong hydrophobicity.40 It is important to modify the PDMS surface property to allow for liquid perfusion as well as to enhance cell adhesion.41 There are many ways of surface treatment on PDMS,42,43 among which oxygen or air plasma is the easiest and most commonly used method.39 When the PDMS substrate is exposed to plasma, silanol bonds are formed on its surface for improving the wettability. In the literature, much effort have been attempted to control the localized plasma treatment, such as using water mask or microplasma generation using high-precision electrode and AC excitation. However, these methods involve nontrivial procedures and expensive materials. An economic solution was proposed44 using a PDMS slab as a stencil to selectively pattern the protein-repelling polymeric network by plasma etching, allowing for modifying surface chemistry to enhance cell attachment. In addition, if a hydrophilic microchannel is peripherally sealed, it can be easily primed with liquid driven by capillary force. However, if the channel is not sealed peripherally, such as in an open burrow, the liquid can hardly fill the channel owing to the surface tension. These unique phenomena inspired us to form temporary or fake channels to selectively change the local surface property of PDMS channel.
For this purpose, we applied a very simple way with Scotch tape (3M, MN, USA), rather than a PDMS slab, as a stencil to mask specific locations on the chip to prevent them from plasma treatment and to create a temporary channel for biochemical functionalization through liquid perfusion. The detailed procedure is illustrated in Figure 1. Briefly, both side channels were coated with 0.1% gelatin (Type B from bovine skin, Sigma-Aldrich, Singapore) to enhance and maintain the wettability after 24 h (Białopiotrowicz and Jańczuk, 2001), while the central channel recovered to be a hydrophobic barrier to isolate two side channels. After the chip was sealed with a substrate by plasma treating, the first layer of fibronectin coating was performed to coat all three channels. After that, the chip was allowed to dry at 50 °C for 24 h to allow hydrophobic recovery. Then, the chip was UV sterilized and a second layer of fibronectin was coated in two side channels, which were hydrophilic owing to gelatin coating. During the second coating, the fibronectin solution was confined within the side channels and could not overflow into the central channel because of surface tension. In this way, the side channels had a double coating of fibronectin, while the central channel was with single coating only. The chip was treated at 50 °C during hydrophobic recovery to minimize protein denaturing under high temperature and thus allows cell migration in subsequent studies.
An optical tensiometer (Attension Theta, Sweden) was used to measure the contact angle of water droplet on PDMS substrates, which gives quantitative characterization of the surface wettability under various conditions.
C. Cell culture
The human bone marrow stromal cell line, HS5 (ATCC, USA), and the human hepatocarcinoma cell line, HuH7, were used as cell models for the co-culture system. Both cell types were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 5% penicillin-streptomycin. Cells were cultured in a T-25 flask till they achieve 80% confluence, following which they were resuspended using 0.25% EDTA-trypsin to density of 106 ml−1. Approximately, 50–60 μl of cell suspension (106 ml−1) was introduced into the side channels of the chip. The cell medium inside the microfluidic chip was changed every 24 h. The flow rate during medium change was maintained at a very low level to prevent the cells from being damaged by the flow shear stress. While for single cell type culture in a 4-well culture plates to characterize the biocompatibility of protein-coated PDMS, the culture medium was changed every 2 days.
D. Protein retention assay
Fibronectin was coated on PDMS to enhance surface biocompatibility. The amount of fibronectin retained on surface was quantified using Pierce BCA protein assay kit (Thermo Scientific, Singapore). Micro BCA assay is a colorimetric assay whereby the peptide bonds in the protein react and reduce the Cu2+ ions to Cu+ ions that can be detected. According to the intensity of the color change, the protein concentration can be determined based on a standard curve. PDMS was casted into a 4-well plate with each well having an area of 1 cm2. The PDMS samples were incubated with 20 μg ml−1 fibronectin (Life technologies, Singapore) at 37 °C for 1 h, followed by washing with 1× phosphate-buffered saline (PBS). The assay was performed based on the protocols supplied by the manufacturer, following which the absorbance at 562 nm was measured using a microplate reader (Infinite M200 Pro, Tecan Asia, Singapore). The results were presented as the protein concentration retained on the PDMS surface with respect to original protein concentration in the solution.
E. Cell adhesion
Cyquant cell proliferation assay (Life Technologies, Singapore) was used to characterize the cell adhesion on the chemically modified PDMS surface in a 4-well plate. Approximately, 104 HS5 cells were seeded into each well and incubated for 90 min followed by washing with 1× PBS, after which the PBS was aspirated and the cells were freezed at −80 °C for 90 min before incubating with the CyQUANT®GR dye prepared as per the manufacturer's instructions. The fluorescence was measured at the excitation of 485 nm and emission of 535 nm using a microplate reader (Infinite M200 series, Tecan Asia, Singapore).
F. Cell viability
The cell viability was quantified with the Prestoblue cell viability reagent (Life Technologies, Singapore). Approximately, 104 HS5 cells were cultured on individual groups of PDMS for at least 12 h. The cells were washed with 1× PBS and then incubated in 10% Prestoblue reagent supplemented in DMEM for 1 h. The absorbance was measured at 570 nm and 600 nm using a microplate reader (Infinite M200 pro, Tecan Asia, Singapore) from which the cell viability can be calculated. This analysis was performed at different time points for up to 5 days.
G. Fluorescence imaging
The cells were fixed using formalin (Sigma-Aldrich, Singapore), stained with TRITC conjugated phalloidin (Life technologies, Singapore) for cytoskeleton and DAPI (Life technologies, Singapore) for nuclei. Cell imaging was under a fluorescence microscope (IX71, Olympus, Singapore).
H. Characterization of reactive oxygen species
The intracellular reactive oxygen species (ROS) were measured using CM-H2DCFDA assay (Life Technologies, Singapore). HS5, HuH7, and a mixture of HS5 and HuH7 cells were seeded into a 96 well plate. 2 μM of CM-H2DCFDA in 1× PBS was incubated with the cells after 24 h for 20 min prior to fluorescence imaging. The fluorescence intensity was calculated using ImageJ software (National Institutes of Health, USA). The assays were performed for up to 5 days to characterize the change of ROS expression levels.
I. Live-dead cell imaging
Live-dead cell staining was performed to demonstrate the cell viability and distribution in the microfluidic chip. Fluorescein diacetate (FDA) (Sigma-Aldrich, Singapore) and ethidium homodimer (EthD-1) (Life Technologies, Singapore) were used to stain the live and the dead cells, respectively, prior to fluorescence imaging.
III. RESULTS AND DISCUSSION
Native PDMS is hydrophobic and thus is not amicable for cell culture. Plasma treatment and protein coating can reduce the hydrophobicity and promote cell growth. In this study, development of a co-culture system primarily depends on the ability of PDMS to recover its hydrophobicity so that different cells can be confined in different compartments on the same chip. We noticed that hydrophobic recovery of PDMS was dependent on both temperature and time. Generally, the hydrophobic recovery becomes faster when temperature increases. However, temperature elevation has an adverse effect on protein retention on PDMS surface. Furthermore, over-recovered hydrophobicity would hinder the fluid flow in microchannels to open the co-culture region. To address these issues, we applied selective plasma treatment, gelatin coating, and double coating of fibronectin to delicately modify the surface wettability and enhance the cell growth.
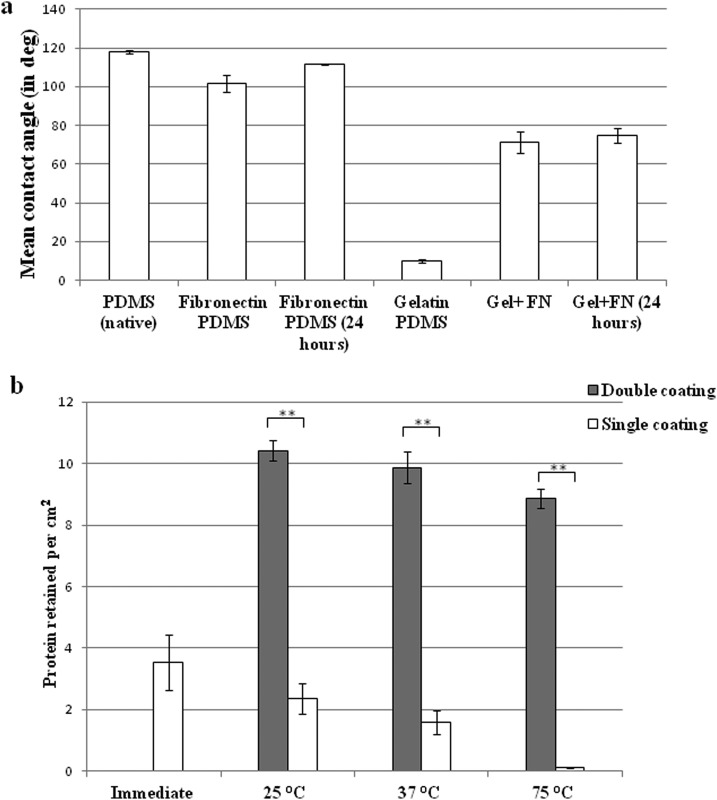
The surface wettability was measured for native and plasma-treated PDMS coated with fibronectin, gelatin, and gelatin + fibronectin combination, respectively. In Figure 2(a), native PDMS showed very large contact angle (∼120°) indicating its poor wettability. For other samples, the contact angles were measured 24 h after surface treatment. The results indicated that although fibronectin reduced the contact angle, the PDMS surface was still in the hydrophobic range (>90°); whereas for gelatin and gelatin + fibronectin coated PDMS samples, the contact angles are much smaller showing excellent wettability. In addition, the gelatin coating helped to maintain the substrate wettability for at least 24 h, by which plasma-treated PDMS would have recovered its hydrophobicity. These results imply that by controlling the area of gelatin coating, distinct hydrophilic and hydrophobic regions could be created on a single substrate.
FIG. 2.
(a) Contact angle measurement of different surface-treated PDMS taken immediately and after 24 h. Statistical analysis was performed using one way ANOVA. p <0.001. (b) Micro-BCA analysis of protein retention immediately after fibronectin coating and after hydrophobic recovery (24 h) under different temperatures. **p <0.005.
The PDMS was coated with fibronectin for 1 h and the protein retained on surface was quantified by micro BCA assay. The absorbed fibronectin immediately after surface coating was approximately 3 μg cm−2 (Figure 2(b)). As discussed earlier, in order to create a hydrophobic barrier between different compartments for initial cell culture, the fibronectin-coated PDMS needs to be subjected to elevated temperature much higher than room temperature for rapid hydrophobic recovery. Therefore, we studied the temperature effect on the protein retention. The fibronectin-coated PDMS samples were subjected to 25 °C, 37 °C, and 75 °C for 24 h, respectively, followed by micro BCA assays. It was found that the protein retention decreased drastically in 24 h to almost zero when the temperature rose to 75 °C (Figure 2(b)). The significant decay of fibronectin adsorption could be due to the protein denaturing under high temperature. To address this issue, we performed a second coating of fibronectin (20 μg ml−1) after 24 h. It could be observed that double coating of fibronectin resulted in an increase of protein retention by at least 3 folds to about 10 μg cm−2 irrespective of the temperature change (Figure 2(b)). This process also rendered a very smooth surface as shown by atomic force microscopy (Figure S2 in the supplementary material52).
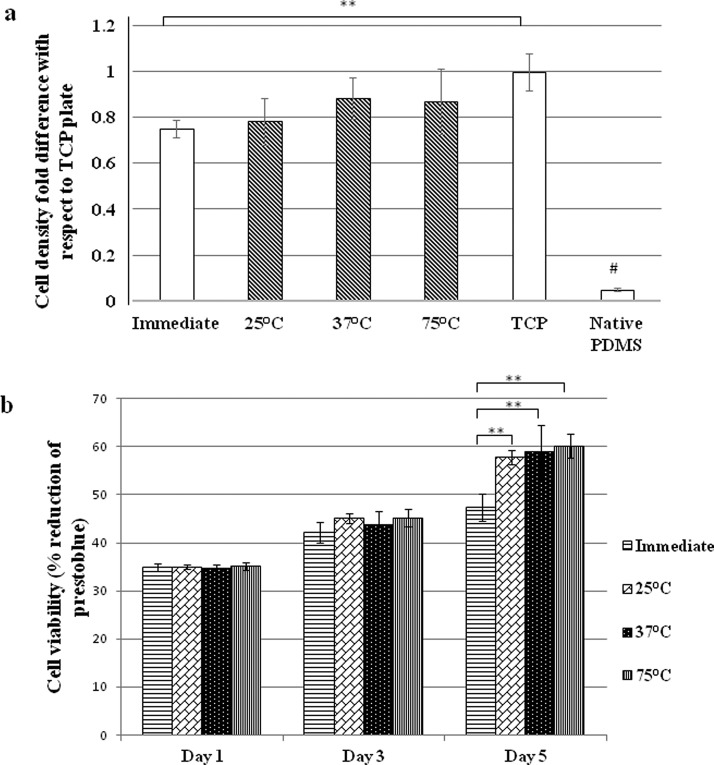
The impact of double coating of fibronectin on cell adhesion and viability was studied using HS5 stromal cells. Cell adhesion on different samples was measured with respect to the control on a tissue culture plate (TCP) (Figure 3(a)). The native PDMS without protein coating exhibited very poor biocompatibility with minimal cell adhesion. Whereas the protein-coated PDMS could support decent level of cell adhesion compared with TCP. The Prestoblue cell viability assay at day 1, day 3, and day 5 further verified the promotional effect of double protein coating on long-term cell culture. It was observed that the cell viability at day 5 was improved significantly on all PDMS surfaces with double fibronectin coating compared to those with single fibronectin coating (Figure 3(b)). These results indicated that double coating of fibronectin on PDMS facilitated both cell adhesion and proliferation in a long term.
FIG. 3.
Characterization of the surface biocompatibility after fibronectin coating using HS5 stromal cells: (a) Cell adhesion. The shaded bars represent the surfaces with double fibronectin coating. (b) Cell viability. “Immediate” refers to the samples on which cells were seeded soon after fibronectin coating. A second coating of fibronectin was performed in all other samples (before cell seeding) after incubation for 24 h under respective temperatures. (**p <0.005; #p <0.001)
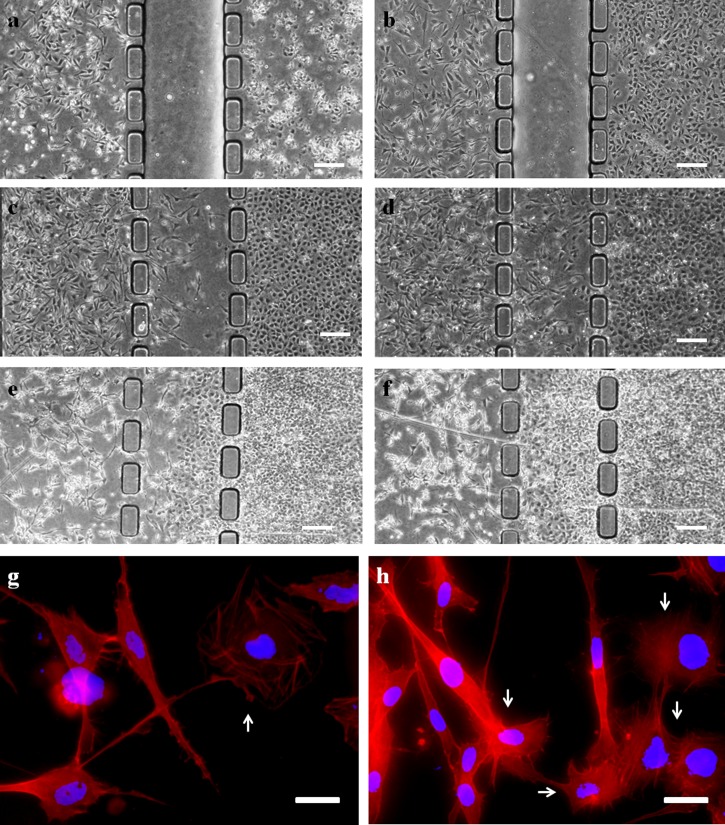
As a demonstration of co-culture on this microfluidic device, HS5 and HuH7 cells were cultured in distinct micro-compartments for 24 h. The cell culture medium was introduced into the central channel by applying a gentle pressure using a micropipette. This procedure removed the hydrophobic barrier and created a co-culture region for cell interaction. The cells were cultured on the chip for up to 9 days. It was observed that highly mobile HS5 cells migrated towards HuH7 cells initially (Figures 4(a)–4(d)). However, when the tumor cells reached confluence, they started to migrate in batches toward stromal cells (Figures 4(e) and 4(f)). After physical interaction between these two cell lines, tumor cells became more aggressive and proliferative resulting in significant death and density drop of stromal cells after day 6. A close-up view using fluorescence microscopy (Figures 4(g) and 4(h)) further revealed that tumor cells generated membrane protrusions toward stromal cells during physical contact. Similar phenomenon was reported previously in a conventional co-culture system, in which the physical contact through membrane protrusion with stromal cells stimulated the papillary thyroid tumor cells and transformed moderately carcinogenic cells to highly aggressive metastatic cells.45 These membrane protrusions are also called tunneling nanotubes (TNTs), which could serve as an intercellular channel for tumor cells to obtain the cytoplasm including mitochondria or other cellular organelles from other tissue cells and hence upregulate the tumor proliferation.45,46
FIG. 4.
(a)–(f) Phase contrast images of co-culture chip taken at 2 h, and 1, 3, 5, 7, and 9 days after cell seeding. HS5 cells in the left compartment and HuH7 cells in the right. The barrier (empty channel in (a) and (b)) between the two compartments was removed after 24 h. By the 9th day, HuH7 cells had occupied the central channel and HS5 cells had retreated into its own compartment and many dead stromal cells were observed. (Scale bar: 200 μm.) (g) and (h) Fluorescence images showed the interaction between HS5 stromal cells and HuH7 cells. The red color strains the actin filaments and the blue color stains the nuclei. The arrows indicate the tumor cells. (Scale bar: 20 μm.)
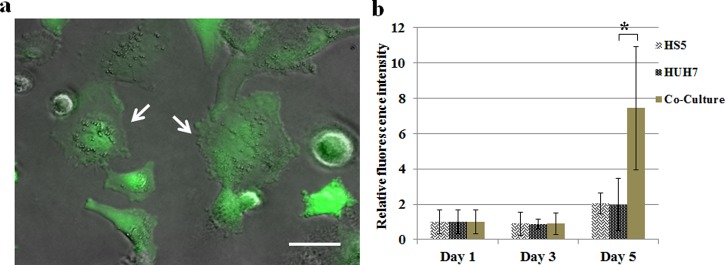
It has also been reported that direct contact of tumor cells with endothelial cells results in generation of ROS that causes oxidation of cell membrane and DNA breakdown of endothelial cells.34,47 ROS is a class of natural byproduct of normal metabolism of oxygen and generated during mitochondrial respiration, and they play important roles in cell signaling and homeostasis. Normally, ROS is quenched by intracellular anti-oxidants. Imbalance of this mechanism leading to dramatic increase in ROS concentration will cause oxidative damage and cell death.48 However, tumor cells usually express higher intrinsic ROS levels than normal cells due to oncogenic stimulation, which implies that the same ROS level to facilitate cancer cell survival could be detrimental to normal cells. In order to study the possible effect of ROS on this HS5-HuH7 co-culture system, we quantified the ROS expression level using CM-H2DCFDA assay. Monocultures of HS5 and HuH7 and their co-culture were seeded in a 96-well plate, respectively, and the ROS assays were performed on day 1, day 3, and day 5 after the initial seeding. Figure 5(a) shows the fluorescence image of the cells indicating the intracellular ROS. Figure 5(b) indicates the fluorescence intensity measured in different cultures showing the dynamic change of the average intracellular ROS levels. It was observed that the ROS level in all cultures increased by day 5, while the ROS elevation in the co-culture system was almost 4 folds greater than that in either monoculture system. These results implied that co-culture of HS5 and HuH7 upregulated ROS level, which could contribute to the ROS-induced stromal cell death in the microfluidic co-culture system.
FIG. 5.
(a) The fluorescence image overlaid with a phase contrast image of HS5-HuH7 co-culture stained with CM-H2DCFDA for ROS expression. Arrows indicate the HuH7 cells. Scale bar: 20 μm. (b) The dynamic change of ROS expression over 5 days in the monoculture and co-culture systems. The fluorescence intensity was normalized with respect to those on day 1, respectively.
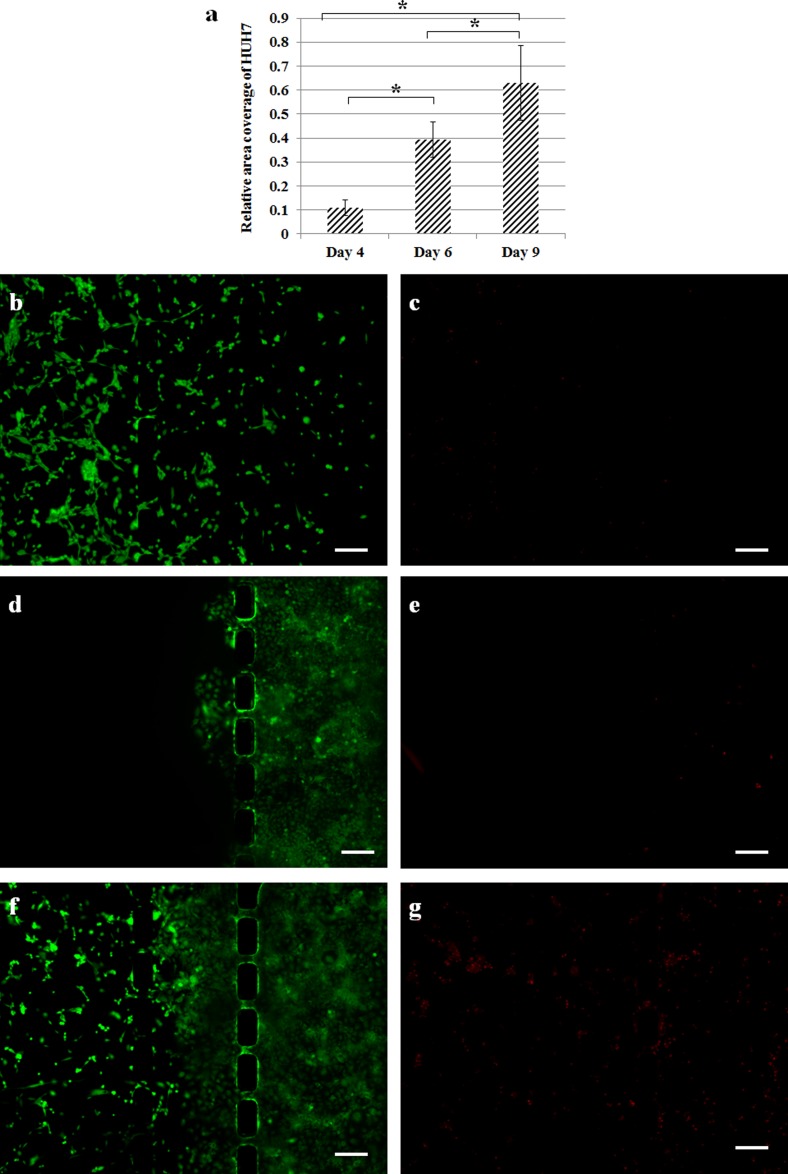
There is reported evidence indicating that increased ROS concentration is able to assist in tumor invasion and progression.49,50 We measured the change of total area covered by HuH7 using phase contrast images of the co-culture chip, which was processed by ImageJ software (detailed procedure illustrated in Figure S3, supplementary material).52 It was observed that the coverage of tumor cells increased significantly after day 4 (Figure 6(a)). Live cell staining using FDA (green) showed that the density and covered area of HS5 both decreased in the co-culture (Figure 6(f)) as compared to the monoculture control (Figure 6(b)). Whereas dead cell staining using EthD-1 (red) revealed an increased death amongst the HS5 cells in the co-culture (Figure 6(g)). These results suggested tumor-induced apoptosis of stromal cells, which could be caused by paracrine signaling of ROS released by tumor cells to control the homeostasis of local microenvironment.34,47,51 On the other hand, the tumor cell migration appeared to be promoted in the co-culture (Figure 6(f)) compared to monoculture (Figure 6(d)).
FIG. 6.
(a) Area covered by HuH 7 cells with respect to the total area of central channel. Live ((b), (d), and (f) in green) and dead ((c), (e), and (g) in red) cell staining of HS5 monoculture, HuH7 monoculture, and HS5-HuH7 co-culture, respectively. All images were taken at day 9. Scale bar: 200 μm.
IV. CONCLUSION
A simple method was applied in this study to create a microfluidic co-culture model for in vitro cell study. Scotch tape was utilized as a stencil for selective plasma treatment on a chip followed by surface coating with gelatin to render distinct hydrophobic and hydrophilic microchannels on the same chip. The fibronectin coating enhanced cell viability on PDMS surface. As a proof-of-concept study, stromal cells (HS5) and tumor cells (HuH7) were co-cultured on this microfluidic chip, and cell migration and interaction were observed. This chip is able to address some key requirements for an effective co-culture system, such as the ability to load cells into different compartments so that they can grow till they reach confluence, the ability to manipulate one cell type in the co-culture model without affecting the other cell type (Figure S4, supplementary material52), and the ability for convenient cell visualization. This chip could be used to develop disease models involving multiple cell types for a synergistic study, or it may be implemented for investigations on drug specificity towards diseased cells in a co-culture environment with healthy cells.
ACKNOWLEDGMENTS
This work was supported by a Tier 2 Academic Research Fund from Singapore Ministry of Education (ARC 22/13) awarded to Y.K. The Ph.D. scholarship from Nanyang Technological University awarded to N.V.M. is gratefully acknowledged. N.V.M. thanks Xue Cao for assistance in cell culture measurement and Chun Kiat Ng for assistance in microscopy.
References
- 1.Clark B. R. and Keating A., Ann. N. Y. Acad. Sci. 770(1), 70–78 (1995). 10.1111/j.1749-6632.1995.tb31044.x [DOI] [PubMed] [Google Scholar]
- 2.Bhatia S. N., Balis U. J., Yarmush M. L., and Toner M., FASEB J. 13(14), 1883–1900 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Chaffer C. L. and Weinberg R. A., Science 331(6024), 1559–1564 (2011). 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 4.Ingber D. E., Differentiation 70(9–10), 547–560 (2002). 10.1046/j.1432-0436.2002.700908.x [DOI] [PubMed] [Google Scholar]
- 5.Grinnell F., J. Cell Sci. 101(1), 1–5 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Kramer N., Walzl A., Unger C., Rosner M., Krupitza G., Hengstschläger M., and Dolznig H., Rev. Mutat. Res. 752(1), 10–24 (2013). 10.1016/j.mrrev.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 7.Shizukuda Y., Tang S., Yokota R., and Ware J. A., Circ. Res. 85(3), 247–256 (1999). 10.1161/01.RES.85.3.247 [DOI] [PubMed] [Google Scholar]
- 8.Brown N. and Bicknell R., in Metastasis Research Protocols, edited by Brooks S. and Schumacher U. ( Humana Press, 2001), Vol. 58, pp. 47–54. [Google Scholar]
- 9.Kozien D., Gerol M., Hendey B., and RayChaudhury A., In Vitro Cell. Dev. Biol.: Anim. 36(9), 555–558 (2000). 10.1007/BF02577521 [DOI] [PubMed] [Google Scholar]
- 10.Shi J., Ahmed D., Mao X., Lin S.-C. S., Lawit A., and Huang T. J., Lab Chip 9(20), 2890–2895 (2009). 10.1039/b910595f [DOI] [PubMed] [Google Scholar]
- 11.Ino K., Ito A., and Honda H., Biotechnol. Bioeng. 97(5), 1309–1317 (2007). 10.1002/bit.21322 [DOI] [PubMed] [Google Scholar]
- 12.Odde D. J. and Renn M. J., Biotechnol. Bioeng. 67(3), 312–318 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Lin R.-Z., Ho C.-T., Liu C.-H., and Chang H.-Y., Biotechnol. J. 1(9), 949–957 (2006). 10.1002/biot.200600112 [DOI] [PubMed] [Google Scholar]
- 14.Yang S.-M., Yu T.-M., Huang H.-P., Ku M.-Y., Hsu L., and Liu C.-H., Opt. Lett. 35(12), 1959–1961 (2010). 10.1364/OL.35.001959 [DOI] [PubMed] [Google Scholar]
- 15.Goubko C. A. and Cao X., Mater. Sci. Eng., C 29(6), 1855–1868 (2009). 10.1016/j.msec.2009.02.016 [DOI] [Google Scholar]
- 16.Roth E. A., Xu T., Das M., Gregory C., Hickman J. J., and Boland T., Biomaterials 25(17), 3707–3715 (2004). 10.1016/j.biomaterials.2003.10.052 [DOI] [PubMed] [Google Scholar]
- 17.Lee K. B., Park S. J., Mirkin C. A., Smith J. C., and Mrksich M., Science 295(5560), 1702–1705 (2002). 10.1126/science.1067172 [DOI] [PubMed] [Google Scholar]
- 18.Tehranirokh M., Kouzani A. Z., Francis P. S., and Kanwar J. R., Biomicrofluidics 7(5), 051502 (2013). 10.1063/1.4826935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inamdar N. K. and Borenstein J. T., Curr. Opin. Biotechnol. 22(5), 681–689 (2011). 10.1016/j.copbio.2011.05.512 [DOI] [PubMed] [Google Scholar]
- 20.Beebe D. J., Mensing G. A., and Walker G. M., Annu. Rev. Biomed. Eng. 4, 261–286 (2002). 10.1146/annurev.bioeng.4.112601.125916 [DOI] [PubMed] [Google Scholar]
- 21.Cimetta E., Cannizzaro C., James R., Biechele T., Moon R. T., Elvassore N., and Vunjak-Novakovic G., Lab Chip 10(23), 3277–3283 (2010). 10.1039/c0lc00033g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosadegh B., Agarwal M., Tavana H., Bersano-Begey T., Torisawa Y. S., Morell M., Wyatt M. J., O'Shea K. S., Barald K. F., and Takayama S., Lab Chip 10(21), 2959–2964 (2010). 10.1039/c0lc00086h [DOI] [PubMed] [Google Scholar]
- 23.Blake A. J., Pearce T. M., Rao N. S., Johnson S. M., and Williams J. C., Lab Chip 7(7), 842–849 (2007). 10.1039/b704754a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felder M., Sallin P., Barbe L., Haenni B., Gazdhar A., Geiser T., and Guenat O., Lab Chip 12(3), 640–646 (2012). 10.1039/c1lc20879a [DOI] [PubMed] [Google Scholar]
- 25.Van Der Meer A. D., Vermeul K., Poot A. A., Feijen J., and Vermes I., Am. J. Physiol.: Heart Circ. Physiol. 298(2), H719–H725 (2010). 10.1152/ajpheart.00933.2009 [DOI] [PubMed] [Google Scholar]
- 26.Shin M. K., Kim S. K., and Jung H., Lab Chip 11(22), 3880–3887 (2011). 10.1039/c1lc20671k [DOI] [PubMed] [Google Scholar]
- 27.Shin Y., Han S., Jeon J. S., Yamamoto K., Zervantonakis I. K., Sudo R., Kamm R. D., and Chung S., Nat. Protoc. 7(7), 1247–1259 (2012). 10.1038/nprot.2012.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong G. S., Han S., Shin Y., Kwon G. H., Kamm R. D., Lee S. H., and Chung S., Anal. Chem. 83(22), 8454–8459 (2011). 10.1021/ac202170e [DOI] [PubMed] [Google Scholar]
- 29.Van Der Meer A. D., Poot A. A., Duits M. H. G., Feijen J., and Vermes I., J. Biomed. Biotechnol. 2009, 10. 10.1155/2009/823148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y., Majumdar D., Jovanovic B., Shaifer C., Lin P. C., Zijlstra A., Webb D., and Li D., Biomed. Microdevices 13(3), 539–548 (2011). 10.1007/s10544-011-9523-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brewer B. M., Shi M., Edd J. F., Webb D. J., and Li D., Biomed. Microdevices 16, 311–323 (2014). 10.1007/s10544-014-9834-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B., Moore J. S., and Beebe D. J., Science 291(5506), 1023–1026 (2001). 10.1126/science.291.5506.1023 [DOI] [PubMed] [Google Scholar]
- 33.Ciofani G., Migliore A., Raffa V., Menciassi A., and Dario P., J. Biosci. Bioeng. 105(5), 536–544 (2008). 10.1263/jbb.105.536 [DOI] [PubMed] [Google Scholar]
- 34.Kaji H., Yokoi T., Kawashima T., and Nishizawa M., Lab Chip 9(3), 427–432 (2009). 10.1039/b812510d [DOI] [PubMed] [Google Scholar]
- 35.Zervantonakis I. K., Kothapalli C. R., Chung S., Sudo R., and Kamm R. D., Biomicrofluidics 5(1), 013406 (2011). 10.1063/1.3553237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei C.-W., Cheng J.-Y., and Young T.-H., Biomed. Microdevices 8(1), 65–71 (2006). 10.1007/s10544-006-6384-8 [DOI] [PubMed] [Google Scholar]
- 37.Hong J. W., Song S., and Shin J. H., Lab Chip 13(15), 3033–3040 (2013). 10.1039/c3lc50163a [DOI] [PubMed] [Google Scholar]
- 38.Esposito M. and Kang Y., Pharmacol. Ther. 141(2), 222–233 (2014). 10.1016/j.pharmthera.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H., Schueller O. J., and Whitesides G. M., Electrophoresis 21(1), 27–40 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Lee J. N., Jiang X., Ryan D., and Whitesides G. M., Langmuir 20(26), 11684–11691 (2004). 10.1021/la048562+ [DOI] [PubMed] [Google Scholar]
- 41.Kuddannaya S., Chuah Y. J., Lee M. H. A., Menon N. V., Kang Y., and Zhang Y., ACS Appl. Mater. Interfaces 5(19), 9777–9784 (2013). 10.1021/am402903e [DOI] [PubMed] [Google Scholar]
- 42.Makamba H., Kim J. H., Lim K., Park N., and Hahn J. H., Electrophoresis 24(21), 3607–3619 (2003). 10.1002/elps.200305627 [DOI] [PubMed] [Google Scholar]
- 43.Zhou J., Khodakov D. A., Ellis A. V., and Voelcker N. H., Electrophoresis 33(1), 89–104 (2012). 10.1002/elps.201100482 [DOI] [PubMed] [Google Scholar]
- 44.Tourovskaia A., Barber T., Wickes B. T., Hirdes D., Grin B., Castner D. G., Healy K. E., and Folch A., Langmuir 19(11), 4754–4764 (2003). 10.1021/la0267948 [DOI] [Google Scholar]
- 45.Castellone M. D., Laatikainen L. E., Laurila J. P., Langella A., Hematti P., Soricelli A., Salvatore M., and Laukkanen M. O., Stem Cells 31(6), 1218–1223 (2013). 10.1002/stem.1361 [DOI] [PubMed] [Google Scholar]
- 46.Vallabhaneni K. C., Haller H., and Dumler I., Stem Cells Dev. 21(17), 3104–3113 (2012). 10.1089/scd.2011.0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paduch R., Walter-Croneck A., Zdzisińska B., Szuster-Ciesielska A., and Kandefer-Szerszeń M., Cell Biol. Int. 29(7), 497–505 (2005). 10.1016/j.cellbi.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 48.Reuter S., Gupta S. C., Chaturvedi M. M., and Aggarwal B. B., Free Radical Biol. Med. 49(11), 1603–1616 (2010). 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiaschi T. and Chiarugi P., Int. J. Cell Biol. 2012, 8. 10.1155/2012/762825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez-Outschoorn U. E., Balliet R. M., Rivadeneira D. B., Chiavarina B., Pavlides S., Wang C., Whitaker-Menezes D., Daumer K. M., Lin Z., Witkiewicz A. K., Flomenberg N., Howell A., Pestell R. G., Knudsen E. S., Sotgia F., and Lisanti M. P., Cell Cycle 9(16), 3256–3276 (2010). 10.4161/cc.9.16.12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin R. Z., Wang T. P., Hung R. J., Chuang Y. J., Chien C. C., and Chang H. Y., J. Cell Physiol. 226(7), 1750–1762 (2011). 10.1002/jcp.22504 [DOI] [PubMed] [Google Scholar]
- 52.See supplementary material at http://dx.doi.org/10.1063/1.4903762E-BIOMGB-8-025406 for additional figure and video.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4903762E-BIOMGB-8-025406 for additional figure and video.