Abstract
To modulate the surface properties of collagen and subsequent cell-surface interactions, a method was developed to transfer protein patterns from glass coverslips to collagen type I hydrogel surfaces. Two proteins and one proteoglycan found in central nervous system extracellular matrix as well as fibrinogen were patterned in stripes onto collagen hydrogel and astrocytes were cultured on these surfaces. The addition of the stripe protein patterns to hydrogels created astrocyte layers in which cells were aligned with underlying patterns and had reduced chondroitin sulfate expression compared to the cells grown on collagen alone. Protein patterns were covalently cross-linked to the collagen and stable over four days in culture with no visible cellular modifications. The present method can be adapted to transfer other types of protein patterns from glass coverslips to collagen hydrogels.
Introduction
Glial cells facilitate neuronal guidance in recovering central nervous system (CNS) tissues and, from among them, astrocytes are known to form aligned networks which coincide with regenerating nerve tracts [1]. The poor recovery of neural functionality after CNS injury has been ascribed to the presence of inhibitory molecules as well as the formation of the disorganized tissue known as the glial scar [2–4]. In response to CNS tissue injury, astrocytes become reactive and begin to express intermediate filaments as they hypertrophy. While this reactive scarring has beneficial roles, astrocytes also upregulate expression of extracellular matrix (ECM) molecules such as chondroitin sulfate proteoglycan (CSPG) that has been shown to inhibit neuronal regeneration [4]. One approach to improve CNS recovery is to treat the glial scar with chondroitinase, a broad enzyme that digests chondroitin sulfate, thus enzymatically removing the inhibitory components of the scar [5]. An alternative to that approach would be to add a biomaterial to the injury site so that injured neuronal cells could grow directionally and eventually re-establish lost connections [6]. Such a biomaterial should both facilitate reestablishing glial organization and promote neuronal regeneration.
Collagen type I has been used extensively in treating injured CNS with devices ranging from dura replacement sheets to nerve guidance conduits. Collagen has been shown to be bioresorbable and easily incorporated by host tissue. Astrocytes themselves have been shown to deposit collagen in the CNS wound environment [7]. Collagen types I, III, and IV have been found in the glial scar, though their presence alone is not inhibitory [8–11]. Collagen is also a permissive substrate for neuronal outgrowth and regeneration in vivo [12,13]. Used as a growth substrate in vitro, collagen has been used to create a scaffolding structure where a quiescent, unreactive astrocyte phenotype is preserved, as opposed to the more reactive, hypertrophic phenotype that occurs in standard 2D astrocyte culture [14]. Collagen has also been used as a scaffold material for cellular therapies including glial precursors [15], Schwann cells [16], and differentiated astrocytes [17].
Rather than implanting a simple biomaterial scaffold to a CNS injury site, the addition of other ECM components to the scaffold has been used to improve CNS recovery as reviewed by Volpato et al. [18]. Collagen has been augmented with various bioactive agents including brain-derived neurotrophic factor (BDNF) [19], neurotrophin-3 [20], fibroblast growth factor (FGF) [21], and chondroitinase [22], which have been crosslinked to collagen gels to improve neuronal outgrowth. Topographical cues, such as the direction of collagen nanofibers, have also been used to align and direct both astrocytes and neurons [23]. Neurons grown at the interface between collagen and ECM components patterned on glass showed that neurons followed associated ECM patterns and largely avoided penetrating deeper into collagen gels [24]. Thus, it seems that both the physical properties of collagen scaffolds and the presence of surface guidance cues are important for determining the CNS cellular response.
Controlling astrocyte alignment and infiltration is a worthy goal for improving CNS regenerative biomaterial potential [21,25]. Astrocytes may serve as a source for guidance cues and growth factors, and, as a large component to the glial scar, their behavior is important for wound healing and neuroprotection [26]. We have shown that even fixed aligned astrocytes will still direct neuronal outgrowth without having to release any active soluble factors [27]. As differentiated astrocytes seeded in collagen constructs have led to functional recovery in vivo [17], the ability of astrocytes to attach to and align on biomaterial constructs may foment subsequent directed neuronal outgrowth and promote regeneration.
The present study aimed at imparting directional guidance cues to collagen gels in a relatively simple way so that astrocytes can attach and align themselves without increasing their reactivity so commonly seen in traditional 2D cultures. The method described here was used to covalently attach patterns of proteins or proteoglycans to collagen hydrogel surfaces. Similar approaches have been proposed for other polymers such as polydimethylsiloxane (PDMS), polyvinyl alcohol, polyacrylamide, and polycaprolactone [28–32]. Recently, microcontact printing of proteins to freeze-dried Matrigel™ and gelatin has been demonstrated [33]. However, to the best of our knowledge, the present report is the first application of protein pattern transfer and covalent attachment to the surface of collagen type I hydrogels and requires no additional, specialized equipment beyond standard materials for creating protein patterns. The techniques described here will be conducive to the transfer of multiple proteins deposited in any desired pattern on glass surface by traditional microcontact printing [34], stamp-off [35], sequential stamping, microscope-aided registration [36], dip-pen nanolithography [37], or inkjet deposition [38], directly to collagen hydrogel surface.
Materials and Methods
Collagen Hydrogel Patterning
To visualize patterns transferred to the surface of collagen gels, proteins were labeled with fluorophores. Human plasma fibrinogen (FBG, plasminogen depleted, Calbiochem) and aggrecan (AGG, A1960, Sigma) were fluorescently labeled with Alexa Fluor 594 (A-20004, Invitrogen). Laminin (LN, L2020, Sigma-Aldrich) and fibronectin (FN, F1141, Sigma-Aldrich) were labeled with Alexa Fluor 488 (A-20000, Invitrogen). To separate labeled protein from free dye, labeled protein solutions were eluted in phosphate buffered saline (PBS) through a PD-10 Sephadex column (GE Healthcare). Collected solutions were then filtered through a 0.20 μm syringe filter (Sarstedt), divided into 100 μL aliquots, and stored at −20°C until use.
To pattern the surface of collagen gels, a two-step process was used. Microcontact printing (μCP) was used first to pattern labeled proteins onto sterilized 18 mm glass coverslips [34,39]. The pattern used in this study consisted of 15 μm stripes separated by 25 μm–wide gaps. This pattern was selected as 15 μm stripes previously aligned astrocytes on glass [27], and the added asymmetry was useful to distinguish printed and non-printed regions while maintaining alignment [36]. In the second step, a process was developed for collagen to undergo simultaneous gelling and cross-linking while the protein patterns were transferred (Figure 1). Bis[sulfosuccinimidyl] suberate (BS3, Thermo Scientific) was prepared immediately before collagen gelling to minimize hydrolysis of its reactive groups. Two mg of BS3 was dissolved in double-distilled, deionized (DDI) water to a concentration of 50 mM. This BS3 solution was placed as a 10 μL droplet on top of the stamped proteins on the coverslip surface and then immediately followed by deposition of 100 μL collagen type I (BD Biosciences) solution at a concentration of 10 mg/mL to the top of the stamped coverslips. NaOH (1 M) was added to bring the gel to neutral pH according to manufacturer specifications. A PDMS sheet (0.005″ thick, Specialty Manufacturing Inc.) was placed on top of the gel to facilitate gel spreading and future handling. Gelling and crosslinking was allowed to proceed for 30 minutes at 37°C. The gel constructs were then placed in a humidified incubator to dry for an additional 48 hours. The patterned collagen was separated from the stamped coverslip with the aid of physical force (i.e., using a cell scraper). To isolate patterned collagen gel alone, the construct was submerged in water and the hydrogel was lifted off the backing substrate using a fine brush. This procedure was successful with collagen gel concentrations ranging from 5 to 10 mg/mL, with higher concentrations being more amenable to physical manipulation. Removal was facilitated by releasing the entire circumference of the gel by scraping under its edge about a millimeter prior to attempting to separate it entirely from the substrate. By using the separated edges, a peeling motion from one side to the other, while ensuring that the gel was separating evenly, was effective. This also prevented damage to the center of the gel where images were later acquired. A collagen concentration of 10 mg/mL, which has been shown to have physical properties relevant to CNS tissue [40], was used in this study to eliminate any further variation in gel stiffness.
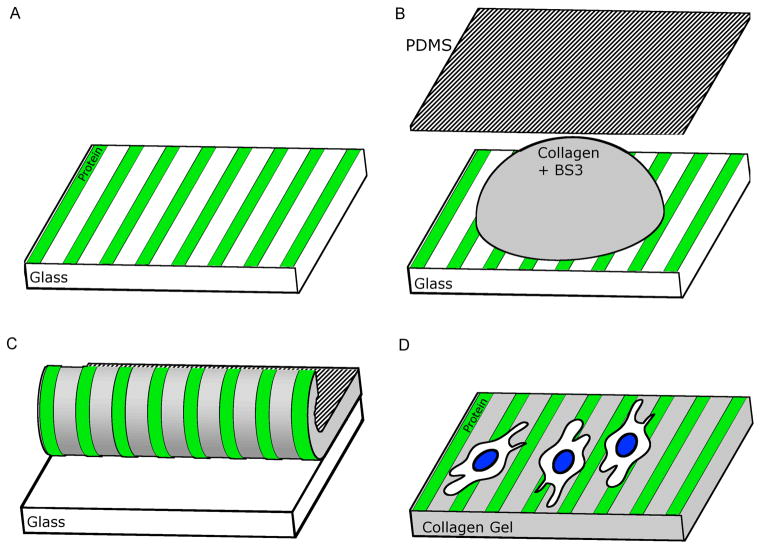
Figure 1.
Schematic of process for transferring protein pattern to collagen. A) The desired pattern is first created on a glass surface after which B) BS3 crosslinker solution and collagen are placed on the pattern and spread with a PDMS sheet. C) The gel is allowed to crosslink with the pattern and then peeled from the glass. The PDMS backing can be removed leaving a D) stand-alone collagen gel with a surface with patterned proteins.
Cell Culture
Primary astrocytes from post-natal day 2 Sprague-Dawley rats were obtained using established protocols [41]. Confluent astrocyte cultures were shaken to remove contaminating cells and frozen. Astrocytes were thawed and cultured for 2 weeks prior to trypsinization and seeding onto collagen gels. Cultures were maintained in a humidified incubator at 37 °C and 5% CO2. Astrocytes were seeded on collagen gels at a density of approximately 25,000 per cm2 in SATO- serum-free media [42] and allowed to attach for 5 hours. After that time, medium was changed to DMEM/F12 (Caisson Labratories) with 10% fetal bovine serum (FBS, Atlanta Biologicals) for the remaining culture period. Medium was exchanged at 48 hours, and the cultures were fixed at 96 hours post-seeding.
Immunocytochemistry for CSPG Quantification
Because real time phase or DIC microscopy of astrocytes on collagen gels were difficult due to extensive light scattering by the gels, cells on gels were fixed in 4% paraformaldehyde (PFA) for 15 minutes and rinsed in PBS with 0.1% sodium azide prior to immunocytochemical staining. All procedures were carried out at room temperature. Samples were blocked with 4% goat serum in PBS for one hour and then rinsed thrice in PBS with sodium azide. Primary anti-chondroitin sulfate (CS-56) antibody (C8035, Sigma) was applied to the astrocytes for one hour in a 1:500 dilution in the block solution. Samples were again rinsed three times in PBS with sodium azide. Secondary goat anti-mouse IgM antibody labeled with Alexa Fluor 488 (A21042, Molecular Probes) or Alexa Fluor 594 (A21044, Molecular Probes) was subsequently applied to the cell samples for 1 hour. Following three more PBS rinses, 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen) in a 1:100 dilution was added for 15 minutes to stain for nuclei. Samples were then rinsed in DDI water and mounted between coverslips (Fisher) with Fluoromount-G (Southern Biotech) for imaging. Astrocytes on plain collagen controls were divided into two batches for use with each type of secondary antibody. Fluorescence images were captured using a Nikon Eclipse E600 epifluorescence microscope with a 20x PlanAPO objective and CCD camera (CoolSNAP, Photometrics).
Quantifying Astrocyte CSPG Expression
CSPG samples were imaged after staining using identical exposure times in Image Pro Plus (Media Cybernetics) software. A blank image was subtracted from each image and the CSPG fluorescence intensity was quantified using ImageJ (NIH). The CSPG fluorescence was integrated over the entire image and divided by the number of cells per image as identified by DAPI staining. Samples with fewer than five nuclei were not included in analysis. Images were taken of five sample sub-areas where protein patterns and cells were visible and on the same focal plane. An ANOVA with a Tukey post hoc test (α = 0.05) was used to determine the significance of data differences.
Astrocyte Alignment Analysis
Astrocyte alignment was assessed by measuring the orientation of their nuclei [27,36]. To do this, the angle of the major axis of the oval-like astrocyte nuclei were quantified using the measure angle function of Image Pro Plus. Figure 2 shows the measured angles (shown as line segments drawn at the measured angle and superimposed over outlined nuclei). Any nuclei that were on image edges that could not be fully resolved were excluded from analysis. The measured nuclei angles were then converted to an alignment angle with respect to the orientation of the underlying protein pattern. For collagen controls lacking protein patterns, the vertical axis was used as reference. The alignment angles of all nuclei across each given sample type were compiled into a histogram in 10 degree bins. A zero degree angle indicated nuclei that were completely parallel with the underlying patterns. Histograms were fit with a Gaussian function using Igor Pro (Wavemetrics) to find the mean alignment angle and the full width at half maximum (FWHM) for each nuclei population (FWHM is approximately equal to 2.4 times the standard deviation of the angle distribution).
Figure 2.
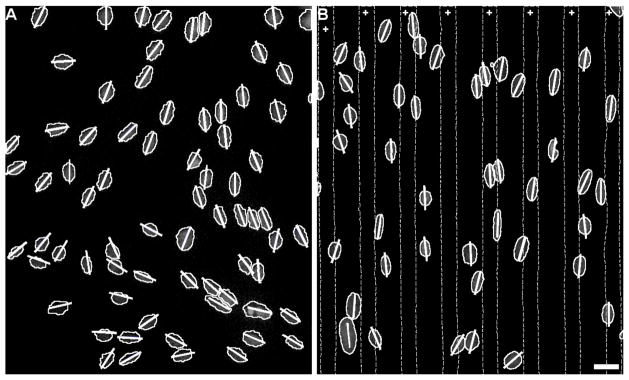
Astrocyte nuclei angle measurements. Representative images of astrocyte nuclei on A) unpatterned collagen control and B) collagen with fibrinogen protein pattern. Areas between dotted lines marked with + indicate regions with patterned FBG on collagen gel. Short lines drawn for each nucleus indicate the angle measured for the major axis of each nucleus. Scale bar = 25 μm.
Results
Protein patterns transferred to collagen gels and resisted removal by astrocytes
The techniques described here created collagen gels with a covalently immobilized pattern of protein transferred from glass to one of the gels’ surfaces. Stripe patterns of aggrecan (AGG), fibrinogen (FBG), fibronectin (FN), and laminin (LN) were all successfully transferred to the surface of collagen type I gels (Figure 3). Fidelity of the pattern on gels required quality protein patterns on glass and careful manual removal of gels to maintain intact gel sheets. As loss of immobilized proteins from the gel surfaces would negate the benefit of patterning these gels, the patterns were imaged prior to and after 96 hours of astrocyte culture to determine pattern integrity. No changes in the protein pattern appearance were observed over that time period. Unlike the removal of glass-adsorbed fibrinogen by astrocytes seen previously [43], proteins patterned on collagen were not removed by cells over the given culture time period. We infer that the protocol used created stable, immobilized protein patterns that did not diffuse into the gel or desorb/detach from the collagen surface.
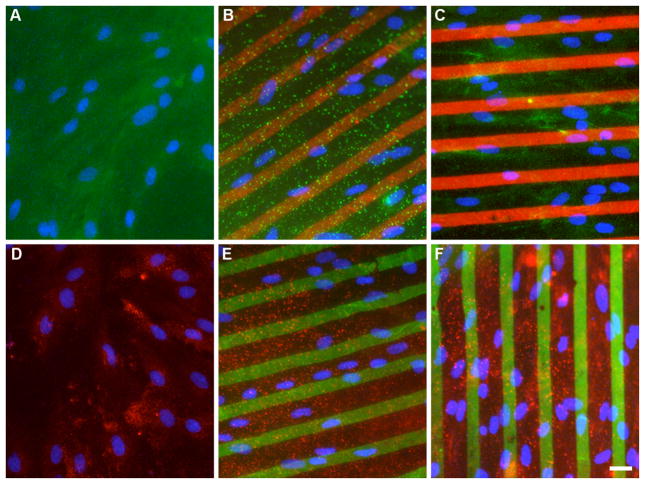
Figure 3.
Representative fluorescence images of astrocytes on patterned collagen gels. A,D) unpatterned collagen control; B) aggrecan (red) on collagen; C) fibrinogen (red) on collagen; E) fibronectin (green) on collagen; and F) laminin (green) on collagen gels were created. In panels A–C, CSPG (green) expression was measured using Alexa Fluor 488 labeled secondary antibody. In panels D–F, CSPG (red) was measured using Alexa Fluor 594 labeled secondary antibody. Nuclei (blue) can be seen in each image (A–F). Scale bar = 25 μm.
Astrocytes align with underlying protein patterns on collagen
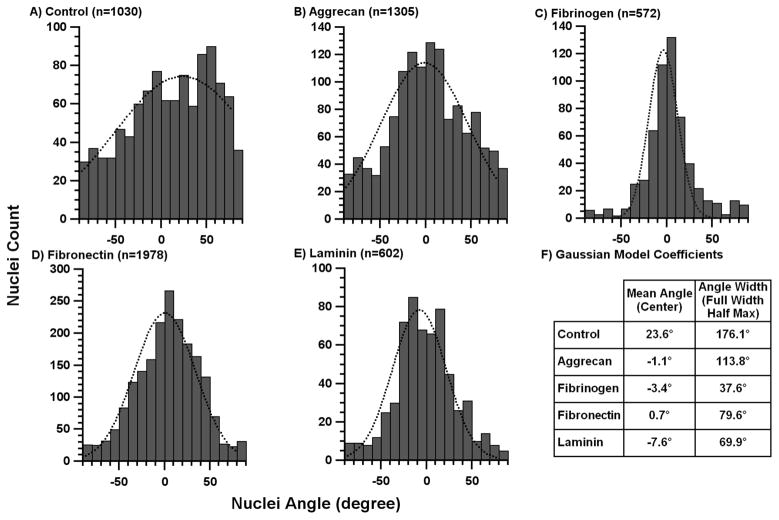
Astrocytes attached and spread onto collagen gel surfaces with and without immobilized protein patterns within the first 5 hours of culture. After the addition of 10% serum and subsequent culture for 4 days, astrocytes oriented themselves on the gels. A degree of polarity and alignment was established locally by the cells even on plain collagen gel controls (Fig 2A) as astrocytes that were close together tended to align with each other in clusters. For samples with immobilized ECM protein patterns, a much stronger alignment was seen (Fig 2B). Only stripes of transferred protein differentiated these surfaces from plain collagen gel controls. A more quantitative measure of the astrocyte alignment is given by the histograms showing the fitted Gaussian function, mean angle, and FWHM values for each sample (Figure 4). On the control collagen gels (Fig 4A), astrocyte alignment was random with angles spread across all possible angles with a FWHM of 176.1°. In contrast, on gels with immobilized ECM protein stripes, the mean angle for astrocyte nuclei was within 8° of the orientation of the underlying pattern for each protein (Fig 4F). The difference between the patterned proteins can be analyzed by the variations in FWHM where the smallest FWHM value indicated better alignment. Nuclei on the FBG pattern showed the narrowest orientation spread (FWHM of 37.6°, equivalent to a standard deviation of ±16°; Fig 4C) FN and LN patterns (Figs 4D,E) were less potent than FBG in aligning cells with FWHMs of 79.6 and 69.9°, respectively. The AGG pattern, which does not strongly align astrocytes on glass [36], was the least effective (Fig 4B; FWHM = 113.8°).
Figure 4.
Astrocyte nuclei angle histograms on patterned collagen gels. The angles of observed astrocyte nuclei were compiled into histogram with 10° bins for A) unpatterned collagen control, B) aggrecan stripes, C) fibrinogen stripes, D) fibronectin stripes, and E) laminin stripes. The dotted curves indicate the Gaussian model fit for each data set. These fitted curves were used to calculate the F) mean angle and full-width-half-max (FWHM) for each pattern.
Astrocyte CSPG reactivity is attenuated with the addition of ECM proteins
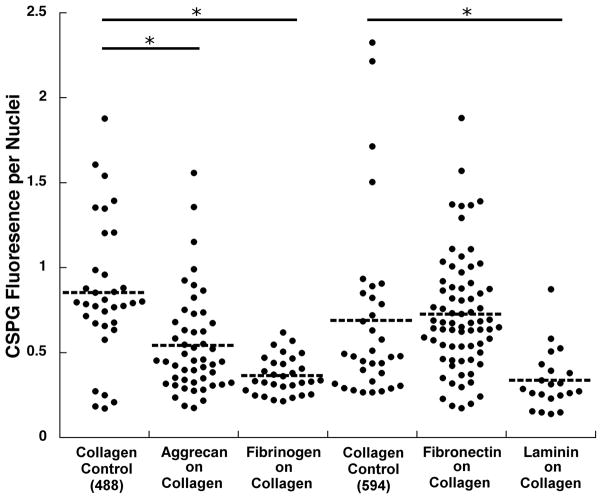
The overall CSPG expression by astrocytes was significantly lower on the ECM protein patterned gels compared with two sets of controls1 (Figure 5). The presence of FBG and LN patterns led to a significant reduction (>50%) of CSPG expression. There was no significant CSPG reduction detected for the FN pattern, while on the AGG pattern the CSPG was reduced to about one-third of the mean CSPG expressed per cell for collagen control. As expected, in the case of AGG patterns, there was some staining of underlying AGG stripes with the anti–CS antibody. This fluorescence was included in the integrated CSPG fluorescence so the actual reduction of CSPG expression by astrocytes on AGG patterns could have been greater than shown in Figure 5.
Figure 5.
CSPG expression by astrocytes on patterned collagen gels. Each dot represents CSPG fluorescence (in arbitrary units) integrated from a single fluorescence image. To account for the variations in cell numbers in each image, the integrated CSPG fluorescence from each image was divided by the number of cells present. CSPG was either visualized using Alexa Fluor 488 labeled secondary antibody for aggrecan and fibrinogen patterns, or Alexa Fluor 594 labeled secondary antibody for fibronectin and laminin patterns. Unpatterned collagen controls for both fluorophores were used to allow for comparison between proteins. Asterisks denote p < 0.005.
Discussion
Collagen gels with stripe patterns aid alignment of astrocytes
As previously shown, neurons send outgrowths that follow aligned astrocytes and Schwann cells, as well as biomimetic artificial surfaces [44]. If one can use a biomaterial surface to align astrocytes and other glia, subsequent neuronal growth and layers of glia could then be guided as well. Using the method developed here for decorating collagen surfaces with ECM proteins, it would be possible to transfer any protein pattern prepared on a glass coverslip surface to collagen. While others have aligned astrocytes in/on collagen via physical forces [25], the current protocol could be readily adapted to transfer multi-protein patterns, thus increasing the repertoire of the molecular cues for astrocyte alignment and/or neuronal outgrowth.
As there were no added topographical cues on these patterned collagen gels, the alignment was caused by the molecules found on the surface of the gels. These could include the patterned proteins, ECM secreted by the cells, proteins from the culture medium, and collagen itself. The presence of adhered serum proteins could not be ruled out as all samples had the access to 10% serum containing medium. The 10% serum medium was introduced after the initial culture period of 5 hours without serum. After this initial attachment period, the composition of the surface proteins might have been altered as both cells and 10% serum were present. Therefore, the actual mechanism as to how the patterned protein stripes functioned to align astrocytes could not be conclusively determined. It is clear, however, that the presence of these protein stripes causes nuclear alignment over plain collagen.
Astrocyte nuclei were seeded randomly across the substrates, and were found to slightly favor the position where the nucleus is partially on the protein stripe pattern and partially off of it after culture. The pattern used in the present study had an unequal distribution of protein vs. collagen-only areas. For any given region, only 37.5% of the area (15/(25+15)) was covered with patterned protein stripes. The likelihood of the cell to attach to collagen alone was 62.5%, (25/(15+25)), almost twice than that for protein stripes. If a fully spread cell were 50 μm wide and was centered in the middle of a collagen stripe, it could sample both neighboring protein patterns on each of its sides. Conversely, if it finds itself in the middle of the protein stripe, it would only sample collagen on both sides. That may explain why some nuclei were found over the collagen regions between the protein stripes. However, the actual count of the nuclei positions showed no clear preference for collagen only regions: the majority of nuclei were located over some portion of both collagen and the protein stripe.
We have previously seen that adsorbed FBG was removed from glass surfaces below adhered astrocytes, most likely because of the protein was simply adsorbed to the surface [43]. In the case of immobilized FBG on collagen gel surface, astrocytes did not remove fibrinogen or any of the other three ECM proteins from the surface, indicating that the transfer procedure created stable crosslinks between the protein molecules and the gel surface. Likewise, crosslinks could have also formed between two collagen molecules or two patterned proteins with this technique. Regardless, the ECM proteins patterned here still maintained alignment potency after this process.
The robust nature of transferred protein patterns, coupled with the tendency for guidance information to be carried from cell to cell, indicates that a collagen biomaterial device with appropriately oriented patterns could provide a scaffold for recovering CNS cells that could combat the default tissue disorganization that occurs in CNS injury. The underlying collagen itself could then be remodeled and degraded leaving potentially superior outcomes post injury.
Astrocyte CSPG reactivity decreases with ECM alignment cues
It is well known that in response to CNS injury, astrocytes acquire a reactive phenotype, which includes increased expression of glial fibrillary acidic protein (GFAP) and CSPG [4]. Although some variants of the chondroitin sulfate (CS) chains, such as CS-C, are inhibitory to neuronal outgrowth and others variants (CS-A and CS-E) are not [45], astrocyte expression of CSPG is correlated with their inhibitive properties to neurons. While CSPG are also shed into the surrounding cellular environment by astrocytes [43,45], the surface expression on their membranes is important for interfacing with neurons. It is therefore informative to determine if a CNS biomaterial scaffold itself is increasing astrocyte surface expression of CSPG. As compared with the collagen control, three ECM proteins--FBG, AGG, and LN—each able to align astrocytes to some degree, were also able to reduce this CSPG expression. The decrease in CSPG reactivity between unpatterned and patterned gels could have been caused by either the ECM molecule itself or the alignment of astrocytes it caused. Based on the previous reports that the introduction of FBG into the CNS after injury triggers CSPG production [46], and that fact that AGG already contains inhibitory CS chains, it appears that the alignment itself was the dominant contributing factor resulting in the decrease in CSPG expression seen here (Fig 5). It has been shown that the presence of topographical cues that aligned astrocytes also reduced astrocyte reactivity [47]. Similarly, astrocytes grown on aligned electrospun fibers of polycaprolactone had decreased GFAP expression compared to those on random fibers [48]. The findings from the present study thus indicate that astrocytes and their surface CSPG expression are sensitive to organized, directional cues.
Conclusions
Creating collagen gels with surfaces patterned with FBG and three ECM proteins was developed and tested on in vitro cultures of astrocytes. Patterns of FBG, AGG, FN, and LN stripes were able to align astrocytes when compared to collagen alone. Additionally, the aligned astrocytes on AGG, FBG, and LN patterns showed reduced overall CSPG expression. Astrocytes survived the entire 96 hours culture period, leaving no indication of toxicity of this process. Other cross-linking agents like genipin [21] and disuccinimidyl-disuccinate-polyethyleneglycol (SS-PEG-SS)[49], which have previously been used to link proteins to collagen, should be amenable to the protein pattern lift-off from glass coverslips. The method developed here will also allow transfer of multiple ECM molecules with any desired spatial presentation on collagen and the investigation of cellular responses. In this way, the method may find its application to biomedical devices where surface-cell interactions can be leveraged to improve patient outcomes. Collagen biomaterials with their surfaces decorated with relevant ECM cues may thus augment regeneration of injured CNS tissue.
Acknowledgments
The authors would like to thank Dr. Elena Budko for astrocyte isolation and the University of Utah Bioengineering Department for its support. This work was partially funded by NIH Grants NS057144 and NS088737.
Footnotes
For imaging both the ECM protein pattern (see Fig. 3) and expressed CSPG, secondary goat anti-mouse IgM antibodies were either labeled with Alexa Fluor 488 or Alexa Fluor 594.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davies SJA, Goucher DR, Doller C, Silver J. Robust Regeneration of Adult Sensory Axons in Degenerating White Matter of the Adult Rat Spinal Cord. J Neurosci. 1999 Jul 15;19(14):5810–22. doi: 10.1523/JNEUROSCI.19-14-05810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu BP, Fournier A, GrandPré T, Strittmatter SM. Myelin-Associated Glycoprotein as a Functional Ligand for the Nogo-66 Receptor. Science. 2002 Aug 16;297(5584):1190–3. doi: 10.1126/science.1073031. [DOI] [PubMed] [Google Scholar]
- 3.Chen MS, Huber AB, van der Haar ME, Frank M, Schnell L, Spillmann AA, et al. Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature. 2000 Jan 27;403(6768):434–9. doi: 10.1038/35000219. [DOI] [PubMed] [Google Scholar]
- 4.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004 Feb;5(2):146–56. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 5.Alilain WJ, Horn KP, Hu H, Dick TE, Silver J. Functional regeneration of respiratory pathways after spinal cord injury. Nature. 2011 Jul 14;475(7355):196–200. doi: 10.1038/nature10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokols S, Tuszynski MH. Freeze-dried agarose scaffolds with uniaxial channels stimulate and guide linear axonal growth following spinal cord injury. Biomaterials. 2006 Jan;27(3):443–51. doi: 10.1016/j.biomaterials.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 7.Heck N, Garwood J, Schütte K, Fawcett J, Faissner A. Astrocytes in culture express fibrillar collagen. Glia. 2003;41(4):382–92. doi: 10.1002/glia.10184. [DOI] [PubMed] [Google Scholar]
- 8.Weidner N, Grill RJ, Tuszynski MH. Elimination of Basal Lamina and the Collagen “Scar” after Spinal Cord Injury Fails to Augment Corticospinal Tract Regeneration. Exp Neurol. 1999 Nov;160(1):40–50. doi: 10.1006/exnr.1999.7200. [DOI] [PubMed] [Google Scholar]
- 9.Joosten EAJ, Dijkstra S, Brook GA, Veldman H, Bär PR. Collagen IV deposits do not prevent regrowing axons from penetrating the lesion site in spinal cord injury. J Neurosci Res. 2000;62(5):686–91. doi: 10.1002/1097-4547(20001201)62:5<686::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10.Okada M, Miyamoto O, Shibuya S, Zhang X, Yamamoto T, Itano T. Expression and role of type I collagen in a rat spinal cord contusion injury model. Neurosci Res. 2007 Aug;58(4):371–7. doi: 10.1016/j.neures.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Klapka N, Müller HW. Collagen Matrix in Spinal Cord Injury. J Neurotrauma. 2006 Apr 21;23(3–4):422–36. doi: 10.1089/neu.2006.23.422. [DOI] [PubMed] [Google Scholar]
- 12.Cholas RH, Hsu H-P, Spector M. The reparative response to cross-linked collagen-based scaffolds in a rat spinal cord gap model. Biomaterials. 2012 Mar;33(7):2050–9. doi: 10.1016/j.biomaterials.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Peulve P, Jin O, Boisset N, Tiollier J, Said G, et al. Axonal regrowth through collagen tubes bridging the spinal cord to nerve roots. J Neurosci Res. 1997;49(4):425–32. doi: 10.1002/(sici)1097-4547(19970815)49:4<425::aid-jnr4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 14.East E, Golding JP, Phillips JB. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. J Tissue Eng Regen Med. 2009;3(8):634–46. doi: 10.1002/term.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketschek AR, Haas C, Gallo G, Fischer I. The roles of neuronal and glial precursors in overcoming chondroitin sulfate proteoglycan inhibition. MicroRNAs—Human Neurobiol Neuropathol. 2012 Jun;235(2):627–37. doi: 10.1016/j.expneurol.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto E, Mukozawa M, Mori H, Hara M. A rolled sheet of collagen gel with cultured Schwann cells: Model of nerve conduit to enhance neurite growth. J Biosci Bioeng. 2010 May;109(5):512–8. doi: 10.1016/j.jbiosc.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Davies SJA, Shih C-H, Noble M, Mayer-Proschel M, Davies JE, Proschel C. Transplantation of Specific Human Astrocytes Promotes Functional Recovery after Spinal Cord Injury. PLoS ONE. 2011 Mar 2;6(3):e17328. doi: 10.1371/journal.pone.0017328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volpato FZ, Führmann T, Migliaresi C, Hutmacher DW, Dalton PD. Using extracellular matrix for regenerative medicine in the spinal cord. Biomaterials. 2013 Jul;34(21):4945–55. doi: 10.1016/j.biomaterials.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 19.Houweling D, van Asseldonk JT, Lankhorst A, Hamers FP, Martin D, Bär P, et al. Local application of collagen containing brain-derived neurotrophic factor decreases the loss of function after spinal cord injury in the adult rat. Neurosci Lett. 1998 Jul 31;251(3):193–6. doi: 10.1016/s0304-3940(98)00536-9. [DOI] [PubMed] [Google Scholar]
- 20.Houweling DA, Lankhorst AJ, Gispen WH, Bär PR, Joosten EAJ. Collagen Containing Neurotrophin-3 (NT-3) Attracts Regrowing Injured Corticospinal Axons in the Adult Rat Spinal Cord and Promotes Partial Functional Recovery. Exp Neurol. 1998 Sep;153(1):49–59. doi: 10.1006/exnr.1998.6867. [DOI] [PubMed] [Google Scholar]
- 21.Macaya DJ, Hayakawa K, Arai K, Spector M. Astrocyte infiltration into injectable collagen-based hydrogels containing FGF-2 to treat spinal cord injury. Biomaterials. 2013 May;34(14):3591–602. doi: 10.1016/j.biomaterials.2012.12.050. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Xu J, Chan BP, Chew SY. Sustained release of neurotrophin-3 and chondroitinase ABC from electrospun collagen nanofiber scaffold for spinal cord injury repair. J Biomed Mater Res A. 2012;100A(1):236–42. doi: 10.1002/jbm.a.33271. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Houle JD, Xu J, Chan BP, Chew SY. Nanofibrous collagen nerve conduits for spinal cord repair. Tissue Eng Part A. 2012;18(9–10):1057–66. doi: 10.1089/ten.tea.2011.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kofron CM, Fong VJ, Hoffman-Kim D. Neurite outgrowth at the interface of 2D and 3D growth environments. J Neural Eng. 2009;6(1):016002. doi: 10.1088/1741-2560/6/1/016002. [DOI] [PubMed] [Google Scholar]
- 25.East E, de Oliveira DB, Golding JP, Phillips JB. Alignment of astrocytes increases neuronal growth in three-dimensional collagen gels and is maintained following plastic compression to form a spinal cord repair conduit. Tissue Eng Part A. 2010;16(10):3173–84. doi: 10.1089/ten.tea.2010.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009 Dec;32(12):638–47. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Hlady V, Tresco PA. Inducing alignment in astrocyte tissue constructs by surface ligands patterned on biomaterials. Biomaterials. 2012 Feb;33(5):1323–35. doi: 10.1016/j.biomaterials.2011.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alford PW, Nesmith AP, Seywerd JN, Grosberg A, Parker KK. Vascular smooth muscle contractility depends on cell shape. Integr Biol. 2011;3(11):1063–70. doi: 10.1039/c1ib00061f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Xiong S, Tay CY, Leong WS, Tan LP. A novel and simple microcontact printing technique for tacky, soft substrates and/or complex surfaces in soft tissue engineering. Acta Biomater. 2012 Mar;8(3):1267–72. doi: 10.1016/j.actbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Rape AD, Guo W, Wang Y. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32(8):2043–51. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polio SR, Rothenberg KE, Stamenović D, Smith ML. A micropatterning and image processing approach to simplify measurement of cellular traction forces. Acta Biomater. 2012 Jan;8(1):82–8. doi: 10.1016/j.actbio.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giannitelli SM, Abbruzzese F, Mozetic P, De Ninno A, Businaro L, Gerardino A, et al. Surface decoration of electrospun scaffolds by microcontact printing. Asia-Pac J Chem Eng. 2014 n/a–n/a. [Google Scholar]
- 33.Castano AG, Hortiguela V, Lagunas A, Cortina C, Montserrat N, Samitier J, et al. Protein patterning on hydrogels by direct microcontact printing: application to cardiac differentiation. RSC Adv. 2014;4(55):29120–3. [Google Scholar]
- 34.Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc. 2010 Mar;5(3):491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 35.Desai RA, Khan MK, Gopal SB, Chen CS. Subcellular spatial segregation of integrin subtypes by patterned multicomponent surfaces. Integr Biol. 2011;3(5):560–7. doi: 10.1039/c0ib00129e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichinger CD, Hsiao TW, Hlady V. Multiprotein Microcontact Printing with Micrometer Resolution. Langmuir. 2011 Dec 28;28(4):2238–43. doi: 10.1021/la2039202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. “Dip-Pen” Nanolithography. Science. 1999 Jan 29;283(5402):661–3. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 38.Calvert P. Inkjet Printing for Materials and Devices. Chem Mater. 2001 Sep 12;13(10):3299–305. [Google Scholar]
- 39.Von Philipsborn AC, Lang S, Bernard A, Loeschinger J, David C, Lehnert D, et al. Microcontact printing of axon guidance molecules for generation of graded patterns. Nat Protoc. 2006 Oct;1(3):1322–8. doi: 10.1038/nprot.2006.251. [DOI] [PubMed] [Google Scholar]
- 40.Elias PZ, Spector M. Viscoelastic characterization of rat cerebral cortex and type I collagen scaffolds for central nervous system tissue engineering. J Mech Behav Biomed Mater. 2012 Aug;12:63–73. doi: 10.1016/j.jmbbm.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980 Jun 1;85(3):890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci. 1979;76(1):514–7. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsiao TW, Swarup VP, Kuberan B, Tresco PA, Hlady V. Astrocytes specifically remove surface-adsorbed fibrinogen and locally express chondroitin sulfate proteoglycans. Acta Biomater. 2013 Jul;9(7):7200–8. doi: 10.1016/j.actbio.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kofron CM, Liu YT, López-Fagundo CY, Mitchel JA, Hoffman-Kim D. Neurite outgrowth at the biomimetic interface. Ann Biomed Eng. 2010;38(6):2210–25. doi: 10.1007/s10439-010-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarup VP, Hsiao TW, Zhang J, Prestwich GD, Kuberan B, Hlady V. Exploiting Differential Surface Display of Chondroitin Sulfate Variants for Directing Neuronal Outgrowth. J Am Chem Soc. 2013 Aug 15;135(36):13488–94. doi: 10.1021/ja4056728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schachtrup C, Ryu JK, Helmrick MJ, Vagena E, Galanakis DK, Degen JL, et al. Fibrinogen Triggers Astrocyte Scar Formation by Promoting the Availability of Active TGF-β after Vascular Damage. J Neurosci. 2010 Apr 28;30(17):5843–54. doi: 10.1523/JNEUROSCI.0137-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ereifej ES, Matthew HW, Newaz G, Mukhopadhyay A, Auner G, Salakhutdinov I, et al. Nanopatterning effects on astrocyte reactivity. J Biomed Mater Res A. 2013;101A(6):1743–57. doi: 10.1002/jbm.a.34480. [DOI] [PubMed] [Google Scholar]
- 48.Lau CL, Kovacevic M, Tingleff TS, Forsythe JS, Cate HS, Merlo D, et al. 3D Electrospun scaffolds promote a cytotrophic phenotype of cultured primary astrocytes. J Neurochem. 2014 doi: 10.1111/jnc.12702. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 49.Koch S, Yao C, Grieb G, Prevel P, Noah EM, Steffens GCM. Enhancing angiogenesis in collagen matrices by covalent incorporation of VEGF. J Mater Sci Mater Med. 2006;17(8):735–41. doi: 10.1007/s10856-006-9684-x. [DOI] [PubMed] [Google Scholar]