Abstract
Background
Osteoporosis, a disease characterized by low bone mineral density (BMD), is an important health problem in Mexico. BMD is a highly heritable trait, with heritability estimates of 50-85%. Several candidate genes have been evaluated to identify those involved in BMD variation and the etiology of osteoporosis. This study investigated the possible association of single-nucleotide polymorphisms (SNPs) in the MEF2C, SOST and JAG1 genes with bone mineral density (BMD) variation in postmenopausal Mexican-Mestizo women.
Methods
Four hundred unrelated postmenopausal women were included in the study. Risk factors were recorded and BMD was measured in total hip, femoral neck and lumbar spine using dual-energy X-ray absorptiometry. In an initial stage, a total of twenty-five SNPs within or near SOST gene and seven SNPs in the JAG1 gene were genotyped using a GoldenGate assay. In a second stage, three MEF2C gene SNPs were also genotyped and SOST and JAG1 gene variants were validated. Real time PCR and TaqMan probes were used for genotyping.
Results
Linear regression analyses adjusted by age, body mass index and ancestry estimates, showed that five SNPs in the SOST gene were significantly associated with BMD in total hip and femoral neck but not lumbar spine. The lowest p value was 0.0012, well below the multiple–test significance threshold (p = 0.009), with mean effect size of -0.027 SD per risk allele. We did not find significant associations between BMD and MEF2C/JAG1 gene variants [rs1366594 “A” allele: β = 0.001 (95% CI -0.016; 0.017), P = 0.938; rs2273061 “G” allele: β = 0.007 (95% CI -0.007; 0.023), p = 0.409].
Conclusions
SOST polymorphisms may contribute to total hip and femoral neck BMD variation in Mexican postmenopausal women. Together, these and prior findings suggest that this gene may contribute to BMD variation across populations of diverse ancestry.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2474-15-400) contains supplementary material, which is available to authorized users.
Keywords: Association study, MEF2C, SOST, JAG1, Genes, Mexican Women
Background
Osteoporosis (OP) is a common skeletal disease characterized by reduced bone mass, microarchitecture deterioration, and a reduction in bone mineral density (BMD), leading to increased bone fragility and susceptibility to fracture [1]. OP is a serious public health problem in Mexico causing up to 30,000 osteoporotic fractures a year, and an annual health service expenditure of about $97 million in 2006 [2]. While multiple risk factors influence the pathogenesis of osteoporosis, genetic factors play an important role in BMD variation. Heritability of this trait is high, as twin and family studies show that genetic factors account for approximately 50% to 85% of BMD [3].
Several candidate genes have been evaluated to assess their role in BMD variation and the etiology of osteoporosis, mainly in populations of European and Asian ancestry [4–6]. Research has focused on the Wnt/β-catenin signaling pathway, which is known to play a major role in bone remodeling [7]. The SOST (sclerostin) gene encodes the sclerostin protein, which binds to LRP5/6 co-receptors and antagonizes Wnt/b-catenin signaling in both osteocytes and osteoblasts [8, 9]. Several SOST single nucleotide polymorphisms (SNPs) have been associated with BMD variation [6, 10–12]. Moreover, the MEF2C (myocyte enhancer factor 2C) gene, another member of the Wnt-signaling pathway [13], is known to play an important role in determining bone density and mediating inflammatory effects in bone [14] and Mef2C is the main transcriptional factor responsible for ECR5-dependent Sost expression in the adult skeleton of mice [15]. Within this gene, rs1366594 has been associated with Femoral Neck (FN) BMD in Europeans, and with Total Hip (TH) BMD in populations from East-Asia [4, 5]; while rs119510131, was found to be significantly associated with forearm BMD in the meta-analysis of 6,584 individuals of European and Mexican American descent [16].
In another pathway, Jagged-1 has been found to induce human mesenchymal stem cell (hMSC) osteoblast differentiation through canonical Notch signaling [17]. A genome-wide association study (GWAS) reported that the rs2273061 polymorphism within the JAG1 (jagged 1) gene was associated with low Lumbar Spine (LS) BMD, FN BMD, and with osteoporotic fractures in subjects of European and Asian descent [18].
To date, most studies examining the relationship of SOST, MEF2C and JAG1 polymorphisms with BMD variation have been performed in populations of European and Asian ancestry [4–6, 10–12, 18]. The aim of this study was to analyze the possible association of polymorphisms in these genes with BMD variation in Mexican-Mestizo postmenopausal women.
Methods
Study population
The study group included only women born in Mexico whose parents and grandparents identified themselves as Mexican Mestizos. This study was performed as part of the third stage of a previously described, long-term cohort study [19, 20]. A total of 400 unrelated postmenopausal women over 45 years of age, without spontaneous menses for at least 1 year, attending the Instituto Mexicano del Seguro Social (IMSS) located in Cuernavaca, Morelos State, were recruited. Exclusion criteria were a history of bone, metabolic or endocrine disease, history of oophorectomy prior to 45 years of age, menopause prior to 40 years of age, or use of medication interfering with bone metabolism. Data on demographic characteristics and information regarding smoking status, menopausal status, estrogen use, medical history, and use of medication were obtained from self-administered questionnaires. The study was approved by the IMSS Research Ethics Committee and all participants provided signed informed consent. Blood samples were obtained and stored at 4°C until use. Genomic DNA was extracted from the peripheral blood of all participants, using a commercial isolation kit (QIAGEN systems Inc., Valencia, CA), according to the manufacturer’s instructions.
Measurement of bone mineral density
BMD at the lumbar spine (L2-4), femoral neck, and total hip were assessed using a dual X-ray absorptiometry (DXA) Lunar DPX NT instrument (Lunar Radiation Corp., Madison WI). BMD was calculated from bone mineral content (g) and bone area (cm2), and then expressed as g/cm2.
Genotyping and single nucleotide polymorphism selection
In an initial phase of the study, a GoldenGate platform with a total of 384 SNPs was used to test for associations with BMD, including twenty-five SOST and seven JAG1 SNPs [21]. These SNPs were selected using the following criteria: minor allele frequency (MAF) >5% in Caucasians, and polymorphisms previously associated with BMD in candidate gene association studies, genome wide association studies (GWAS) and meta-analyses [4, 6, 10–12, 18]. Genotype data were extracted for further analysis.
In the present study, we extended the analysis to seek other possible associations between SNPs and BMD variation in another member of the Wnt signaling pathway, the MEF2C gene, analyzing three SNPs (rs12521522, rs11951031 and rs13665949). Furthermore, we genotyped five SOST SNPs (rs9911277, rs4793018, rs1983490, rs1881107 and rs4792909), and three JAG1 SNPs (rs2273061, rs6040061 and rs2235811) in 30% of randomly selected samples. Genotyping of these eleven SNPs was performed using commercial predesigned TaqMan Probes (Applied Biosystems, Foster City, CA, USA) in a StepOne Plus RT PCR system. Information on the parental frequencies and genotype data for a panel of 96 Ancestry Informative Markers (AIMs) was extracted from the initial phase genotypes, in order to control for the effect of false associations due to population stratification.
Statistical analyses
All data from the population in the study are showed as mean ± SD (standard deviation) for quantitative variables and absolute and relative frequencies for qualitative variables. Hardy-Weinberg equilibrium was tested for each SNP using the standard χ2 test. Linear regression analyses were used in order to test for associations between BMD and genotype using an additive genetic model adjusting for age and body mass index (BMI). Ancestry from principal component analysis (PCA) was estimated using the smartpca program in the Eigensoft 3.0 package [22], and ancestry estimates were included as confounding factors for correction of population stratification. All statistical analyses were performed with Statistical Package for Social Sciences software (SPSS 20.0; SPSS Inc.; Chicago, IL, USA). P-values <0.05 were considered statistically significant. Linkage disequilibrium (LD) and haplotype frequencies were estimated using Haploview 4.2 [23]. Statistical power was calculated with Quanto 1.1 software, for a significance level of 0.05 and MAF of 5% in 400 postmenopausal women with a minimal power of 80% to detect differences in BMD, under an additive model. The significance threshold after multiple test correction for each gene was estimated using the single nucleotide spectral decomposition software (SNPSpD) [24]. This approach can be applied to obtain an effective number of independent marker loci (Meff). The application of SNPSpD to our nine SNP sample set gave a Meff of 5.48 SNPs, leading to a significance threshold of 0.05/5.48 = 0.009.
Results
The quality control (QC) criteria for subject and SNP genotyping for the initial phase have been previously described [21]. Briefly, data from 14 individuals whose DNA did not genotype successfully were excluded and 11 more participants were discarded from subsequent analysis because their African ancestry exceeded the mean reported for the Mexican mestizo population [25]. In the second phase, the overall genotype call rate was 98%, and the genotypes showed 100% reproducibility with those obtained in the initial phase. Two MEF2C SNPs (rs12521522 and rs11951031) were not observed in this sample of Mexican women, and were thus excluded from the analysis.
The general characteristics of the participating women are presented in Table 1. Mean age was 62.25 ± 9.11 years, and a mean BMI was 28.08 ± 4.76 kg/m2. Mean bone parameters were within normal range, although mean LS BMD was 0.990 ± 0.151 (mild osteopenia), which is expected given the age of study participants.
Table 1.
Demographic characteristics and BMD of postmenopausal Mexican Mestizo women
| Variable | Mean (SD) |
|---|---|
| Age (Yr) | 62.25 (9.11) |
| Height (cm) | 152.98 (5.74) |
| Weight (Kg) | 65.73 (11.74) |
| BMIa (Kg/m2) | 28.08 (4.76) |
| BMDb total hip (g/cm2) | 0.929 (0.133) |
| BMDb femoral neck (g/cm2) | 0.880 (0.125) |
| BMDb lumbar spine (g/cm2) | 0.990 (0.151) |
| Age of Menarche | 12.94 (1.55) |
| Number of children | 3.12 (2.13) |
| Duration of breastfeeding (months) | 15.90 (19.86) |
| Years since menopause | 17.84 (11.59) |
| Estrogen remplacement therapyc | 110 (26.00) |
| Tobacco usec | 137 (34.33) |
| Alcohol intakec | 45 (11.26) |
| Carbonated beverage consumptionc | 315 (78.75) |
N = 400 Postmenopausal women.
a BMI = Body Mass Index.
b BMD = Bone Mineral Density.
cn (%); n, number of women; SD, standard deviation.
In the initial stage, twenty-five SOST and seven JAG1 SNPs were tested for single-marker allelic association, adjusting for age, body mass index (BMI) and ancestry estimates. In the second phase, five SOST and three JAG1 SNPs showing significant associations with BMD were further analyzed. SNP details are described in Table 2. Genotype and allele frequency distributions of all SNPs did not differ from Hardy-Weinberg equilibrium (Table 3).
Table 2.
Information on the SNPs analyzed in this study
| Gene | SNP | Allele | Position | Location in the gene |
|---|---|---|---|---|
| MEF2C | rs1366594 | C/A | 5:88376061 | Intergenic |
| SOST | rs9911277 | G/A | 17:41780484 | Intergenic |
| rs4793018 | C/T | 17:41781069 | Intergenic | |
| rs1983490 | G/A | 17:41782957 | Intergenic | |
| rs1881107 | A/G | 17:41786076 | Intergenic | |
| rs4792909 | G/T | 17:41798824 | Intergenic | |
| JAG1 | rs2273061 | A/G | 20:10639543 | Intron variant |
| rs6040061 | A/C | 20:10640306 | Intron variant | |
| rs2235811 | C/T | 20:10644158 | Intron variant |
Table 3.
Genotype/allele frequencies and Hardy-Weinberg equilibrium (HWE) P values for the analyzed SNPs
| aSNP reference number | Genotype | N | % | bHWE |
|---|---|---|---|---|
| rs1366594 MEF2C | C/C | 186 | 46.6 | p = 0.30 |
| C/A | 166 | 41.6 | ||
| A/A | 47 | 11.8 | ||
| Allele frequency | C | 538 | 67.4 | |
| A | 260 | 32.6 | ||
| rs9911277 SOST | G/G | 113 | 28.6 | p = 0.70 |
| G/A | 193 | 48.9 | ||
| A/A | 89 | 22.5 | ||
| Allele frequency | G | 419 | 53.0 | |
| A | 371 | 47.0 | ||
| rs4793018 SOST | C/C | 117 | 29.5 | p = 0.58 |
| C/T | 192 | 48.4 | ||
| T/T | 88 | 22.2 | ||
| Allele frequency | C | 426 | 53.7 | |
| T | 368 | 46.3 | ||
| rs1983490 SOST | G/G | 121 | 30.3 | p = 0.55 |
| G/A | 192 | 48.1 | ||
| A/A | 86 | 21.6 | ||
| Allele frequency | G | 434 | 54.4 | |
| A | 364 | 45.6 | ||
| rs1881107 SOST | A/A | 122 | 30.7 | p = 0.43 |
| A/G | 189 | 47.6 | ||
| G/G | 86 | 21.7 | ||
| Allele frequency | A | 433 | 54.5 | |
| G | 361 | 45.5 | ||
| rs4792909 SOST | G/G | 116 | 29.2 | p = 0.27 |
| G/T | 187 | 47.1 | ||
| T/T | 94 | 23.7 | ||
| Allele frequency | G | 419 | 52.8 | |
| T | 375 | 47.2 | ||
| rs2273061 JAG1 | A/A | 132 | 33.2 | p = 0.43 |
| A/G | 201 | 50.5 | ||
| G/G | 65 | 16.3 | ||
| Allele frequency | A | 465 | 58.4 | |
| G | 331 | 41.6 | ||
| rs6040061 JAG1 | A/A | 147 | 37.1 | p = 0.21 |
| A/C | 198 | 50.0 | ||
| C/C | 51 | 12.9 | ||
| Allele frequency | A | 492 | 62.1 | |
| C | 300 | 37.9 | ||
| rs2235811 JAG1 | C/C | 151 | 38.1 | p = 0.43 |
| C/T | 193 | 48.7 | ||
| T/T | 52 | 13.1 | ||
| Allele frequency | C | 495 | 62.5 | |
| T | 297 | 37.5 |
a SNP = single nucleotide polymorphism; b HWE = Hardy-Weinberg Equilibrium.
In the second phase, linear regression analyses adjusted by age, BMI, and ancestry estimates showed that SOST gene SNPs rs9911277, rs4793018, rs1983490, rs1881107 and rs4792909 were significantly associated with TH-BMD and NF-BMD (mean effect size of -0.027 per risk allele; lowest p value = 0.0012) but not with LS-BMD (Table 4). MEF2C gene rs1366594 failed to show a significant association with any BMD estimation (p value range 0.938 - 0.542). On the other hand, JAG1 gene SNPs rs2235811 and rs2275811 showed a trend to association with FN-BMD and LS-BMD (p = 0.051 and 0.089, respectively); well above the multiple test-significance threshold estimated at 0.009. After correction for multiple testing, only the association of BMD with SOST polymorphisms remained significant.
Table 4.
Association of MEF2C, SOST and JAG1 polymorphisms with BMD in postmenopausal Mexican-women
| Gene | dbSNP | β (95% IC)d | p adjust e | |
|---|---|---|---|---|
| BMD-TH a (g/ cm 2 ) | MEF2C | rs1366594 | 0.001 (-0.016; 0.017) | 0.938 |
| SOST | rs9911277 | -0.031 (-0.046; -0.016) | 0.000072 | |
| rs4793018 | -0.027 (-0.043; -0.012) | 0.00035 | ||
| rs1983490 | -0.027 (-0.042; -0.012) | 0.00040 | ||
| rs1881107 | -0.027 (-0.042; -0.011) | 0.00055 | ||
| rs4792909 | -0.025 (-0.040; -0.010) | 0.0012 | ||
| JAG1 | rs2273061 | 0.007 (-0.009; 0.023) | 0.409 | |
| rs6040061 | 0.009 (-0.007; 0.026) | 0.280 | ||
| rs2235811 | 0.011 (-0.005; 0.027) | 0.189 | ||
| BMD-FN b (g/ cm 2 ) | MEF2C | rs1366594 | 0.004 (-0.011; 0.019) | 0.638 |
| SOST | rs9911277 | -0.029 (-0.043; -0.015) | 0.000048 | |
| rs4793018 | -0.026 (-0.040; -0.012) | 0.00024 | ||
| rs1983490 | -0.027 (-0.041; -0.013) | 0.00018 | ||
| rs1881107 | -0.026 (-0.040; -0.012) | 0.00027 | ||
| rs4792909 | -0.025 (-0.039; -0.011) | 0.00053 | ||
| JAG1 | rs2273061 | 0.007 (-0.008; 0.022) | 0.331 | |
| rs6040061 | 0.013 (-0.003; 0.028) | 0.109 | ||
| rs2235811 | 0.013 (-0.002; 0.028) | 0.089 | ||
| BMD-LS c (g/ cm 2 ) | MEF2C | rs1366594 | 0.007 (-0.015; 0.028) | 0.542 |
| SOST | rs9911277 | -0.006 (-0.027; 0.014) | 0.539 | |
| rs4793018 | -0.007 (-0.027; 0.013) | 0.498 | ||
| rs1983490 | -0.007 (-0.027; 0.014) | 0.514 | ||
| rs1881107 | -0.003 (-0.023; 0.018) | 0.806 | ||
| rs4792909 | -0.002 (-0.022; 0.019) | 0.865 | ||
| JAG1 | rs2273061 | 0.021 (0.000; 0.042) | 0.051 | |
| rs6040061 | 0.008 (-0.014; 0.030) | 0.484 | ||
| rs2235811 | 0.007 (-0.015; 0.028) | 0.557 |
aBMD-TH = Total hip bone mineral density.
bBMD-FN = Femoral neck bone mineral density.
cBMD-LS = Lumbar spine bone mineral density.
dData are shown for the additive model.
e p-adjust values were adjusted by age, BMI, and ancestry estimates.
Linkage disequilibrium analysis revealed significant relationships among the SOST gene polymorphisms, identifying a single high LD block (pair-wise r2 values >0.89; Figure 1). Significant LD was observed across the gene, and we inferred the existence of two main haplotypes: ACAC and CACA, with frequencies of 0.49 and 0.48, respectively.
Figure 1.
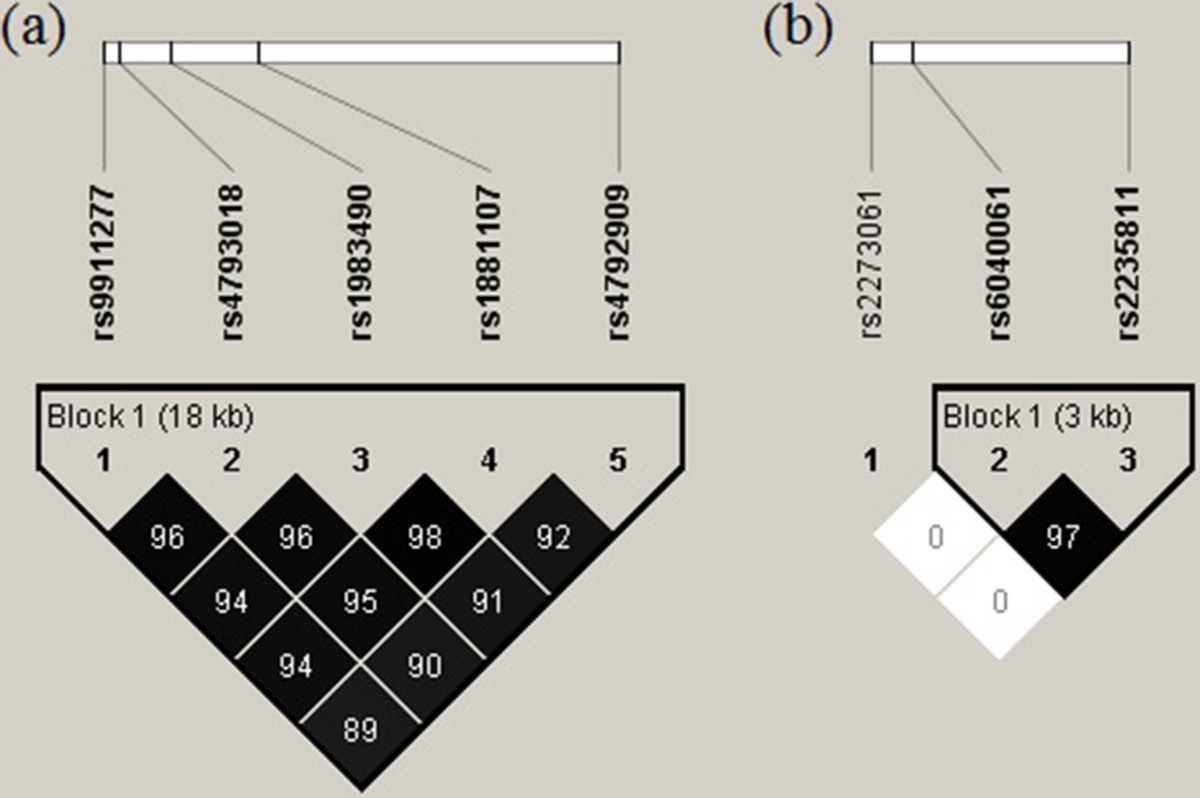
Location and pair-wise linkage disequilibrium values of SOST and JAG1 polymorphisms in Mexican postmenopausal women. Darker color indicates higher LD and lighter color indicates less LD. Values of the pair-wise (r2) are shown to SOST (a) and JAG1 (b) genes.
Discussion
OP constitutes a serious and increasing public health problem in the Mexican population [2]. To date, GWAS and candidate gene studies have revealed 70 genetic loci associated with BMD variation, but these studies have been performed mostly in populations of European and Asian ancestry [5, 6]. In the present study, we analyzed the association of polymorphisms of the MEF2C, SOST and JAG1 genes with BMD variation in postmenopausal Mexican-mestizo women.
We found that SNPs of the SOST gene were associated with total hip and femoral neck, but not with lumbar spine BMD. These results are consistent with recent genome-wide association studies [4–6, 11], and may be explained at least partly by a lower heritability for BMD in the femoral neck site. This is supported by a previous study reporting that hip BMD of grandmothers is better predictor of hip BMD in mothers (beta = 0.46); while maternal LS BMD was less predictive of LS BMD in grandchildren (beta = 0.30) [19].
Together, our findings support the association of Wnt signaling pathway gene polymorphisms with BMD variation in the Mexican-mestizo population [21]. SOST is a known Wnt signaling inhibitor, which acts suppressing LRP function. In vitro, SOST inhibits osteoblast development and is expressed primarily in osteocytes [26]. SOST polymorphisms have been associated with BMD at different skeletal sites and with osteoporotic fractures in various populations [10–12]. Our present finding of a SOST-BMD association in the Mexican-mestizo population is in agreement with a similar association found by Estrada et al. [6] and Richards et al. [11] in postmenopausal women of European ancestry.
The MAFs of SOST gene SNPs rs9911277, rs4793018, rs1983490, and rs1881107 observed in the present study were very similar to those reported in the HapMap in individuals of Mexican ancestry of Los Angeles (0.45 to 0.47), but were less frequent than in European populations (MAF: 0.35 to 0.38). On the other hand, the rs4792909 “G” allele, most widely reported and associated with fracture risk in a large meta-analysis, was less frequent in our population (0.53) than in Europeans (0.62) [6], but very similar to that observed in a Los Angeles population of Mexican ancestry (0.53), (http://hapmap.ncbi.nlm.nih.gov).
Conversely, we did not find the associations between specific MEF2C and JAG1 SNPs and BMD variation previously identified in populations of European and Asian ancestry [4, 5, 18]. The MEF2C rs1366594 “A” allele was not associated with BMD variation in the present study, and was less frequent in Mexican postmenopausal women (0.33) than in Europeans (0.55), East-Asians (0.42) and Han Chinese individuals (0.40) [4, 5, 27]. Moreover, the MEF2C rs11951031 previously associated with forearm BMD in Europeans and individuals of Mexican-American descent was not informative in this sample of Mexican menopausal women. This may be partially explained by our sample size being too small to detect a risk allele with a frequency of 0.06 [16].
Similarly, we failed to observe associations of JAG1 polymorphisms with BMD variation. JAG1 rs2273061 was initially found to be associated with BMD variation (LS and FN) in a GWAS performed in Hong Kong Chinese individuals, and this association was afterwards confirmed in several European populations [18]. However, this SNP was not associated with BMD variation in a Mexican-Mestizo cohort [28], in consistency with our present observations. Moreover, the JAG1 rs2273062 allelic and genotypic frequencies reported here are virtually identical to those described previously in Mexicans [28], and the “G” allele frequency (0.42) was similar to that reported in Europeans (0.40), but higher than in the population of Hong Kong (0.31) [18]. The rs2235811 and rs6040061 MAFs observed in Mexican postmenopausal women were similar to those published in the dbSNP database for a population of Asian descent (http://www.ncbi.nlm.nih.gov/SNP/). However, it is notable that although Kung et al. reported that rs2273061, rs6040061 and rs2235811 are in high LD in individuals of both Chinese (r2 > 0.9) and European (r2 > 0.7, CEU) descent [18], in our population rs2273061 was clearly not in LD with the other JAG1 polymorphisms (Figure 1). This may explain why rs2273061 was not associated with BMD in this study. Further studies including other JAG1 SNPs will define whether this gene is associated with BMD or not in the Mexican population.
The Mexican population is admixed and has a complex genetic structure including European, Native American and a small proportion of African genes [29, 30]. Spurious association signals produced by differences in ancestral background can be a confounding factor in genetic association studies [31, 32]. Since population stratification may be a source of spurious associations, we used the first and second principal component from PCA, described previously [21] to account for population stratification. Our results suggest that population stratification was most likely not a confounding factor in this study.
Our study has some limitations. First, MEF2C, JAG1 and SOST SNPS were selected based on previous reports from European and Asian populations, and thus other SNPs within the same genes may contribute to BMD variation in Mexican-mestizos. Second, the failure to observe associations of MEF2C and JAG1 SNPs with BMD in our population may be due to the small sample size and reduced statistical power of the study [33]. Although our sample size (400 individuals) was large enough to identify the effect of SOST gene variants on BMD, in order to replicate the previously observed association of MEF2C rs1366594, considering a MAF of 0.33 and an effect size of 0.08 (in standard deviations of BMD), a total of 2,769 samples would be needed to achieve 80% statistical power to detect a significant association. The statistical power of the present study to find a significant association with this SNP was only 19%. Similarly, we would require 6,909 samples to obtain adequate statistical power to detect an association of JAG1 rs2273061 with BMD variation, considering a MAF of 0.42 and an effect size of 0.048 SDs. Thus, even the combined results of the present study with those of Rojano-Mejía et al., who genotyped the rs2273061 in 750 Mexican individuals, would not reach adequate statistical power to replicate the association of this SNP with BMD. These results should be interpreted with caution, and should be considered as uncertain rather than null associations. Additional studies in larger samples are required to understand the role of MEF2C and JAG1 gene variation in BMD variation in the Mexican population.
Conclusions
In conclusion, our results suggest that SOST polymorphisms contribute to total hip and femoral neck BMD variation in postmenopausal Mexican-mestizo women. Taken together, these and other previously reported findings in the Mexican population suggest that variation in genes located in the Wnt pathway may be important contributors to BMD variation in the Mexican-mestizo population. Further investigation is required to understand interactions of genes located in this pathway with other factors, such as hormones and nutrition.
Acknowledgements
The authors would like to thank all the study participants for their participation in this study. Grants from the Consejo Nacional de Ciencia y Tecnología (SALUD-2008-C01-87331) and INMEGEN (CI/26/2010) supported this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RVC participated in the conception and design of the study, and wrote the manuscript. RFJO, AYPT and NP performed genotyping, interpreted and analyzed the data. MCL contributed with statistical expertise. MQ is involved in biological and anthropometric measurements. TVM guided and critically revised the manuscript. JS obtained funding for recruitment of individuals of the cohort. All authors critically revised the manuscript and approved the final version.
Contributor Information
Rafael Velázquez-Cruz, Email: rvelazquez@inmegen.gob.mx.
Rogelio F Jiménez-Ortega, Email: frankunam_1@hotmail.com.
Alma Y Parra-Torres, Email: yoli_379@hotmail.com.
Manuel Castillejos-López, Email: mcastillejos@gmail.com.
Nelly Patiño, Email: lnpatino@inmegen.gob.mx.
Manuel Quiterio, Email: mquitero@insp.mx.
Teresa Villarreal-Molina, Email: mvillareal@inmegen.gob.mx.
Jorge Salmerón, Email: jsalme@prodigy.net.mx.
References
- 1.NIH Consensus Development Panel on osteoporosis prevention, diagnosis, and therapy Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285(6):785–795. doi: 10.1001/jama.285.6.785. [DOI] [PubMed] [Google Scholar]
- 2.Clark P, Fernando C, Vazquez JL. Epidemiology, costs and burden of osteoporosis in Mexico. Arch Osteoporos. 2010;8(5):9–17. doi: 10.1007/s11657-010-0042-8. [DOI] [Google Scholar]
- 3.Özbaş H, Tutgun S, Özdamar K. Genetic and environmental factors in human osteoporosis. Mol Biol Rep. 2012;39(12):11289–11296. doi: 10.1007/s11033-012-2038-5. [DOI] [PubMed] [Google Scholar]
- 4.Rivadeneira F, Styrkársdottir U, Estrada K, Halldórsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HA, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FM, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, Tang NL, Koh JM, Xiao SM, Kwok TC, Kim GS, Chan JC, Cherny S, Lee SH, Kwok A, Ho S, Gretarsdottir S, Kostic JP, Palsson ST, Sigurdsson G, Sham PC, Kim BJ, Kung AW, Kim SY, Woo J, Leung PC, Kong A, Thorsteinsdottir U, Stefansson K. European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS One. 2010;5(10):e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–503. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monroe DG, McGee ME, Oursler MJ, Westendorf JJ. Update on Wnt signaling in bone cell biology and bone disease. Gene. 2012;492(1):1–18. doi: 10.1016/j.gene.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang MK, Kramer I, Keller H, Gooi JH, Collett C, Jenkins D, Ettenberg SA, Cong F, Halleux C, Kneissel M. Reversing LRP5-dependent osteoporosis and SOST deficiency-induced sclerosing bone disorders by altering WNT signaling activity. J Bone Miner Res. 2014;29(1):29–42. doi: 10.1002/jbmr.2059. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Li Y, Zhou Z, Wu C, Xing Y, Zou X, Tian W, Zhang C. HIF-1α inhibits Wnt signaling pathway by activating Sost expression in osteoblasts. PLoS One. 2013;8(6):e65940. doi: 10.1371/journal.pone.0065940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims AM, Shephard N, Carter K, Doan T, Dowling A, Duncan EL, Eisman J, Jones G, Nicholson G, Prince R, Seeman E, Thomas G, Wass JA, Brown MA. Genetic analyses in a sample of individuals with high or low BMD shows associatin with multiple Wnt pathway genes. J Bone Miner Res. 2008;23(4):499–506. doi: 10.1359/jbmr.071113. [DOI] [PubMed] [Google Scholar]
- 11.Richards JB, Kavvoura FK, Rivadeneira F, Styrkársdóttir U, Estrada K, Halldórsson BV, Hsu YH, Zillikens MC, Wilson SG, Mullin BH, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra BA, Pols HA, Sigurdsson G, Thorsteinsdottir U, Soranzo N, Williams FM, Zhou Y, Ralston SH, Thorleifsson G, Van CM, Kiel DP, et al. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann Intern Med. 2009;151(8):528–537. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valero C, Zarrabeitia MT, Hernández JL, Pineda B, Cano A, García MA, Riancho JA. Relationship of sclerostin and secreted frizzled protein polymorphisms with bone mineral density: an association study with replication in postmenopausal women. Menopause. 2011;18(7):802–807. doi: 10.1097/gme.0b013e3182091664. [DOI] [PubMed] [Google Scholar]
- 13.Kramer I, Baertschi S, Halleux C, Keller H, Kneissel M. Mef2c deletion in osteocytes result in increased bone mass. J Bone Miner Res. 2012;27(2):360–373. doi: 10.1002/jbmr.1492. [DOI] [PubMed] [Google Scholar]
- 14.Johnson ME, Deliard S, Zhu F, Xia Q, Wells AD, Hankenson KD, Grant S. A ChIP-seq-defined genome-wide map of MEF2C binding reveals inflammatory pathways associated with its role in bone density determination. Calcif Tissue Int. 2014;94(4):396–402. doi: 10.1007/s00223-013-9824-5. [DOI] [PubMed] [Google Scholar]
- 15.Collete NM, Genetos DC, Economides AN, Xie L, Shahnazari M, Yao W, Lane NE, Harland RM, Loots GG. Targeted deletion of Sost distal enhancer increases bone formation and bone mass. Proc Natl Acad Sci U S A. 2012;109(35):14092–14097. doi: 10.1073/pnas.1207188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng HF, Duncan EL, Yerges LM, Eriksson J, Bergström U, Leo PJ, Leslie WD, Goltzman D, Blangero J, Hanley DA, Carless MA, Streeten EA, Lorentzon M, Brown MA, Spector TD, Pettersson U, Ohlsson C, Mitchell BD, Richards JB. Meta-analysis of genome-wide studies identifies MEF2C SNPs associated with bone mineral density at forearm. J Med Genet. 2013;50(7):473–478. doi: 10.1136/jmedgenet-2012-101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta/Jagged1-activated Notch1 signaling. J Biol Chem. 2005;280(16):15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- 18.Kung AW, Xiao SM, Cherny S, Li GH, Gao Y, Tso G, Lau KS, Luk KD, Liu JM, Cui B, Zhang MJ, Zhang ZL, He JW, Yue H, Xia WB, Luo LM, He SL, Kiel DP, Karasik D, Hsu YH, Cupples LA, Demissie S, Styrkarsdottir U, Halldorsson BV, Sigurdsson G, Thorsteinsdottir U, Stefansson K, Richards JB, Zhai G, Soranzo N, et al. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 2010;86(2):229–239. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazcano E, Tamayo J, Diaz R, Burquete A, Salmeron J. Correlation trends for bone mineral density in Mexican women: evidence of familiar predisposition. Salud Publica Mex 51 Suppl. 2009;5(1):93–99. doi: 10.1590/S0036-36342009000700012. [DOI] [PubMed] [Google Scholar]
- 20.Denova E, Castañón S, Talavera JO, Flores M, Macías N, Rodríguez S, Flores YN, Salmerón J. Dietary patterns are associated with different indexes of adiposity and obesity in an urban Mexican population. J Nutr. 2011;141(5):921–927. doi: 10.3945/jn.110.132332. [DOI] [PubMed] [Google Scholar]
- 21.Velázquez-Cruz R, Garcia H, Castillejos M, Quiterio M, Valdés M, Orozco L, Villareal T, Salmerón J. WNT3A gene polymorphisms are associated with bone mineral density variation in postmenopausal mestizo women of an urban Mexican population: findings of a pathway-based high-density single nucleotide screening. Age (Dordr) 2014;36(3):1483–1492. doi: 10.1007/s11357-014-9635-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2004;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Nyholt R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva I, Hidalgo A, Estrada J, Fernandez JC, Uribe L, Contreras A, Balam E, del Bosque L, Velazquez D, Lara C, Goya R, Hernandez E, Davila C, Barrientos E, March S, Jimenez G. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A. 2009;106(21):8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semënov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280(29):26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 27.Hua YH, Zhao L, Zhang MJ, Pan MJ, Pan CM, Zhao SX, Zhao HY, Sun LH, Tao B, Song HD, Wang WQ, Ning G, Liu JM. The influence of the genetic and non-genetic factors on bone mineral density and osteoporotic fractures in Chinese women. Endocrine. 2013;43(1):127–135. doi: 10.1007/s12020-012-9726-8. [DOI] [PubMed] [Google Scholar]
- 28.Rojano D, Coral RM, Espinosa LC, López G, Aguirre MC, Coronel A, Canto P. JAG1 and COL1A1 polymorphisms and haplotypes in relation to bone mineral density variations in postmenopausal Mexican-Mestizo Women. Age (Dordr) 2013;35(2):471–478. doi: 10.1007/s11357-011-9363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz A, Groop L, Saetta AA, Korkolopoulou P, Seligsohn U, Waliszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein DB, Reich D, Hirschhorn JN, Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80(6):1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno A, Gignoux CR, Fernández JC, Zakharia F, Sikora M, Contreras AV, Acuña V, Sandoval K, Eng C, Romero S, Ortiz P, Robles V, Kenny EE, Nuño I, Barquera R, Macín G, Granados J, Huntsman S, Galanter JM, Via M, Ford JG, Chapela R, Rodriguez W, Rodríguez JR, Romieu I, Sienra JJ, del Rio NB, London SJ, Ruiz A, Garcia R, et al. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344(6189):1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardon L, Palmer L. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 32.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36(5):512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 33.Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 2012;10(2):117–122. doi: 10.5808/GI.2012.10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2474/15/400/prepub


