Abstract
It is controversial whether dietary fiber protects against colorectal cancer because of conflicting results from human epidemiologic studies. However, these studies and mouse models of colorectal cancer have not controlled the composition of gut microbiota, which ferment fiber into short-chain fatty acids such as butyrate. Butyrate is noteworthy because it has energetic and epigenetic functions in colonocytes and tumorsuppressive properties in colorectal-cancer cell lines. We utilized gnotobiotic mouse models colonized with wild-type or mutant strains of a butyrate-producing bacterium to demonstrate that fiber does have a potent tumor-suppressive effect but in a microbiota- and butyrate-dependent manner. Furthermore, due to the Warburg effect, butyrate was metabolized less in tumors where it accumulated and functioned as an HDAC inhibitor to stimulate histone acetylation and affect apoptosis and cell proliferation. To support the relevance of this mechanism in human cancer, we demonstrate that butyrate and histone-acetylation levels are elevated in colorectal adenocarcinomas compared to normal colonic tissues.
INTRODUCTION
It is highly controversial whether dietary fiber protects against colorectal cancer (CRC) because of conflicting results from human cohort-based epidemiologic studies (1-5). These studies have been complicated by the participants’ genetic heterogeneity, differences in the composition of their gut microbiota, and the utilization of different fiber sources (6-8). It also has not been established how dietary fiber might protect against CRC, but two general mechanisms have been proposed that are not mutually exclusive. First, insoluable fiber bulks luminal contents and may speed colonic transit to minimize the exposure of the colonic epithelium to ingested carcinogens such as nitrosamines from charred meat. Second, bacteria in the lumen of the colon ferment soluable fiber into short-chain fatty acids (SCFAs) and other metabolites with potentially beneficial properties. Butyrate is an abundant (up to >10 mM) SCFA that is transported into the colonic epithelium and localizes within two subcellular compartments (9,10). It undergoes β-oxidation inside the mitochondria and accounts for ≥70% of the energy used by normal colonocytes (11), and it also functions as a histone deacetylase (HDAC) inhibitor inside the nucleus to epigenetically regulate gene expression (12). Butyrate is a plausible candidate for tumor suppression because it inhibits cell proliferation and induces cell differentiation or apoptosis when added to tumor-derived cell lines (9,10). However, butyrate can also have contradictory effects (13,14), and it is crucial that we move beyond “factor dump” experiments where relatively high doses of butyrate are added to CRC cell lines. It must be demonstrated that dietary fiber and gut microbiota can modulate butyrate levels in the colonic lumen, and that this, in turn, can inhibit colorectal tumorigenesis in vivo where the colonic crypt architecture is intact and functions in the presence of stromal cells.
RESULTS
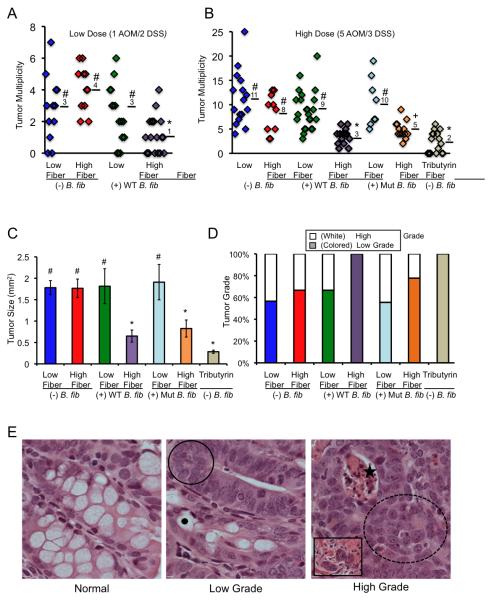
Our experiments were designed to test the hypothesis that dietary fiber protects against CRC in a microbiota- and butyrate-dependent manner (Supplementary Fig. S1). To control the genetics and microbiota in our experiments, we maintained a colony of BALB/c inbred mice polyassociated with 4 commensal bacteria [a subset of the altered Schaedler flora or ASF (15)] plus or minus the butyrate-producing bacterium Butyrivibrio fibrisolvens (16) in gnotobiotic isolators (Supplementary Figs. S1 and S2). These mice were provided low- or high-fiber (6% fructo-oligosaccharide/inulin) diets that were otherwise identical and calorically matched (Supplementary Figs. S1 and S3). As expected, mice colonized with the ASF plus B. fibrisolvens that were also provided high-fiber (“prebiotic”) diets (herein referred to as the experimental group) had significantly higher luminal butyrate levels than the other 3 groups of mice (herein referred to as the control groups) based on LC-MS measurements (Supplementary Fig. S4). This difference was not observed for acetate or propionate, which are the other two major SCFAs (Supplementary Table 1). We treated these mice with azoxymethane (AOM) and dextran sodium sulfate (DSS) to induce CRC and analyzed them 5 months later (17). Following 1 injection of AOM and 2 cycles of DSS, the experimental group had significantly fewer tumors than each control group. The experimental group had a mean of 1 tumor per mouse, whereas the control groups had 3-4 tumors per mouse (Fig. 1A). These results indicate that neither B. fibrisolvens nor high fiber had a protective effect on their own, whereas they did have a protective effect when combined.
Figure 1.
A high-fiber diet protects against colorectal tumors in a microbiota- and butyrate-dependent manner. (A, B) Scatter plots showing tumor multiplicity for mice in each treatment group following low (A) and high (B) carcinogen exposures. Means are shown, and symbols denote groups with statistically-significant differences: * versus #, p < 0.01; * versus +, p = 0.05; # versus +, p < 0.05. (C) Tumor size presented as mean ± SE with significant differences indicated (* versus #, p < 0.01). (D) Percentage of low-grade versus high-grade tumors for each treatment group based on H&E characteristics of dysplasia within tumors as described in panel E. (E) H&E-stained sections representative of normal colonic epithelium, low-grade tumors, and high-grade tumors. Compared to normal colon, low-grade tumors exhibit hyperplasia (circled region) and are less differentiated with fewer goblet cells that are identified by mucous vacuoles (black dot). High-grade tumors exhibit these features plus a loss of polarity (dotted circled region) and have crypt abscesses with apoptotic material (star). A subset of high-grade tumors display signs of potential invasion based on tumor cells within the muscularis (inset).
Next, we treated mice with 5 injections of AOM and 3 cycles of DSS, which increased the tumor incidence and multiplicity as expected (Fig. 1B). More importantly, the B. fibrisolvens and high-fiber diet still had a protective effect in combination (3 tumors per mouse), but not in isolation (8-11 tumors per mouse) (Fig. 1B). The experimental group also had significantly smaller (Fig. 1C) and less advanced (Fig. 1D-E) tumors based on their histopathologic characteristics (18). Tumors from the experimental and control groups had β-catenin gain-of-function mutations (Supplementary Fig. S5A-S5C) and were positive for nuclear β-catenin staining (Supplementary Fig. S6), which indicates that the mechanism of tumor initiation was due to aberrant Wnt signaling. These findings were expected based on previous AOM studies and confirm that the mechanism of tumor initiation is similar to human CRC (17). For the 5 AOM/3 DSS dose regimen, we also identified squamous cell tumors derived from the surface ectoderm or glandular epithelium of the peri-anal region rather than the colonic mucosa (Supplementary Fig. S7A-S7B). In contrast to the colorectal tumors, the incidence, size, and histopathologic progression of the squamous tumours were not significantly different between the experimental and control groups (Supplementary Fig. S7A-S7B). Therefore, the squamous tumors, which are known to occur in the AOM/DSS model (17), served as an internal control. They demonstrate that mice from the different treatment groups received the same exposure to carcinogen, as expected, and that the tumor-suppressive effect of the fiber-microbiota-butyrate axis is specifically localized to the colonic mucosa and does not influence tumor formation outside of the lumen even at nearby sites. This dichotomy is compatible with the bioavailability of butyrate being primarily restricted to the colonic mucosa (9,10).
To confirm that the tumor-suppressive mechanism for B. fibrisolvens and high fiber is due to butyrate production, we exploited a mutant B. fibrisolvens strain with a 0.8-kb deletion in the butyryl-CoA synthesis operon (16) that produces ≤5-fold less butyrate when cultured (19) (Supplementary Fig. S8A-S8C). To validate this deficiency in butyrate production, we cultured the mutant and wild-type B. fibrisolvens strains in media supplemented with fructo-oligosaccharides/inulin (the same fiber source provided to the mice) and observed 7-fold lower butyrate levels in the media from mutant cultures compared to wild-type cultures (Supplementary Fig. S8A-S8C) Next, we colonized BALB/c mice with ASF plus the mutant B. fibrisolvens strain and evaluated AOM/DSS-induced tumorigenesis. These microbiota combined with a high-fiber diet conferred an attenuated protective effect with significantly more tumors than mice colonized with the wild-type bacterium (Fig. 1B). The mutant B. fibrisolvens combined with a high-fiber diet also had a diminished protective effect on tumor size and progression (Fig. 1C-E). These results demonstrate that bacterial butyrate production is commensurate with the tumor-suppressive effect. As an additional control, we colonized BALB/c mice with ASF only (in the absence of wild-type or mutant B. fibrisolvens) and provided them a tributyrin-fortified diet, which increases colonic butyrate levels in a microbiota-independent manner (20) (see Supplementary Fig. S3 for the formulation of this diet and the salient attributes of tributyrin and Supplementary Fig. S4 for luminal butyrate levels in these mice). Following the 5 AOM/3 DSS dose regimen, these mice had fewer (mean of 2) colorectal tumors than any other treatment group (Fig. 1B). These tumors were small and low grade similar to the previous experimental group (Fig. 1C-E). Therefore, exogenous butyrate recapitulated the tumor-suppressive effect of wild-type B. fibrisolvens and high fiber. Taken together, these results demonstrate unequivocally that butyrate is a causative factor in the tumor-suppressive mechanism.
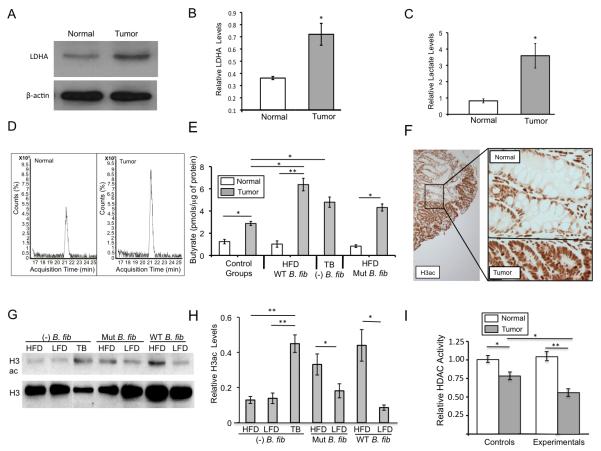
Our previous analysis of cell-culture models suggested that the molecular mechanism of tumor suppression might involve metabolic differences between normal and cancerous colonocytes (21,22). Unlike normal colonocytes, which utilize butyrate as their primary energy source (11), CRC cells primarily rely on glucose and undergo increased glycolysis with a concomitant decrease in mitochondrial oxidative metabolism because of the Warburg effect (23). Consequently, butyrate is not metabolized inside of mitochondria to the same extent and accumulates as an HDAC inhibitor in CRC cells (22). To test the validity of this model in vivo, we confirmed that tumors dissected from our gnotobiotic mice exhibited certain characteristics of the Warburg effect such as increased expression of lactate dehydrogenase A (LDHA) (Fig. 2A-B) and other markers (Supplementary Fig. S9A-S9C). Furthermore, compared to normal colonic tissue, the tumors produced increased levels of lactate (, which is a glycolytic end product that is catalyzed by LDHA and known to be elevated in cancer cells undergoing the Warburg effect. Based on LC-MS measurements, butyrate levels were significantly higher inside of tumors than normal colonic tissue (Fig. 2D-E). To investigate the basis for this difference, we performed flux experiments that measured butyrate oxidation. Consistent with the Warburg effect and our previous analysis of cell-culture models, butyrate oxidation was significantly diminished in tumors compared to normal colonic tissues (Supplementary Fig. S10A-S10E). We also analyzed several monocarboxylate transporters (MCTs) responsible for butyrate influx, but they did not show consistent expression differences between tumors and normal colonic tissues (Supplementary Fig. S10A-S10E). These findings suggest that butyrate accumulates in tumors because of diminished oxidation rather than increased uptake.
Figure 2.
Colorectal tumors exhibit characteristics of the Warburg effect, accumulate butyrate, and have increased H3ac levels. (A) Representative western blot of LDHA in normal colonic tissue and a colorectal tumor. β-actin serves as a loading control. (B) Quantification of LDHA levels normalized to β-actin in normal colonic tissues and tumors based on western blot data. Each histogram shows the mean ± SE based on 3 biological replicates (i.e. colonic tissue and tumors from 3 separate mice) with significant differences indicated (*, p < 0.05). (C) Lactate levels in normal colonic tissue and colorectal tumors based on LC-MS measurements. Results are normalized to μg of protein, and each histogram shows the mean ± SE of 4-5 biological replicates (i.e. colonic tissue and tumors from 4-5 separate mice) with significant differences indicated (*, p < 0.05). (D) Representative LC-MS chromatograms showing butyrate levels in normal colonic tissue and a colorectal tumor. (E) Butyrate levels in normal colonic tissues and tumors from mice in control and experimental treatment groups. Values are based on LC-MS measurements, and each histogram shows the mean ± SE of 5 biological replicates (i.e. colonic tissue and tumors from 5 separate mice) with significant differences indicated (*, p < 0.05; **, p < 0.01). (F) Representative IHC image showing H3ac staining intensity in tumor cells and adjacent normal colonic epithelial cells. (G) Western blot analysis of H3ac and total H3 levels in tumors from mice of the different treatment groups. (H) Quantification of western blot data with H3ac levels normalized to total H3 in tumors from mice of each treatment group. LFD, low-fiber diet; HFD, high-fiber diet. Each histogram shows the mean ± SE based on 5 biological replicates for each treatment group with significant differences indicated (*, p < 0.05; **, p < 0.01). (I) HDAC activity levels normalized to protein levels in normal colonic tissue and tumors from control and experimental treatment groups. Each histogram shows the mean ± SE for 5 independent experiments with significant differences indicated (*, p < 0.05; **, p < 0.01).
The butyrate concentration inside tumors from mice in the experimental treatment groups was calculated to be >100 μM, which is consistent with it acting as an HDAC inhibitor based on its IC50 (24). Therefore, we performed immunohistochemistry (IHC) to analyze pan-histone 3 acetylation (H3ac) levels. H3ac levels were markedly higher in tumor cells than normal adjacent colonocytes (Fig. 2F). Because the tumor cells and normal colonocytes were present in close physical proximity on the same slide, our IHC experiments were internally controlled. These results were confirmed by western blot analyses (Fig. 2G), and quantification demonstrated H3ac levels were significantly higher in tumors from the experimental groups compared to grade-matched tumors from the control groups (Fig. 2H). The observed differences in H3ac were inversely correlated with significant differences in HDAC activity (Fig. 2I), which is compatible with butyrate functioning as an HDAC inhibitor.
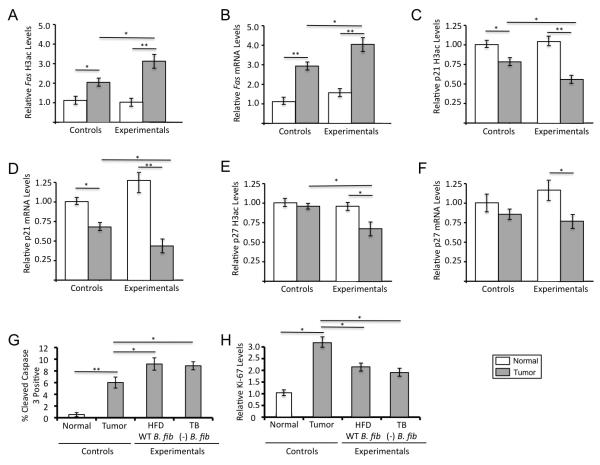
Butyrate can induce histone acetylation and expression of pro-apoptotic genes such as FAS and cell-cycle regulators such as p21 and p27 to stimulate apoptosis and inhibit proliferation in CRC cell lines (10). Therefore, we performed quantitative ChIP assays and RT-qPCR to evaluate promoter H3ac levels and mRNA levels of these genes. H3ac and mRNA levels were significantly higher in tumors from experimental mice than control mice, which, in turn, were significantly higher than normal colonic tissues (Fig. 3A-F). Furthermore, cleaved caspase 3 assays and Ki-67 assays demonstrated that the experimental tumors exhibited increased apoptosis and decreased cell proliferation (Fig. 3G-H and Supplementary Fig. S11). These results provide insight into gene targets and the cell biological basis of the tumor-suppressive effect.
Figure 3.
Tumors from mice provided a high-fiber diet and colonized with B. fibrisolvens (experimental treatment group) have increased H3ac and expression levels of target genes and undergo increased apoptosis and decreased cell proliferation. (A,C,E) Quantitative ChIP assays showing H3ac enrichment at the promoters of the Fas, p21, and p27 genes in normal colonic tissues and tumors as indicated. qPCR results for each ChIP were normalized to input, and the values of normal colonic tissues from control mice were set at 1.0. Each histogram shows the mean ± SE for 3-4 independent experiments with significant differences indicated (*, p < 0.05; **, p < 0.01). (B,D,F) RT-qPCR analysis of Fas, p21, and p27 mRNA levels normalized to Gapdh levels in normal colonic tissues and tumors. The values of normal colonic tissues from control mice were set at 1.0. Each histogram shows the mean ± SE for 5 independent experiments with significant differences indicated (*, p < 0.05; **, p < 0.01). (G) Quantification of cleaved caspase 3 positive cells in normal colonic epithelial cells and tumors from the control and experimental treatment groups. Each histogram shows the mean ± SE based on 10 biological replicates for each group with significant differences indicated (*, p < 0.05; **, p < 0.01). (H) Quantification of Ki-67 levels in normal colonic epithelial cells and tumors from the control and experimental treatment groups. The values of normal colonic tissues from control mice were set at 1.0. Each histogram shows the mean ± SE based on 10 biological replicates for each group with significant differences indicated (*, p < 0.05).
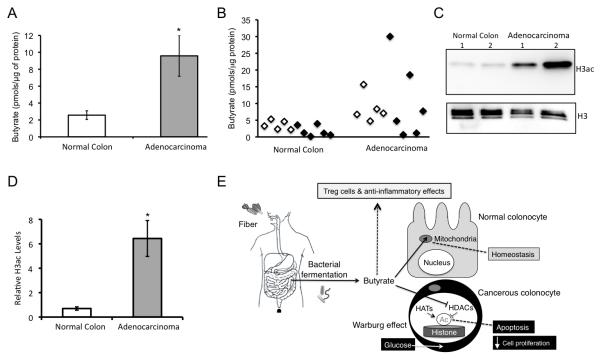
To provide support for the relevance of the proposed molecular mechanism in human tumor suppression, we analyzed human colorectal adenocarcinomas and adjacent macroscopically normal mucosa (see Supplementary Table 2 for clinical information). We detected significantly elevated levels of butyrate (Fig. 4A-B) and H3ac (Fig. 4C-D) in the adenocarcinomas compared to the macroscopically normal mucosa samples. Although these clinical samples were obtained from two different medical centers, there was a high level of concordance between them (Fig. 4B).
Figure 4.
A model depicting the mechanism of the fiber-microbiota-butyrate axis in tumor suppression and evidence to support the relevance of this model to human cancer prevention. (A- D) Relevance of the model to human cancer. (A) Butyrate levels in macroscopically normal colonic tissue and colorectal adenocarcinomas from humans based on LC-MS measurements. Each histogram shows the mean ± SE from 11 biological replicates of each with significant differences indicated (*, p < 0.05). (B) A scatter plot of the samples from panel A. The samples were obtained from the University of North Carolina at Chapel Hill (white, left) and Colorado State University (black, right) tissue procurement facilities. (C) Western blot analysis of H3ac and total H3 levels in macroscopically normal colonic tissue and colorectal adenocarcinomas from 2 pairs of human samples. (D) Quantification of western blot data with H3ac levels normalized to total H3 in macroscopically normal colonic tissue and colorectal adenocarcinomas. Each histogram shows the mean ± SE from 5 biological replicates of each with significant differences indicated (*, p < 0.05). (E) Final working model.Dietary fiber is fermented by microbiota into butyrate, which is a short-chain fatty acid that is abundant (mM levels) in the lumen of the colon. In normal colonocytes, butyrate undergoes oxidative metabolism in the mitochondria to promote homeostasis. In the cancerous colonocyte, due to the Warburg effect, butyrate is metabolized to a lesser extent and accumulates in the nucleus where it functions as an HDAC inhibitor to induce apoptosis and inhibit cell proliferation. The importance of the Warburg effect in this model draws upon previous work (22). In addition to these cell-autonomous effects within the cancer cell, some of the tumor suppression could be mediated by butyrate promoting Treg cell differentiation and having anti-inflammatory effects in the presence of a complex gut microbiota.
DISCUSSION
The findings presented here strongly support the hypothesis that dietary fiber protects against CRC. Our data support a general mechanism that includes microbial fermentation of fiber rather than fiber exclusively speeding colonic transit to minimize the exposure of colonocytes to ingested carcinogens. Our data also support a molecular mechanism where microbial fermentation of fiber yields butyrate, which serves as the preferred energy source of normal colonocytes and supports homeostasis (11,25) but accumulates in cancerous colonocytes due to the Warburg effect and functions as an HDAC inhibitor to inhibit cell proliferation and stimulate apoptosis (21,22) These general and molecular mechanisms are depicted as a final working model in Fig. 4E.
Butyrate may have additional effects independent of HDAC inhibition in tumor suppression. For example, both fiber and butyrate can be metabolized by certain clades of bacteria, and this can influence the microbiome and host metabolism (26). Butyrate is also a ligand for certain G-protein-coupled receptors (GPRs) and has anti-inflammatory effects (9,10), which include the ability to induce the differentiation and expansion of immunosuppressive regulatory T (Treg) cells (27-30). However, we did not observe a significant difference in the number of Treg cells (Supplementary Fig. S12A-S12E) or the abundance of associated cytokines such as IL-10 (Supplementary0 Fig. S13) based on flow-cytometry and cytokine-profiling experiments, respectively. In fact, there was minimal inflammation in our gnotobiotic models following DSS or AOM/DSS treatments (Supplementary Figs. S12-14A-C), which was a consequence of our experimental design. In order to rigorously control butyrate levels, our gnotobiotic mice were colonized with 4-5 commensal bacteria that have not been implicated in inflammation (including 2 Lactobacillus species), and they were unable to induce a robust inflammatory response. We know that this is the case based on control experiments where we transferred a subset of our AOM-treated gnotobiotic mice to a specific pathogen free (SPF) facility where they became “conventionalized” with diverse microbiota. When these conventionalized mice were subsequently treated with DSS in the SPF facility, the inflammatory response was markedly more severe and the tumor burden was >10-fold higher than the same AOM-treated mice that received an identical DSS treatment while still maintained in gnotobiotic isolators (Supplementary Fig. S15). Butyrate is an agonist for at least one GPR (GPR109A) expressed on the apical surface of colonocytes that can mediate butyrate tumor-suppressive effects (30,31). Although our data support a model where butyrate enters tumor cells via MCTs to inhibit HDACs (Fig. 4E), butyrate could also be activating GPR109A signaling. We did not detect significant differences in the expression of Il18 (Supplementary Fig. S16), which is a GPR109A target gene in the colonic epithelium that is relevant to tumor suppression (30), although this does not exclude a role for GPR109A or other GPRs.
Our gnotobiotic mouse model was polyassociated with several species of bacteria and provided a homogeneous diet that included a single type of fiber. This reductionist approach was necessary to demonstrate that fiber protects against CRC and interrogate butyrate function. However, it does not reflect the complex microbiota that exist within the human GI tract or our more varied diets that include multiple sources of fiber. Other bacterial metabolites undoubtedly participate in tumor suppression but were beyond the scope of our study. Nevertheless, it is tempting to speculate that many of the conclusions drawn from our mouse model are relevant to human health as discussed below.
As human populations have shifted away from traditional, high-fiber diets towards processed foods containing less complex carbohydrates and more refined sugars, CRC incidence has increased markedly. Yet the link between fiber and CRC prevention has been tenuous because of conflicting results from cohort-based epidemiologic studies. By rigorously controlling genetics, the composition of gut microbiota, and other dietary factors such as fat that may mask a beneficial fiber effect, we can conclude from this study that fiber does, in fact, protect against colorectal tumorigenesis. An important aspect of this work is the central role of gut microbiota and the fermentation product butyrate. Consequently, we propose that cohort-based epidemiologic studies should be revisited and integrated with microbiome studies. We predict that if microbiome differences of participants were taken into account, then it would be possible to discriminate between those individuals who respond to the anticancer chemoprotective effect of fiber from non-responders. This would resolve some of the conflicting results from previous human studies and possibly confirm butyrate as an important molecule in human chemoprotection. This idea is supported by several microbiome studies that have reported fewer butyrate-producing bacteria in human colorectal cancer cases than controls even without diet being taken into consideration (32-36).
Metagenomic sequencing projects have made a number of observations regarding the microbiome and the incidence of certain cancers. However, these studies are correlative so it is difficult or impossible to know whether the microbiome differences are a cause or a consequence of the disease state. In contrast, relatively few studies have investigated the function of microbiota in gnotobiotic mouse models of cancer (37,38). Furthermore, most or all of these studies have focused on bacteria that promote oncogenesis by causing inflammation or genotoxicity (37,38). In contrast, butyrate-producing bacteria confer a tumor-suppressive effect, which arguably has greater translational potential for chemoprevention via probiotics or prebiotics.
The molecular mechanism described here involves butyrate functioning as an HDAC inhibitor, which is noteworthy because synthetic HDAC inhibitors are being used as anticancer chemotherapeutic agents with some having already received FDA approval and others being evaluated in phase 3 clinical trials (4). The translational implication of our model is attractive because diet (prebiotics-fiber) and possibly microbiota supplementation (probiotics-butyrate-producing bacteria) modulate the levels of an endogenous HDAC inhibitor (butyrate) for the purpose of chemoprevention. This approach has several advantages compared to the systemic delivery of synthetic HDAC inhibitors for chemotherapy (4). First, it should be easier to modulate the epigenome and transcriptome profiles of cells at an early stage of tumorigenesis; later-stage tumor cells are more likely to have accumulated mutations or epimutations that make them refractory to HDAC inhibition. Second, because the bioavailability of butyrate is primarily restricted to the colon, it will not have adverse effects in other tissues. Third, unlike synthetic HDAC inhibitors, butyrate is a naturally occurring fatty acid readily metabolized by normal cells so it does not have adverse effects even in the colon. The ability of butyrate to specifically target tumor cells in the colon is due to the Warburg effect (22). Because cancerous colonocytes rely on glucose as their primary energy source, butyrate is not metabolized in the mitochondria to the same extent and is able to accumulate as a tumor-suppressive metabolite (analogous to the oncometabolite 2-hydroxyglutarate) in the nucleus where it functions as an HDAC inhibitor to stimulate histone acetylation, induce apoptosis, and inhibit cell proliferation.
METHODS
Mice
BALB/c mice were maintained in isolators (Class Biologically Clean, Madison, WI) and bred in house at the National Gnotobiotic Rodent Resource Center (NGRRC) at the University of North Carolina at Chapel Hill. All mouse experiments were approved by the Institutional Animal Care and Use Committees (IACUC) review board at the University of North Carolina at Chapel Hill and were performed in accordance with federal guidelines.
Gnotobiotic Mouse Husbandry
Food, water, and all other materials including cages and bedding were autoclaved and imported into the isolators using aseptic technique following standard procedures. Each diet was from Test Diet (Richmond, IN). The low-fiber diet (5SRZ, Cat. No. 1813680) contained 2% cellulose while the high-fiber diet (5SVL, Cat. No. 1813901) contained 2% cellulose plus 6% fructo-oligosaccharide/inulin (Sigma-Aldrich #F8052 and 12255, St. Louis, MO). The tributyrin diet (5AVC, Cat. No. 1814961) contained 2% cellulose plus 6% tributyrin (Sigma-Aldrich #W222305). The initial colonization of germfree BALB/c mice with specific bacteria (which were the only imported materials not autoclaved except filter-sterilized AOM/DSS) was performed by oral gavage following standard procedures.
Bacteria
Bacteria were cultured in BBL Schaedler broth containing vitamin K (Cat. No. 221541) that was supplemented with 5% FBS in an anaerobic chamber (ThermaForma) filled with an anaerobic mixture (Airgas #750333 consisting of 5% CO2 and 10% H2 balanced with N2) at 37°C. For experiments measuring butyrate production in culture, the Schaedler broth was supplemented with 0.5% or 5% FOS/inulin (the same additive that was used for the high-fiber mouse diet). The following bacteria were cultured: ASF360 (Lactobacillus acidophilus), ASF 361 (Lactobacillus salivvarius), ASF457 (Flexistipes phylum), and ASF519 (Bacteroides distasonis), which were obtained from Taconic, and Butyrivibrio fibrisolvens (type I, ATCC 19171; type II, ATCC 51255) obtained from ATCC.
AOM/DSS
AOM (Sigma-Aldrich #A5486) was dissolved in PBS at a concentration of 50 mg/mL and stored as aliquots at -80°C. Aliquots were subsequently thawed and diluted in sterile saline (0.9% NaCl) at a concentration of 1.25 mg/mL. This final AOM solution was then filter sterilized in a tissue-culture hood, imported into gnotobiotic isolators, and delivered by i.p. injection at a final concentration of 10 mg per kg body weight. AOM injections were performed on a weekly basis for a total of up to 5 injections. 5 days after the final AOM treatment, the mice were provided drinking water containing 2.5% DSS (ICN, MW 36-50 kDa) for 5 days. The DSS-containing water was then replaced by normal drinking water for 16 days. This cycle was repeated 1 or 2 more times resulting in either 2 or 3 DSS treatments. The DSS was prepared by dissolving it in autoclaved water followed by filter sterilization in a tissue-culture hood and importation into gnotobiotic isolators.
Scoring of Tumors
Tumor scoring was performed in a blinded manner by two veterinary pathologists. Colons were flushed and then splayed open. Colonic mucosal masses counted as tumors grossly appeared as variably sized, irregular, asymmetrical, shiny to roughened, tan to red, sessile to pedunculated, occasionally coalescing nodular masses. For coalescing tumors, the number of individual tumors was estimated. Colonic mucosal masses considered lymphoid nodules and not included in the tumor count were approximately 0.5 cm in diameter, 0.2 cm in height, and grossly appeared as symmetrical, shiny, translucent, flat, plaque-like masses.
Histopathology and IHC
Swiss-rolled colons and other tissues (e.g., lymph nodes and liver) were fixed in 4% paraformaldehyde or 10% formalin and processed for the production of 5-micron paraffin sections. Sections were stained with hematoxylin and eosin (H&E) or processed for IHC using standard procedures. Antibodies included pan-acetyl H3 (Millipore #06-599), total H3 (Millipore #05-928), LDHA (Cell Signaling #3582), Cleaved Caspase-3 (Asp175) (Cell Signaling #9661), and Ki-67 (Abcam #15580). IHC quantification was performed using Image J to count the number of normal colonic epithelial cells or tumor cells that were positive for Cleaved Caspase-3 and hematoxylin in randomly selected crypts. The cleaved caspase-3 positive cells were identified by their distinctive brown cytoplamic staining. The number of cleaved caspase-3 cells counted per crypt was divided by the total number of hematoxylin-stained cells counted in the exact same crypt. At least 10 randomly selected normal crypts or tumorigenic crypts were quantified per animal times 5 animals per treatment group (i.e., biological replicates). A similar approach was done for Ki-67 and H3ac except the H3ac results were normalized to total H3.
Sample Acquisition
After each mouse was sacrificed, colons were removed starting at a point immediately distal to the junction between the cecum and the ascending colon and ending at the anus. The entire colon was splayed open, and luminal contents were removed from the proximal third of the colon for LC-MS experiments. Fecal pellets were not used as proxy for luminal contents. After luminal contents were removed, colons were rinsed twice in PBS. Macroscopically normal colonic tissue and tumors were dissected for LC-MS experiments and other experiments (e.g., western blots, HDAC activity assays, ChIP, RT-qPCR, flux experiments). Tumor dissections were performed to minimize the amount of normal adjacent tissue.
LC-MS
For detection of butyrate, samples were treated with 13C1 or 13C4-butyrate (Isotech #292656) at a final concentration of 10 mM (as an internal control to assess recovery efficiency and as a standard to calculate endogenous butyrate levels) and then homogenized in 0.1% ammonium hydroxide. Macromolecules were removed by centrifugation of lysates through 3-kDa spin-filters (Pall Corporation, Cat. #OD003C33). Flow-throughs were then analyzed for exogenous (13C4) and endogenous (12C4) butyrate by HPLC separation with subsequent detection by an Agilent 6520 AccurateMass Q-TOF mass spectrometer operating in negative mode (Santa Clara, CA). Peak areas were calculated using MassHunter Workstation software. Chromatographic peaks were integrated for samples and areas were compared to peak area for standards (10 mM) for each compound. For detection of other SCFAs, the procedure was the same except the appropriate 13C4-labeled acetate and propionate were used as standards.
Western Blot Analyses
Western blots were performed following standard procedures, and antibodies that were used included pan-acetyl H3 (Millipore #06-599), total H3 (Millipore #05-928), LDHA (Cell Signaling #3582), α-tubulin (Sigma-Aldrich #T6793), P-AKT (Ser473) (Cell Signaling #4060), total AKT (Cell Signaling #2965), and P-PDH (Ser293 of E1α isoform)(Calbiochem #AP1062).
HDAC Assays
Nuclear extracts were prepared from frozen tissues using a kit (Sigma #NXTRACT) dounce homogenization. HDAC colorimetric assays were performed using a kit (Epigentek #P-4034). HDAC activities were normalized to protein levelsfor each sample.
RT-qPCR
RNA was prepared using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and SuperScript II RT (Invitrogen) according to standard procedures. 20 μl reactions that included 10 μl of either 2× TaqMan or 2× Power SYBR Green master mix (Applied Biosystems) with 50-100 ng of cDNA and the appropriate primers (all assays were previously published) were run on an ABI 7300 instrument. Product accumulation was monitored by FAM or SYBR Green fluorescence. Control reactions lacking reverse transcriptase yielded very low to no signal. Relative expression levels were determined using the ΔΔCt method and normalized to Gapdh.
ChIP Assays
Tissues were pulverized in liquid nitrogen using a mortar and pestle and then crosslinked in pre-warmed 0.4% formaldehyde in PBS for 10 minutes at 37°C. The crosslinking reaction was stopped by adding glycine to a final concentration of 0.125 M and then sonicated with four 10-sec pulses at 30% of maximum power. IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris at pH 8.1, 167 mM NaCl, protease inhibitors) was added, 5% of the volume was removed and used as input while the remainder was incubated overnight at 4°C with the appropriate antibody: pan-acetyl H3 (Millipore #06-599), total H3 (Millipore #05-928), or rabbit IgG (Santa-Cruz) as a negative control. Protein A/G agarose beads (Santa Cruz) were added and incubated for at least 2 h at 4°C, and then washed and eluted according to standard procedures.
qPCR was performed using Power SYBR Green Master Mix (Applied Biosystems (Applied Biosystems) on an ABI 7300 instrument under default cycling conditions (95°C 15 sec followed by 60°C 1 min for 45°C cycles). Dissociation curves and agarose gels demonstrated a single PCR product in each case without primer dimers. Relative enrichment was determined from a standard curve of serial dilutions of input samples.
Flux Experiments
~1×106 cells were incubated inside Exetainer breath storage tubes (Labco Limited) in 1 mL of PBS containing 0.5 mM 13C1-butyrate and 5 mM 12C-glucose for 1 hr at 37°C. Reactions were stopped with sodium azide treatment. Butyrate oxidation was assessed by analyzing isotopic CO2 derived from butyrate to non-isotopic CO2 derived from glucose using a 20/20 gas isotope ratio mass spectrometer (Europa Scientific) at Metabolic Solutions (Nashua, NH). Dissolved CO2 in solution was liberated into the tube headspace by the addition of 100 μL of saturated citric acid. The ratio of 13CO2 to 12CO2 (mass 45 to 44) was measured directly from the sample tube headspace. All samples were compared to an internal reference gas (5% CO2, balance 75% N2 and 20% O2) that had been calibrated against the International standard PeeDeeBelmnite (PDB). The results were expressed as %13C = 13CO2 / (13CO2 + 12CO2) and normalized to normal colon. The analytical precision of the instrument is 0.0001 atom % 13C.
Fetal Human Colonocytes (FHC) and HCT116 cells were obtained from ATCC and split into Seahorse 24-well cell culture plates seeded at 20k cells per well. The next day the normal growth media for each cell was replaced with fatty acid oxidation media, which contains 110 mM NaCl, 4.7 mM KCl, 2 mM MgSO4, 1.2 mM Na2HPO4, 0.5 mM, and 2.5 mM glucose adjusted to a pH of 7.4. FHC and HCT116 cells were incubated in this media for 1 hr in a non-CO2 incubator at 37°C and then run on the XF24 Analyzer (Seahorse Bioscience, North Billerica, MA) After three baseline measurements of the oxygen consumption rate (OCR) butyrate was injected into each well. 6 more OCR measurements were then made and 2DG (final concentration of 5 mM) was injected into each well. 6 more OCR measurements were then made. OCR was then suppressed by injecting 10% sodium azide into each well. Contribution of butyrate oxidation to OCR was determined after injection of 2DG, which inhibited glucose oxidation, and before sodium azide treatment.
Flow Cytometry
Colonic epithelial cells were isolated by incubating splayed-open colons in PBS containing 5 mM EDTA at 37°C. After a 30-minute incubation while being rotated, the colonic tissue was transferred to a new tube for the isolation of lamina propria cells, and the epithelial cells were pelleted and washed. Lamina propria cells were isolated by incubating the colonic tissue minus the delaminated epiethelial cells in pre-warmed 0.5 mg/mL Collagenase Type 4 (Worthington #LS004186) in 1X PBS with DNAse. Samples were rotated at 37°C for 20 minutes, filtered through a 40-μm cell strainer and repeated twice more. After pre-digest and digest steps, collected cells were immediately washed in PBS containing 2% FBS and kept at 4°C to maintain viability. The colonic epithelial cells and lamina propria cells were then combined. These cells as well as single-cell suspensions from spleens were stained for flow cytometric analysis using a cocktail of PE-Texas Red anti mouse CD11c (clone N418) from Molecular Probes, PerCP anti-mouse CD45R [B220] (RA3-6B2) from BioLegend, and AF488 anti-mouse CD4 (GK 1.5), PE anti-mouse CD69 (H1.2F3), PE-Cy7 anti-mouse Gr-1 (RB6-8C5), eF450 anti-mouse CD11b (M1/70), APC anti-mouse CD8α (53-6.7), and APC-eF780 anti-mouse CD45 [Leukocyte common antigen] (30-F11), all from eBioscience. A separate set of reactions was used to detect Treg cells using a kit from eBioscience (#88-8118-40). Stained samples were read on a CyAn cytometer (Dako Cytomation), and the listmode files analysed with the Summit Software package from Beckman-Coulter. Abbreviations for this section: PE, phycoerythrin; AF488, Alexa fluor 488; APC, allophycocyanin; PerCP, Peridinin-chlorophyll-protein complex.
Luminex-Based Cytokine Profiling
In order to measure cytokine and chemokine levels, serum and colonic sections were isolated from experimental and control mice. Blood was centrifuged at 2,500 rpm at 4°C for 5 minutes, and the separated serum was collected. 30 mg of colonic tissue was homogenized in 0.5 mL of PBS buffer containing 0.1% Tween 20 with protease inhibitors. Tissue homogenates were then centrifuged at 14,000 rpm for 10 minutes at 4°C to pellet debris, and the supernatant was used for cytokine/chemokine detection. The levels of cytokines and chemokines were detected in the serum using a Milliplex Mouse Cytokine/Chemokine Immunoassay (Millipore) per the manufacturer’s instructions. The assay included reagents to measure the following: Eotaxin, G-CSF, GM-CSF, IFNγ, IL-1α, IL-1β;, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IL-17, IP-10, LIF, LIX, MCP-1, M-CSF, MIG, MIP-1β;, MIP-1α, MIP-2, RANTES, TNFα, and VEGF. The Milliplex protocol was altered for the colonic tissue, substituting the provided matrix solution with the buffer used for colonic homogenization. All samples were analyzed in triplicate with detection and analysis performed on a on a Bio-Plex 200 detection system (Bio-Rad).
Human Clinical Samples
Following institutional review board (IRB) review and approval, normal and cancerous colonic tissue was obtained as frozen samples and paraffin-embedded sections from the Tissue Procurement Facility at the University of North Carolina at Chapel Hill and Poudre Valley Hospital (Fort Collins, CO) in collaboration with Colorado State University. Each specimen was distributed with a unique identification number (no patient identifying information was distributed) in accordance with patient privacy and confidentiality guidelines.
Statistics
Supplementary Table S3 lists the statistical test used for each figure. In experiments where a hypothesis was tested by comparing 2 groups of samples, a two-tailed t test was used to determine significant differences. For more than 2 groups of samples, ANOVA followed by a Tukey post-hoc test was used. In experiments where the data did not follow a normal distribution, a Kruskal-Wallis non-parametric statistical test was used. It was used instead of a Mann-Whitney test because more than 2 groups of samples were compared in our experiments.
Supplementary Material
SIGNIFICANCE.
These results, which link diet and microbiota to a tumor-suppressive metabolite, provide insight into conflicting epidemiologic findings and suggest that probiotic/prebiotic strategies can modulate an endogenous HDAC inhibitor for anticancer chemoprevention without the adverse effects associated with synthetic HDAC inhibitors used in chemotherapy.
Acknowledgments
We acknowledge L. Paterson and R. Tether for their advice on the anaerobic culture conditions for B. fibrisolvens. We also acknowledge the staff of the National Gnotobiotic Resource Center (supported by P40RRO118603 and P30 DK 34987) for maintaining the gnotobiotic mice, C. Suitt at the UNC CGIBD Histopathology Core (also supported by P30 DK 034987) for tissue sectioning, B. Bagnell and V. Madden at the UNC Microscopy Services Laboratory for use of the laser-capture dissection microscope. We would also like to acknowledge M. Huang at the UNC Tissue Procurement Facility and Poudre Valley Hospital (Fort Collins, CO) for providing human clinical samples.
Grant Support
This work was supported with grants from the NIH (CA125237 to SJB), the American Institute of Cancer Research (S.J.B.) and the Prevent Cancer Foundation (D.R.D. and S.J.B). The Biomarker Mass Spectrometry Facility is supported by NIH grant P30-ES10126.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
Authors’ Contributions
S.J.B., D.S.T, and D.W.T. conceived the overall study; S.J.B. and D.R.D. designed specific experiments that were part of the study; D.R.D. and D.H. took primary responsibility for performing the experiments; K.P.C. and S.W.R. also performed experiments; S.A.M. and V.G. supervised the necropsies, scored tumour numbers, and performed histopathologic analysis of tissue sections; L.B.C. and J.A.S. were responsible for the LC-MS experiments; A.H. and D.W.T. performed the cytokine-profiling experiments; A.C.W. and M.T.H. provided advise and reagents for the flow cytometry experiments and contributed to the data analyses; E.P.R. provided human clinical samples; S.J.B. took primary responsibility for writing the manuscript, which was approved by all authors.
REFERENCES
- 1.Bingham SA, Day NE, Luben R, Ferrari P, Slimani N, et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): an observational study. Lancet. 2003;361:1496–501. doi: 10.1016/s0140-6736(03)13174-1. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs CS, Giovannucci EL, Colditz GA, Hunter DJ, Stampfer MJ, Rosner B, et al. Dietary fiber and the risk of colorectal cancer and adenoma in women. The New England Journal of Medicine. 1999;340:169–76. doi: 10.1056/NEJM199901213400301. [DOI] [PubMed] [Google Scholar]
- 3.Park Y, Hunter DJ, Spiegelman D, Bergkvist L, Berrino F, van den Brandt PA, et al. Dietary fiber intake and risk of colorectal cancer: a pooled analysis of prospective cohort studies. The Journal of the American Medical Association. 2005;294:2849–57. doi: 10.1001/jama.294.22.2849. [DOI] [PubMed] [Google Scholar]
- 4.Ahuja N, Easwaran H, Baylin SB. Harnessing the potential of epigenetic therapy to target solid tumors. The Journal of Clinical Investigation. 2014;124:56–63. doi: 10.1172/JCI69736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatzkin A, Mouw T, Park Y, Subar AF, Kipnis V, Hollenbeck A, et al. Dietary fiber and whole-grain consumption in relation to colorectal cancer in the NIH-AARP Diet and Health Study. The American Journal of Clinical Nutrition. 2007;85:1353–60. doi: 10.1093/ajcn/85.5.1353. [DOI] [PubMed] [Google Scholar]
- 6.Baron JA. Dietary fiber and colorectal cancer: an ongoing saga. The Journal of the American Medical Association. 2005;294:2904–6. doi: 10.1001/jama.294.22.2904. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson LR, Harris PJ. The dietary fibre debate: more food for thought. Lancet. 2003;361:1487–8. doi: 10.1016/S0140-6736(03)13219-9. [DOI] [PubMed] [Google Scholar]
- 8.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung KY, Cosgrove L, Lockett T, Head R, Topping DL. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. The British Journal of Nutrition. 2012;108:820–31. doi: 10.1017/S0007114512001948. [DOI] [PubMed] [Google Scholar]
- 10.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Alimentary Pharmacology & Therapeutics. 2008;27:104–19. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 11.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–8. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davie JR. Inhibition of histone deacetylase activity by butyrate. The Journal of Nutrition. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 13.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. The Journal of Nutrition. 2004;134:479–82. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta S, Muir JG, Gibson PR. Does butyrate protect from colorectal cancer? Journal of Gastroenterology and Hepatology. 2006;21:209–18. doi: 10.1111/j.1440-1746.2006.04213.x. [DOI] [PubMed] [Google Scholar]
- 15.Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Applied and Environmental Microbiology. 1999;65:3287–92. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asanuma N, Ishiwata M, Yoshii T, Kikuchi M, Nishina Y, Hino T. Characterization and transcription of the genes involved in butyrate production in Butyrivibrio fibrisolvens type I and II strains. Current Microbiology. 2005;51:91–4. doi: 10.1007/s00284-005-4477-x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg DW, Giardina C, Tanaka T. Mouse models for the study of colon carcinogenesis. Carcinogenesis. 2009;30:183–96. doi: 10.1093/carcin/bgn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003;124:762–777. doi: 10.1053/gast.2003.50094. [DOI] [PubMed] [Google Scholar]
- 19.Shane BS, Gouws L, Kistner A. Cellulolytic bacteria occurring in the rumen of sheep conditioned to low-protein teff hay. Journal of General Microbiology. 1969;55:445–57. doi: 10.1099/00221287-55-3-445. [DOI] [PubMed] [Google Scholar]
- 20.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PloS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber Induction of proliferation and apoptosis via butyrate for cancer prevention. Clinical Cancer Research. 2014;20:799–803. doi: 10.1158/1078-0432.CCR-13-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular Cell. 2012;48:612–26. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekhavat A, Sun JM, Davie JR. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochemistry and Cell biology. 2007;85:751–8. doi: 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- 25.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metabolism. 2011;13:517–26. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. Journal of Applied Microbiology. 2007;102:1197–208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 27.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 29.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–39. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Research. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. Journal of Gastroenterology and Hepatology. 2008;23:1298–303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 33.Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. The American Journal of Clinical Nutrition. 2013;98:111–20. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME Journal. 2012;6:320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial Ecology. 2013;66:462–70. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 37.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–55. doi: 10.1093/carcin/bgt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwabe RF, Jobin C. The microbiome and cancer. Nature Reviews. Cancer. 2013;13:800–12. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.