Abstract
Glucogon-like peptide 1 receptor (GLP1R) signaling has been shown to have antipsychotic properties in animal models and to impact glucose-dependent insulin release, satiety, memory, and learning in man. Previous work has shown that two coding mutations (rs6923761 and rs1042044) are associated with altered insulin release and cortisol levels. We identified four frequently occurring haplotypes in Caucasians, haplotype 1 through haplotype 4, spanning exons 4-7 and containing the two coding variants. We analyzed response to antiapsychotics, as defined as predicted change in PANSS-Total (dPANSS) at 18 months, in Caucasian subjects from Clinical Antipsychotic Trial of Intervention Effectiveness treated with (olanzapine, n=139; perphenazine, n=78; quetiapine, n=14; risperidone, n=143; and ziprasidone, n=90). Haplotype trend regression analysis revealed significant associations with dPANSS for olanzapine (best p=0.002), perphenazine (best p=0.01), quetiapine (best p=0.008), risperidone (best p=0.02), and ziprasidone (best p=0.007). We also evaluated genetic models for the two most common haplotypes. Haplotype 1 (uniquely including the rs1042044 [Leu260] allele) was associated with better response to olanzapine (p=0.002), and risperidone (p=0.006), and worse response to perphenazine (p=.03), and ziprasidone (p=0.003), with a recessive genetic model providing the best fit. Haplotype 2 (uniquely including the rs6923761 [Ser168] allele) was associated with better response to perphenazine (p=0.001) and worse response to olanzapine (p=.02), with a dominant genetic model providing the best fit. However, GLP1R haplotypes were not associated with antipsychotic-induced weight gain. These results link functional genetic variants in GLP1R to antipsychotic response.
Keywords: olanzapine, perphenazine, quetiapine, risperidone, ziprasidone, pharmacogenetics, schizophrenia
1 Introduction
While antipsychotics have the potential to greatly reduce psychotic symptoms, no drug is safe and efficacious for all patients. Indeed, the Clinical Antipsychotic Trial of Intervention Effectiveness (CATIE) demonstrated that only about a third of patients achieved a clinically meaningful response when treated with commonly used antipsychotics, and even fewer patients were able to maintain the response for extended periods of time (Das et al., 2012). Despite this lack of universal success for any one drug, some patients did experience significant and sustained symptom relief. The high prevalence of side-effects, particularly weight gain and metabolic syndrome, further complicates treatment. Two of the most efficacious antipsychotics, clozapine and olanzapine, also have the highest incidence and severity of weight gain and associated metabolic side-effects (Das et al., 2012). For both clozapine and olanzapine, symptom improvement correlates with weigh gain, i.e. individuals that gain significant amounts of weight are more likely to respond or have a more robust response than those who do not experience significant gain weight (Ascher-Svanum et al., 2005;Bai et al., 2006). Data from CATIE extend this observation to include other drugs as well (Hermes et al., 2011). Thus, identifying which drug will provide the optimal blend of efficacy and minimal side-effects for a given patient remains challenging.
The glucogon-like peptide 1 (GLP1) and the GLP1 receptor (GLP1R) play multiple roles in modulating weight gain, metabolic status, and stress response. In the intestine and pancreas, activation of GLP1R by GLP1 stimulates insulin release and slows gastric emptying (Shah and Vella, 2014). However, in addition to its prevalence in the gut and pancreas, GLP1R is also expressed in the brain (Brunetti et al., 2008). Animal data suggest that both peripheral and central administration of either GLP1 or GLP1R agonists can induce decreased appetite and weight loss (Rupprecht et al., 2013;Hayes, 2012). Hayes provides a thorough review of the potential GLP1R-mediated CNS signaling mechanisms that may impact weight gain through appetite suppression and satiety (Hayes, 2012). Data from human studies supports the use of GLP1R agonists as modifiers of antipsychotic induced weight gain (AIWG). Exenatide and liraglutide, both GLP1R agonists, have been approved for use in the United States. These medications have promise as adjunct therapy for the management of AIWG (Ebdrup et al., 2012;Lykkegaard et al., 2008;Ishoy et al., 2013).
In addition to the appetite control mechanisms, GLP1R may play a role in other pathways and mechanisms important for psychosis and antipsychotic response. Specifically, GLP1 signaling impacts hypothalamic-pituitary-adrenal axis (HPA) activation, autonomic stress response and anxiety-related behaviors (Ghosal et al., 2013). For example, GLP1R interacts with several neurotransmitter-related proteins in the brain such as gamma amino butyric acid receptor B2; calcium related proteins neurogranin and calmodulin; and two proteins, synaptogyrin and GPR37 that interact with the dopamine transporter (http://www.ncbi.nlm.nih.gov/gene/2740 ). Both GLP1 and exenatide stimulate serotonin release from rat hypothalamus in vitro, which may partially explain weigh loss through 5HT2C receptor agonism (Brunetti et al., 2008).
Beyond biochemical interactions, in vivo data from animal studies offer intriguing observations regarding the potential utility of GLP1R agonists for treating psychiatric disorders. For example, Graham et al. report that pre-treatment with exenatide can mitigate conditioned place preference (a model for addiction) for cocaine in mice (Graham et al., 2013). Additionally, GLP1R agonists mitigate the effects of both alcohol and amphetamine (Erreger et al., 2012). GLP1R agonists have demonstrated neuroprotective action in model systems for Alzhemier's disease and Parkinson's disease (Holscher, 2014). Moreover, the GLP1R agonist liraglutide shows remarkable antipsychotic-like properties comparable to haloperidol in a mouse model of psychosis (Dixit et al., 2013).
In humans, coding variants in GLP1R have been shown to impact both insulin release and morning cortisol levels. In non-diabetic subjects, genetic variation in GLP1R impacts insulin secretion following exogenous administration of GLP1 (Sathananthan et al., 2010). Subjects homozygous for the major allele of rs6923761 (Gly168) secreted significantly more insulin than subjects containing at least one copy of the minor allele. Another coding variant, rs1042044, has been linked to increased morning cortisol levels in children (Sheikh et al., 2010). Subjects homozygous for the minor allele (Leu260) displayed higher morning cortisol levels, which have been linked to increased risk for major depressive disorder in youths and adults (Bhagwagar et al., 2005;Goodyer et al., 2009;Koole et al., 2011).
Previously, many studies have been published on the efficacy, side-effects, and pharmacogenetics of the CATIE study (Adkins et al., 2011;Hermes et al., 2011;Lieberman et al., 2005;Liu et al., 2012a;Liu et al., 2012b;McClay et al., 2011b;McClay et al., 2011a;Need et al., 2009;Ramsey et al., 2013;Stroup et al., 2007). Given the potential for both metabolic and CNS effects of GLP1R signaling as well as in vitro and in vivo data suggesting that genetic variation in GLP1R impacts potentially relevant pharmacogenetic phenotypes, we investigated whether genetic variation in GLP1R impacted response to antipsychotics in the CATIE trial. Here we report the findings with a focus on the potential impact of coding variation in the gene.
2 Materials and Methods
2.1 Subjects and data
CATIE Sample
The design of the CATIE study has been described in detail elsewhere (Stroup et al., 2003;Stroup et al., 2007). Only self described “white” or Caucasian patients were included in the current analysis. Furthermore, only the blinded phases of the study, Phases 1, 1B and 2 were included in the current analysis. In Phase 1, schizophrenia patients were randomly assigned to one of four atypical antipsychotic drugs, olanzapine, quetiapine, risperidone or olanzapine, or to perphenazine, a typical antipsychotic drug. Investigators could elect to switch medication at any time during the 18 month study duration. A drug switch triggered a new randomization to a previously unused study drug. National Institute of Mental Health Center for Collaborative Genetic Studies on Mental Disorders (CGSMD) (https://www.nimhgenetics.org/) provided the genotype and phenotype data used in this study. Table 1 details the distribution of Caucasian subjects by drug across phases 1, 1B, and 2.
Table 1.
Number of subjects in each phase by drug
| Phase 1/1A | Phase 1B | Phase 2 | Total | Mean Age Yrs (SD) | Percent male | |
|---|---|---|---|---|---|---|
| Olanzapine | 93 | 10 | 41 | 144 | 40.3 (11.5) | 74 |
| Perphenazine | 78 | 78 | 41.2 (11.1) | 81 | ||
| Quetiapine | 95 | 16 | 30 | 141 | 41 (10.3) | 80 |
| Risperidone | 97 | 12 | 34 | 143 | 41.5 (11) | 76 |
| Ziprasidone | 49 | 42 | 91 | 39.2 (11.5) | 74 | |
| Total | 412 | 38 | 147 | 597 | 40.7 (11) | 77 |
2.2 Haplotype determination
The CATIE data included genotypes from 13 SNPs within the promoter and transcribed region of GLP1R: rs10305416, rs910171, rs926674, rs10305439, rs9296283, rs7766275, rs2300615, rs1042044, rs932443, rs2268645, rs10305492, rs1126476, and rs2300612. We determine blocks of linkage disequilibrium (LD) using the default settings of Haploview 4.2 (Broad Institute, Cambridge Ma) for haplotype block determination using the confidence interval method (upper CI 0.98, lower CI 0.7, and excluded markers with minor allele frequency < 0.05) (Barrett et al., 2005). To assign haplotypes to individuals, we used the expectation maximization (EM) algorithm function of helix tree version 6.4.3 (GoldenHelix, Bozeman, MT) on the identified haplotype block (Barrett et al., 2005).
The HapMap CEU data set was used to determine if haplotype blocks identified in the CATIE genotype data (which was genotyped using an older technology) tagged additional coding variant(s) beyond rs1042044. GLP1R Hapmap data (Chromosome 6 39,048 KB to 39,088 KB GRCh38 primary assembly) was downloaded through Haploview using Hapmap version 3 release R2. We determined haplotype blocks using the default settings of Haploview (Broad Institute, Cambridge Ma) for haplotype block definition as described above. As described in the Results, one allele of rs6923761, which was not included in the genotypes provided by CGSMD, is associated exclusively with a particular haplotype in the region ranging from 39,142 KB (rs6923761) to 39,149.5 KB (rs1042044). By comparing the HapMap data to the CATIE data we determined that the coding variant rs6923761 could be uniquely assigned to haplotype 2.
2.3 Definition of response
In order to provide consistent and unbiased definition of response, we used predicted changed in Positive and Negative Syndrome Scale Total scores (dPANSS) from using the 30-day lag model developed by Van den Oord et al (response model) (van den Oord et al., 2009). This response model is a mixed model repeated measures (MMRM) model that incorporates baseline, treatment, and response at each time point into the predicted 18 month response. The model provided 18 month predicted dPANSS values for each subject. As baseline response was already included in the response model, it was not used as covariate in the analysis, which predicted dPANSS as the dependent variable.
2.4 Haplotype trend regression
We used the haplotype trend regression (HTR) function in HelixTree to evaluate whether or not haplotypic variation in GLP1R was associated with differential response to the CATIE drugs. HelixTree assigns haplotypes to subjects using EM algorithm and performs a linear regression with the various haplotypes as independent variables. The dependent variable was dPANSS. To control for multiple testing, we used the Benjamini graphically sharpened false discovery rate method (Benjamini and Hockberg, 2000).
2.5 Genetic model analysis
The four GLP1R Haplotypes were assigned names haplotype 1 through 4 based on descending frequency. Following assignments of diplotypes (phased haplotypes) to each individual using EM algorithm maximization, the genotype for the subject was coded to reflect the number of copies of the particular haplotype (0, 1, or2). For the additive model, we used linear regression analysis of the dependent variable dPANSS with the number of copies of the haplotype (0, 1, or 2) as the independent variable. For the dominant and recessive analysis, we created a binary variable. For the dominant genetic model one or two copies of the particular haplotype was coded as “1” and no copies was coded as “0”. For the recessive model two copies of the haplotype was coded as “1” and fewer than two copies was coded as “0”. These binary variables then served as independent variables in a linear regression with dPANSS as the dependent variable. In each case, for the genetic model showing the most significant results, an effect size (Cohen's d) was calculated (*Mean of group with binary score of 1 - Mean of group with binary score of 0] / the standard deviation of the group mean) (Cohen, 1988).
2.6 Weight gain
CGSMD provided a variable for change in weight from baseline of a given phase (C_WT). This weight change was divided by in-phase baseline weight to derive percent weight change. Since subjects could have variable amounts of time on drug, ranging from less than 30 days to over 18 months, we normalized the percentage weight gain by dividing total weight gain percent by months on drug to derive monthly percent weight change. The monthly weight change was used as the dependent variable in linear regression to determine potential association of GLP1R haplotypes with weight gain.
3 Results
3.1 Halotype structure of GLP1R in CATIE and HAPMAP
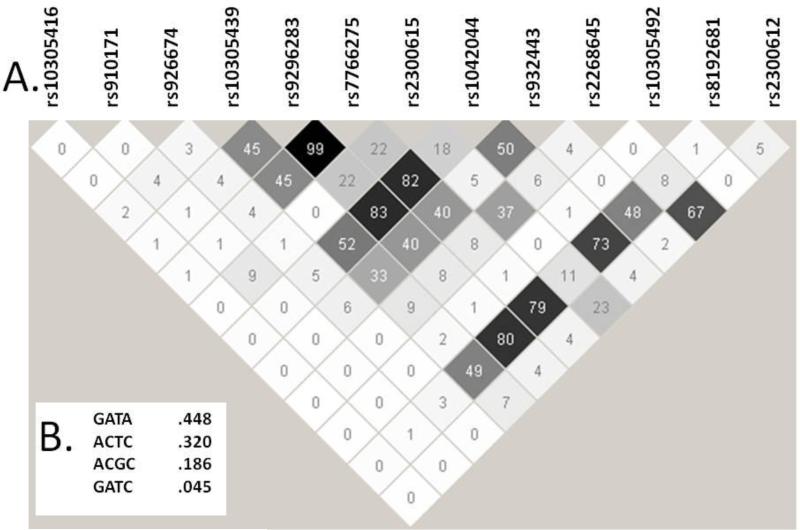
The affymetrix 5.0 Human Genome SNP chip (Affymetrix, Santa Clara, CA) that was used to genotype the CATIE samples includes 13 SNPs in or near the GLP1R gene. One of these SNPs, rs1042044 (Leu260), is a frequent coding variant. Figure 1A displays the linkage disequilibrium structure of these 12 SNPs from haploview. Figure 1B shows the four common haplotypes that this LD block identifies.
Figure 1.
Haploview analysis of the GLP1R SNPs available in the CATIE data set. A) Pairwise r2 values for the 12 SNPs. The area highlighted by the thick black lines defines the core haplotype block identified by Haploview. B) The Haploview-reported haplotypes and frequencies for the core haplotype block with alleles for the SNPs listed in the following order: rs9296283, rs7766275, rs2300615, and rs1042044
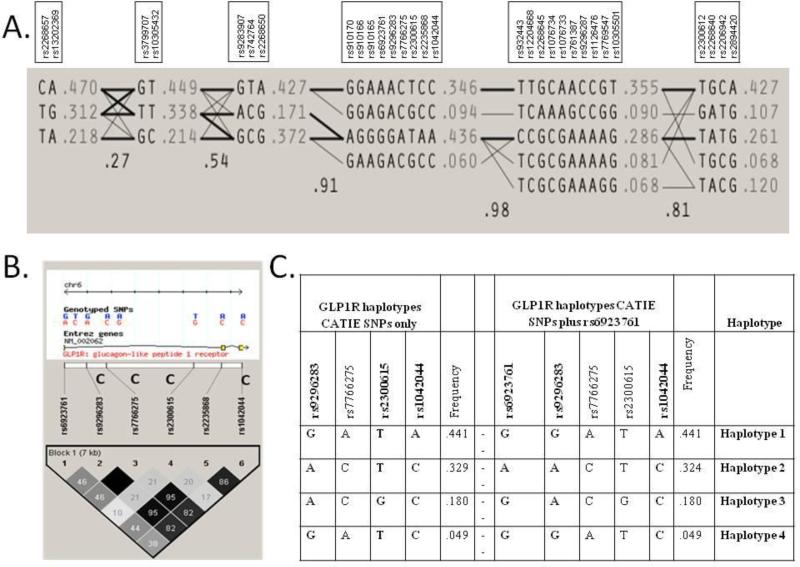
HAPMAP data show that GLP1R has extensive LD across the gene. LD as defined by r2 > 0.5 for the tagging SNPS of the two most frequent haplotypes from CATIE covers exons 3 through 12. The haplotype blocks defined using the Haploview settings described in the methods are shown in Figure 2A. Haplotype block 4, the core haplotype, is the same haplotype as the block identified in CATIE samples with the addition of the coding variant rs6923761 (Ser168) and other SNPs. The coding variant rs1042044 (Leu260) tags the most frequent haplotype of haplotype block 4, hereinafter designated as haplotype 1. We have labeled the second most frequent haplotype of haplotype block 4 as haplotype 2. Despite the coding SNP rs6923761 not being available in the CATIE data, haplotype analysis is able to capture this coding variant indirectly. Figure 2B shows Haploview output of HAPMAP CEU data, Utah residents of northern and western European ancestry, using the haplotype block that extends from rs6923761 to rs1042044. This block includes the four SNP haplotype (rs9296283, rs7766275, rs2300615, and rs1042044) from the CATIE data. Among haplotypes with a frequency > 0.01, rs6923761 is exclusively associated with the ACTC haplotype (haplotype 2) from the 4 SNP CATIE haplotype. This LD relationship can be seen in Figure 2C.
Figure 2.
Halplotype assignments from the full gene HAPMAP data for GLP1R. A) Haploview identifies six haplotype blocks with strong LD between the two most frequent haplotypes in block 4 and frequently occurring haplotypes in other blocks spanning the gene. B) The LD map for GLP1R SNPs from haplotype block 4 between rs6923761 and rs1042044 with the CGSMD-provided CATIE SNPs identified with a C in the figure. C) Haplotypes from the CATIE data and HAPMAP. As can be seen, the minor allele of rs6923761 is uniquely associated with haplotype 2.
Using D'≥ 0.5, the core haplotype, including the SNPs from the haplotype defined in the CATIE data, displays strong LD across the entire gene (Supplemental Figure 1). Thus, this core haplotype block produces two very common haplotypes, Haplotype 1 (44.1%) and Haplotype 2 (32.4%), that are tagged by coding SNPS in LD with SNPs from the promoter to the 3’ end of the gene. In the CATIE genotype data, Haplotype 1 is uniquely tagged by the coding SNP rs1042144 (A), while defining Haplotype 2 requires both rs9296283 (A) and rs2300615 (T).
3.2 Haplotype trend regression of antipsychotic response
Table 2 shows the results HTR of the four GLP1R haplotypes (defined by rs9293283, rs7766275, rs2300615, and rs1042044) in HelixTree using the response model to provide predicted dPANSS from the blinded Phases of CATIE. The rs9293283(G), rs7766275(A), rs2300615(T), and rs1042044(A) haplotype, (haplotype 1) tagged by the rs1042044 (Leu260) allele, correlated with better response to olanzapine and risperidone and worse response to ziprasidone. Haplotype 2, which uniquely includes the rs6923761 (Ser168) allele, correlated with better response to perphenazine and worse response to olanzapine and quetiapine. Haplotype ACGC (haplotype 3), which is contains the major allele at both rs6923761 (GLY168) and rs1042044 (Phe260), correlated with better response to quetiapine and worse response to risperidone. In contrast, no significant associations were found for haplotype 4.
Table 2.
GLP1R Haplotype trend regression of dPANSS for CATIE drugs
| Drug | Haplotype | Number | Frequencya | Relative Betab | P-valuea | Corrected P-valuec |
|---|---|---|---|---|---|---|
| Olanzapine | ||||||
| GATA | 1 | 0.46 | −11.5 | 0.002 | 0.020 | |
| ACTC | 2 | 0.32 | −1.9 | 0.043 | 0.060 | |
| ACGC | 3 | 0.18 | −1.7 | 0.258 | 0.218 | |
| GATC | 4 | 0.03 | 0.0 | 0.486 | 0.314 | |
| Perphenazine | ||||||
| GATA | 1 | 0.45 | −0.7 | 0.100 | 0.122 | |
| ACTC | 2 | 0.33 | −10.4 | 0.015 | 0.033 | |
| ACGC | 3 | 0.17 | 0.0 | 0.324 | 0.255 | |
| GATC | 4 | 0.05 | −8.1 | 0.514 | 0.314 | |
| Quetiapine | ||||||
| GATA | 1 | 0.51 | −6.7 | 0.611 | 0.354 | |
| ACTC | 2 | 0.28 | 0.0 | 0.008 | 0.029 | |
| ACGC | 3 | 0.15 | −15.8 | 0.012 | 0.033 | |
| GATC | 4 | 0.05 | −10.9 | 0.490 | 0.314 | |
| Risperidone | ||||||
| GATA | 1 | 0.41 | −16.2 | 0.038 | 0.060 | |
| ACTC | 2 | 0.35 | −13.4 | 0.687 | 0.378 | |
| ACGC | 3 | 0.18 | −3.0 | 0.018 | 0.033 | |
| GATC | 4 | 0.06 | 0.0 | 0.111 | 0.122 | |
| Ziprasidone | ||||||
| GATA | 1 | 0.39 | 0.0 | 0.007 | 0.029 | |
| ACTC | 2 | 0.38 | −9.3 | 0.155 | 0.155 | |
| ACGC | 3 | 0.18 | −7.4 | 0.200 | 0.183 | |
| GATC | 4 | 0.04 | −5.5 | 0.443 | 0.314 |
Frequency and p-value from Helixtree haplotype trend regression
Relative Beta is the beta weight (in dPANSS) assigned in the HTR normalized such that the maximum beta value is set to 0 and all other values adjust accordingly (relative value = HTR reported value for each haplotype– (the highest value from the group of four haplotypes). Thus the haplotype with the worst response has a beta value of 0.
FDR p-values derived using the graphically sharpened method (Benjamini and Hockberg, 2000).
3.3 Analysis of genetic models of response
In order to evaluate which genetic models provided the best explanation for the HTR results seen above, we assigned haplotypes to each subject using EM algorithm in HelixTree. Each subject was coded for the number of copies (0, 1, or 2) of the two most common haplotypes, Haplotype 1 (GATA) or Haplotype 2 (ACTC). We were able to assign haplotypes to all subjects with greater than 98% certainty. After assigning the haplotypes to individuals, the two rarer haplotypes, haplotypes 3 and 4, had insufficient subjects with two copies of those haplotypes for analysis beyond the HTR described above.
Tables 3 and 4 show the mean and standard deviation of the predicted dPANSS for the three genotypic groups for each haplotype. For each drug by haplotype combination, we evaluated the additive, dominant and recessive genetic models. As expected, the additive model closely resembled the results from the HTR analysis. For haplotype 1 (corresponding to Leu260), a recessive genetic model provided the best fit, with four of the five drugs showing a significant difference in dPANSS between subjects that had two copies of haplotype 1 and subjects with fewer than two copies. For haplotype 2 (corresponding to Ser168), a dominant model provided the best fit for olanzapine and perphenazine response, while a recessive model best described quetiapine response. Table 5 lists the haplotype and model with most significant p-value for response for each drug, the differences in the means of the two groups described by the model, and the effect sizes (Cohen's D).
Table 3.
Antipsychotic response to CATIE drugs segmented by number of copies of haplotype 1
| 0a | 1a | 2a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | N | Meanb | SDc | N | Meanb | SDc | N | Meanb | SDc | ADd | Dome | Recf |
| Olanzapine | 44 | −10.1 | 9.9 | 68 | −11.9 | 13.1 | 32 | −19.4 | 15.8 | 0.003 | 0.08 | 0.002 |
| Perphenazine | 28 | −11.0 | 11.0 | 30 | −10.6 | 15.6 | 20 | −3.8 | 9.4 | 0.07 | 0.31 | 0.03 |
| Quetiapine | 29 | −5.7 | 16.7 | 78 | −4.2 | 11.8 | 34 | −7.0 | 9.2 | 0.64 | 0.79 | 0.33 |
| Risperidone | 48 | −8.4 | 11.8 | 72 | −8.4 | 14.3 | 23 | −17.0 | 15.6 | 0.04 | 0.38 | 0.006 |
| Ziprasidone | 36 | −6.5 | 10.3 | 39 | −4.9 | 9.8 | 16 | 3.3 | 13.6 | 0.007 | 0.09 | 0.003 |
| Grand Total | 185 | −8.4 | 11.9 | 287 | −7.8 | 13.2 | 125 | −10.2 | 15.1 | NS | ||
Number of copies of the haplotype present
Mean of predicted delta PANSS from the Response model
Standard deviation of the mean
P-value for the additive genetic model
P-value for the dominant genetic model - (0 copies) versus (1 Copy + 2 copies)
P-value for the recessive model genetic model - (0 copies + 1 Copy) vs 2 copies
Table 4.
Antipsychotic response to CATIE drugs segmented by number of copies of haplotype 2
| 0a | 1a | 2a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | N | Meanb | SDc | N | Meanb | SDc | N | Meanb | SDc | ADd | Dome | Recf |
| Olanzapine | 69 | −15.7 | 14.7 | 57 | −10.7 | 11.9 | 18 | −10.2 | 9.5 | 0.03 | 0.02 | 0.33 |
| Perphenazine | 38 | −4.4 | 10.5 | 29 | −14.2 | 14.6 | 11 | −11.4 | 10.4 | 0.01 | 0.001 | 0.51 |
| Quetiapine | 74 | −6.7 | 10.6 | 56 | −5.0 | 13.7 | 11 | 4.4 | 13.2 | 0.02 | 0.11 | 0.007 |
| Risperidone | 60 | −9.3 | 15.3 | 67 | −9.9 | 13.6 | 16 | −11.2 | 10.3 | 0.66 | 0.74 | 0.68 |
| Ziprasidone | 34 | −2.5 | 13.0 | 44 | −4.1 | 10.0 | 13 | −8.1 | 9.6 | 0.15 | 0.31 | 0.16 |
| Grand Total | 275 | −8.7 | 13.8 | 253 | −8.5 | 13.2 | 69 | −7.9 | 11.6 | NS | ||
Number of copies of the haplotype present in a given subject
Mean of predicted delta PANSS from the Response model
Standard deviation of the mean
P-value for the additive genetic model
P-value for the dominant genetic model - (0 copies) versus (1 Copy + 2 copies)
P-value for the recessive model genetic model - (0 copies + 1 Copy) vs 2 copies
Table 5.
Effect size for most significant model for each of the Phase 1 CATIE drugs
| Treatment | Modela | Differenceb | Effect sizec |
|---|---|---|---|
| Olanzapine | Haplotype 1 recessive | −8.23 | −0.62 |
| Perphenazine | Haplotype 2 dominant | −9.06 | −0.70 |
| Quetiapine | Haplotype 2 recessive | 10.45 | 0.84 |
| Risperidone | Haplotype 1 recessive | −8.62 | −0.62 |
| Ziprasidone | Haplotype 1 recessive | 9.01 | 0.81 |
Model from Tables 2A or 2B with lowest p-value
Difference between the means of predicted delta PANSS in the two (binary) groups described by the model
Effect size is Cohen's d, calculated as (difference in means) / (standard deviation of group mean)
3.4 Weight gain
As shown in Tables 6 and 7, we used the same genotypic model segmentation to evaluate whether variation in GLP1R was associated with weight gain. Interestingly, none of the haplotypes displayed even nominal significance in any genetic model. Overall, percent monthly change in weight did correlate with the model-predicted dPANSS, p = 0.001. However, despite the significance level, weight change only explained 1.5% of the variance.
Table 6.
Change in weight for each treatment group segmented by number of copies of haplotype 1
| 0a | 1a | 2a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | N | Meanb | SDc | N | Meanb | SDc | N | Meanb | SDc |
| Olanzapine | 42 | 0.62 | 2.13 | 66 | 1.32 | 3.10 | 31 | 1.14 | 2.09 |
| Perphenazine | 28 | 0.58 | 2.22 | 30 | −0.31 | 1.77 | 20 | 0.08 | 1.28 |
| Quetiapine | 28 | 0.69 | 1.52 | 77 | 0.24 | 2.42 | 34 | 1.47 | 5.73 |
| Risperidone | 48 | 0.27 | 2.66 | 70 | 0.48 | 2.51 | 23 | 0.45 | 2.36 |
| Ziprasidone | 35 | 0.13 | 3.02 | 35 | −1.24 | 3.27 | 16 | −0.41 | 2.20 |
| Grand Total | 181 | 0.44 | 2.39 | 278 | 0.31 | 2.77 | 124 | 0.73 | 3.48 |
Number of copies of the haplotype present
Mean of percent weight change per month
Standard deviation of the mean
Table 7.
Change in weight for each treatment group segmented by number of copies of haplotype 2
| 0a | 1a | 2a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | N | Meanb | SDc | N | Meanb | SDc | N | Meanb | SDc |
| Olanzapine | 67 | 0.98 | 1.81 | 54 | 1.24 | 3.54 | 18 | 0.88 | 2.01 |
| Perphenazine | 38 | −0.02 | 1.19 | 29 | 0.03 | 1.92 | 11 | 0.77 | 3.26 |
| Quetiapine | 74 | 0.97 | 4.40 | 55 | 0.10 | 1.77 | 10 | 1.08 | 1.32 |
| Risperidone | 59 | 0.11 | 2.49 | 66 | 0.81 | 2.63 | 16 | −0.20 | 1.94 |
| Ziprasidone | 32 | −0.82 | 3.26 | 42 | −0.47 | 3.22 | 12 | 0.03 | 1.19 |
| Grand Total | 270 | 0.43 | 3.04 | 246 | 0.44 | 2.79 | 67 | 0.48 | 2.06 |
Number of copies of the haplotype present in a given subject
Mean of percent weight change per month
Standard deviation of the mean
4 Discussion
While the role of GLP1R system in energy metabolism and homeostasis has long been known, an increasing body of evidence indicates that GLP1R signaling may impact neuropsychiatric diseases. For example, GLP1 signaling impacts hypothalamic-pituitary-adrenal axis (HPA) activation, autonomic stress response and anxiety-related behaviors (Ghosal et al., 2013). GLP1R also has direct connection with neurotransmitter receptors, calcium signaling molecules, and indirect connection with the dopamine transporter (http://www.ncbi.nlm.nih.gov/gene/2740). Both the native ligand GLP1 and the GLP1R-agonist exenatide stimulate serotonin release in vitro. Furthermore, animal data support a potential role for GLP1R signaling in antipsychotic response. For example, in animal models GLP1R agonists attenuate condition place preference for cocaine; mitigate the effects of alcohol and amphetamine; demonstrate neuroprotective activity in Alzheimer's and Parkinson's diseases; and show direct antipsychotic-like effect comparable to haloperidol (Erreger et al., 2012;Graham et al., 2013;Holscher, 2014;Dixit et al., 2013). Recently, Sharma and colleagues demonstrated that the GLP1R agonist liraglutide can ameliorate some of the negative metabolic and behavioral effects of olanzapine treatment in rats (Sharma et al., 2014a). Finally, the DPP-IV inhibitor sitagliptin, which increases GLP1 concentrations by slowing GLP1 breakdown by DPP-IV, extends the time to tolerance of the anxiolytic effect of ethanol and delays onset of ethanol withdrawal induced anxiety in rats (Sharma et al., 2014b).
In this study, we provide the first report that genetic variation in GLP1R may impact response to commercially available antipsychotics. We identified a large area of high LD spanning exons 3 through 12 of the GLP1R that contains 4 common haplotypes, two of which occur with sufficient frequency to evaluate recessive genetic models. Two coding variants, rs1042044 Leu260 and rs6923761 Ser168, define the two largest haplotypes, haplotype 1 and 2 respectively.
At least one haplotype of GLP1R correlated with altered response to each of five of the antipsychotics evaluated in CATIE. Interestingly, some haplotypes showed associations in opposite directions for different antipsychotics. While more work will be needed to determine the underlying reasons for this, directionally different responses to different medications could relate to heterogeneity in receptor binding profiles for the various drugs.
For each of the five Phase 1 CATIE drugs, the haplotype with the most significant correlation had a “large” effect size as defined as their having absolute values of > 0.5 (Cohen, 1988). Indeed, as seen in Table 5, there is a haplotype associated with each drug with an effect size greater 0.60. That threshold provides an important point of reference for interpreting the clinical meaning of the effect sizes see here, as meta-analysis places the effect size for the comparison of olanzapine versus placebo at 0.59 (Leucht et al., 2009). Thus, we have identified GLP1R haplotypes for each CATIE drug that are potential pharmacogenetic markers to distinguish patient populations with differential response to these antipsychotics.
Both of the coding variants, rs6923761 and rs1042044, have been associated with phenotypic variation in previous studies. Both coding variants encode amino acid residues that are located on intracellular loops of the GLP1R protein, with the rs6923761 variant being in loop 1 and the rs1042044 variant being in loop 2 (Koole et al., 2011). Sheikh et al. reported that children with zero or one copy of the minor allele of rs1042044 (Leu260) had higher morning cortisol, a depression risk factor, than those with two copies, indicating a recessive genetic model for lower morning cortisol (Sheikh et al., 2010). Interestingly, we too find that a recessive model, best explains the impact of haplotype 1 on antipsychotic response. Sathananthan and coworkers reported that healthy subjects possessing at least one copy of the minor allele of rs6923761 (Ser168) have decreased insulin release in response to exogenous GLP-1 (Sathananthan et al., 2010), indicating a dominant genetic model. Correspondingly, a dominant genetic model also provides the best fit for the antipsychotic response data for haplotype 2 in the current study.
In vitro data from Koole et al. may provide some insight for the underlying mechanism for these observed phenotypic differences (Ghosal et al., 2013;Koole et al., 2011). In protein expression assays, cells containing either of the minor alleles for the coding variants, Ser168 or Leu260, produced significantly lower protein levels on the cell surface, approximately 30 to 40 percent of the levels for the other alleles. Calcium mobilization in response to both GLP1 and exenatide was significantly lower in the Ser168 cells (Ghosal et al., 2013;Koole et al., 2011). Thus, given that insulin release is calcium dependent, lower protein levels provide a plausible explanation of reduced insulin secretion through lower signal strength. Interestingly, however, despite significantly lower cell surface expression, cells expressing Leu260 had numerically greater calcium mobilization than those expressing (Phe260), potentially indicating a three-fold greater response to agonist activity per receptor (Ghosal et al., 2013;Koole et al., 2011). Either the lower cell surface expression of Ser168 and Leu260 and / or the enhanced calcium signaling of Leu260 could play important roles in the differential response to antipsychotics through potential interactions with second messengers or transcription factors that modulate activity or expression of cell surface receptors for the various antipsychotics.
Recently, genetic variation in the GLP1R ligand GLP1 has been associated with altered AIWG (Brandl et al., 2014). However, genetic variation in GLP1R was not associated with AIWG in that study. Our observations in the CATIE study agree with that finding, as none of the genetic models tested had a significant association with AIWG. Given the role that GLP1R and its agonists play in weight management broadly and in response to exogenous GLP1 analogue therapy, this lack of association is somewhat unexpected. However, it should be noted that most of the CATIE subjects were being treated with antipsychotics at study entry, which may have blunted any impact by GLP1R genetic variation on weight gain. This potential impact would be studied best in antipsychotic naïve patients. It does suggest, however, that if GLP1R haplotype status is used to select antipsychotic treatment, there should be no particular disadvantage in relation to AIWG.
It will be of interest to determine if the results found here for Caucasians will replicate in other populations, though such comparisons will prove challenging in some cases. Preliminary results do indicate that the poor response to perphenazine and ziprasidone in subjects homozygous haplotype 1 (corresponding to Leu260) may replicate in African Americans (Ramsey and Brennan, unpublished). On the other hand, haplotype 2 (corresponding to Ser168) though present in 54% the Caucasian subjects in CATIE occurs so rarely in African-Americans that it is not practical to evaluate it in this population.
The combination of biological rationale and observed pharmacogenetic effect across a wide range of antipsychotics indicates that GLP1R haplotypes have potential as pharmacogenetic markers of antipsychotic response. Interestingly, the SV2C gene, which controls glucose dependent insulin release as well as GABA vesicle transport in the brain, also appears to impact response to antipsychotic drugs (Ramsey et al., 2013). This connection between metabolic systems and neurological systems and their interplay with antipsychotic response deserves further exploration, at both the genetic and biochemical levels.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Mental Health grant MH078437 to MDB. The principal investigators of the CATIE (Clinical Antipsychotic Trials of Intervention Effectiveness) trial were Jeffrey A. Lieberman, M.D., T. Scott Stroup, M.D., M.P.H., and Joseph P. McEvoy, M.D. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Prior genotyping for the CATIE study was funded by Eli Lilly and Company.
The funder had no role in study design, data analysis, writing of the report, or the decision to submit a manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts
TLR is an employee of SureGene, LLC. MDB is a consultant for SureGene, LLC. Both authors are equity holders in SureGene, LLC.
Contributors
TLR designed the study, with input from MDB, and wrote the first draft of the manuscript. MDB reviewed and approved study design, had oversight of the scientific execution, and participated with writing and editing the manuscript. Both authors have contributed to and approve the final manuscript.
Reference List
- Adkins DE, Aberg K, McClay JL, Bukszar J, Zhao Z, Jia P, Stroup TS, Perkins D, McEvoy JP, Lieberman JA, Sullivan PF, van den Oord EJ. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Mol Psychiatry. 2011;16:321–332. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascher-Svanum H, Stensland M, Zhao Z, Kinon BJ. Acute weight gain, gender, and therapeutic response to antipsychotics in the treatment of patients with schizophrenia. BMC Psychiatry. 2005;5:3. doi: 10.1186/1471-244X-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai YM, Lin CC, Chen JY, Lin CY, Su TP, Chou P. Association of initial antipsychotic response to clozapine and long-term weight gain. Am J Psychiatry. 2006;163:1276–1279. doi: 10.1176/ajp.2006.163.7.1276. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hockberg Y. On the adaptve control of the false discovery rate in multiple testing with independent statistics. Journal of Educational and Behavioral Statistics. 2000;25:60–83. [Google Scholar]
- Bhagwagar Z, Hafizi S, Cowen PJ. Increased salivary cortisol after waking in depression. Psychopharmacology (Berl) 2005;182:54–57. doi: 10.1007/s00213-005-0062-z. [DOI] [PubMed] [Google Scholar]
- Brandl EJ, Tiwari AK, Chowdhury NI, Zai CC, Lieberman JA, Meltzer HY, Kennedy JL, Muller DJ. Genetic variation in the GCG and in the GLP1R genes and antipsychotic-induced weight gain. Pharmacogenomics. 2014;15:423–431. doi: 10.2217/pgs.13.247. [DOI] [PubMed] [Google Scholar]
- Brunetti L, Orlando G, Recinella L, Leone S, Ferrante C, Chiavaroli A, Lazzarin F, Vacca M. Glucagon-like peptide 1 (7-36) amide (GLP-1) and exendin-4 stimulate serotonin release in rat hypothalamus. Peptides. 2008;29:1377–1381. doi: 10.1016/j.peptides.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Chohen J. Statistical Power Analysis for the Behavioral Sciences Lawrence Erlbaum Associates. 1988 [Google Scholar]
- Das C, Mendez G, Jagasia S, Labbate LA. Second-generation antipsychotic use in schizophrenia and associated weight gain: a critical review and meta-analysis of behavioral and pharmacologic treatments. Ann Clin Psychiatry. 2012;24:225–239. [PubMed] [Google Scholar]
- Dixit TS, Sharma AN, Lucot JB, Elased KM. Antipsychotic-like effect of GLP-1 agonist liraglutide but not DPP-IV inhibitor sitagliptin in mouse model for psychosis. Physiol Behav. 2013;114-115:38–41. doi: 10.1016/j.physbeh.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Ebdrup BH, Knop FK, Ishoy PL, Rostrup E, Fagerlund B, Lublin H, Glenthoj B. Glucagon-like peptide-1 analogs against antipsychotic-induced weight gain: potential physiological benefits. BMC Med. 2012;10:92. doi: 10.1186/1741-7015-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Davis AR, Poe AM, Greig NH, Stanwood GD, Galli A. Exendin-4 decreases amphetamine-induced locomotor activity. Physiol Behav. 2012;106:574–578. doi: 10.1016/j.physbeh.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal S, Myers B, Herman JP. Role of central glucagon-like peptide-1 in stress regulation. Physiol Behav. 2013;122:201–207. doi: 10.1016/j.physbeh.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer IM, Bacon A, Ban M, Croudace T, Herbert J. Serotonin transporter genotype, morning cortisol and subsequent depression in adolescents. Br J Psychiatry. 2009;195:39–45. doi: 10.1192/bjp.bp.108.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Erreger K, Galli A, Stanwood GD. GLP-1 analog attenuates cocaine reward. Mol Psychiatry. 2013;18:961–962. doi: 10.1038/mp.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MR. Neuronal and intracellular signaling pathways mediating GLP-1 energy balance and glycemic effects. Physiol Behav. 2012;106:413–416. doi: 10.1016/j.physbeh.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes E, Nasrallah H, Davis V, Meyer J, McEvoy J, Goff D, Davis S, Stroup TS, Swartz M, Lieberman J, Rosenheck R. The association between weight change and symptom reduction in the CATIE schizophrenia trial. Schizophr Res. 2011;128:166–170. doi: 10.1016/j.schres.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holscher C. Insulin, incretins and other growth factors as potential novel treatments for Alzheimer's and Parkinson's diseases. Biochem Soc Trans. 2014;42:593–599. doi: 10.1042/BST20140016. [DOI] [PubMed] [Google Scholar]
- Ishoy PL, Knop FK, Vilsboll T, Glenthoj BY, Ebdrup BH. Sustained weight loss after treatment with a glucagon-like peptide-1 receptor agonist in an obese patient with schizophrenia and type 2 diabetes. Am J Psychiatry. 2013;170:681–682. doi: 10.1176/appi.ajp.2013.12101344. [DOI] [PubMed] [Google Scholar]
- Koole C, Wootten D, Simms J, Valant C, Miller LJ, Christopoulos A, Sexton PM. Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol Pharmacol. 2011;80:486–497. doi: 10.1124/mol.111.072884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht S, Arbter D, Engel RR, Kissling W, Davis JM. How effective are second-generation antipsychotic drugs? A meta-analysis of placebo-controlled trials. Mol Psychiatry. 2009;14:429–447. doi: 10.1038/sj.mp.4002136. [DOI] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- Liu Q, Jamba M, Patrick C, III, Padmanabhan S, Brennan MD. Targeted pharmacogenetic analysis of antipsychotic response in the CATIE study. Pharmacogenomics. 2012a;13:1227–1237. doi: 10.2217/pgs.12.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ramsey TL, Meltzer HY, Massey BW, Padmanabhan S, Brennan MD. Sulfotransferase 4A1 Haplotype 1 (SULT4A1-1) Is Associated With Decreased Hospitalization Events in Antipsychotic-Treated Patients With Schizophrenia. Prim Care Companion CNS Disord. 2012b;14 doi: 10.4088/PCC.11m01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykkegaard K, Larsen PJ, Vrang N, Bock C, Bock T, Knudsen LB. The once-daily human GLP-1 analog, liraglutide, reduces olanzapine-induced weight gain and glucose intolerance. Schizophr Res. 2008;103:94–103. doi: 10.1016/j.schres.2008.05.011. [DOI] [PubMed] [Google Scholar]
- McClay JL, Adkins DE, Aberg K, Bukszar J, Khachane AN, Keefe RS, Perkins DO, McEvoy JP, Stroup TS, Vann RE, Beardsley PM, Lieberman JA, Sullivan PF, van den Oord EJ. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011a;36:616–626. doi: 10.1038/npp.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, Lieberman JA, Sullivan PF, van den Oord EJ. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry. 2011b;16:76–85. doi: 10.1038/mp.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, Goldstein DB. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet. 2009;17:946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey TL, Liu Q, Massey BW, Brennan MD. Genotypic variation in the SV2C gene impacts response to atypical antipsychotics the CATIE study. Schizophr Res. 2013;149:21–25. doi: 10.1016/j.schres.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht LE, Mietlicki-Baase EG, Zimmer DJ, McGrath LE, Olivos DR, Hayes MR. Hindbrain GLP-1 receptor-mediated suppression of food intake requires a PI3K-dependent decrease in phosphorylation of membrane-bound Akt. Am J Physiol Endocrinol Metab. 2013;305:E751–E759. doi: 10.1152/ajpendo.00367.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan A, Man CD, Micheletto F, Zinsmeister AR, Camilleri M, Giesler PD, Laugen JM, Toffolo G, Rizza RA, Cobelli C, Vella A. Common genetic variation in GLP1R and insulin secretion in response to exogenous GLP-1 in nondiabetic subjects: a pilot study. Diabetes Care. 2010;33:2074–2076. doi: 10.2337/dc10-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Vella A. Effects of GLP-1 on appetite and weight. Rev Endocr Metab Disord. 2014 doi: 10.1007/s11154-014-9289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AN, Ligade SS, Sharma JN, Shukla P, Elased KM, Lucot JB. GLP-1 receptor agonist liraglutide reverses long-term atypical antipsychotic treatment associated behavioral depression and metabolic abnormalities in rats. Metab Brain Dis. 2014a doi: 10.1007/s11011-014-9591-7. [DOI] [PubMed] [Google Scholar]
- Sharma AN, Pise A, Sharma JN, Shukla P. Dipeptidyl-peptidase IV (DPP-IV) inhibitor delays tolerance to anxiolytic effect of ethanol and withdrawal-induced anxiety in rats. Metab Brain Dis. 2014b doi: 10.1007/s11011-014-9603-7. [DOI] [PubMed] [Google Scholar]
- Sheikh HI, Dougherty LR, Hayden EP, Klein DN, Singh SM. Glucagon-like peptide-1 receptor gene polymorphism (Leu260Phe) is associated with morning cortisol in preschoolers. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:980–983. doi: 10.1016/j.pnpbp.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroup TS, Lieberman JA, McEvoy JP, Swartz MS, Davis SM, Capuano GA, Rosenheck RA, Keefe RS, Miller AL, Belz I, Hsiao JK. Effectiveness of olanzapine, quetiapine, and risperidone in patients with chronic schizophrenia after discontinuing perphenazine: a CATIE study. Am J Psychiatry. 2007;164:415–427. doi: 10.1176/ajp.2007.164.3.415. [DOI] [PubMed] [Google Scholar]
- Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM, McGee MF, Simpson GM, Stevens MC, Lieberman JA. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull. 2003;29:15–31. doi: 10.1093/oxfordjournals.schbul.a006986. [DOI] [PubMed] [Google Scholar]
- van den Oord EJ, Adkins DE, McClay J, Lieberman J, Sullivan PF. A systematic method for estimating individual responses to treatment with antipsychotics in CATIE. Schizophr Res. 2009;107:13–21. doi: 10.1016/j.schres.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.