Abstract
N-RAS mutation at codon 12, 13 or 61 is associated with transformation; yet, in melanoma, such alterations are nearly exclusive to codon 61. Here, we compared the melanoma susceptibility of an N-RasQ61R knock-in allele to similarly designed K-RasG12D and N-RasG12D alleles. With concomitant p16INK4a inactivation, K-RasG12D or N-RasQ61R expression efficiently promoted melanoma in vivo, whereas N-RasG12D did not. Additionally, N-RasQ61R mutation potently cooperated with Lkb1/Stk11 loss to induce highly metastatic disease. Functional comparisons of N-RasQ61R and N-RasG12D revealed little difference in the ability of these proteins to engage PI3K or RAF. Instead, N-RasQ61R showed enhanced nucleotide binding, decreased intrinsic GTPase activity and increased stability when compared to N-RasG12D. This work identifies a faithful model of human N-RAS mutant melanoma, and suggests that the increased melanomagenecity of N-RasQ61R over N-RasG12D is due to heightened abundance of the active, GTP-bound form rather than differences in the engagement of downstream effector pathways.
Keywords: NRAS, melanoma, codon-specific oncogene
Introduction
One-third of all human cancers harbor activating K-, H- or N-RAS mutations, which localize predominantly to codons 12, 13 or 61 (1, 2). RAS proteins function as canonical GTPase switches, binding to effectors in the presence of GTP and activating downstream signaling pathways to influence cellular proliferation, differentiation and survival. Return of RAS to an inactive, GDP-bound state is catalyzed by GTPase activating proteins (GAPs), which stimulate the weak, intrinsic GTPase activity of these proteins. Mutations at codons 12 or 13 render RAS proteins insensitive to GAP activity, resulting in constitutive, oncogenic signaling (3). Similarly, mutation of Q61, a catalytic residue required for efficient GTP hydrolysis, impedes the return of RAS to an inactive GDP-bound state (4).
Historically, RAS proteins with codon 12, 13 or 61 alterations have been considered oncogenic equivalents; however recent clinical observations suggest functional differences for each RAS mutation. For example, in colorectal cancer, K-RAS mutational status is used as a prognostic indicator of resistance to therapy with EGFR antibodies (e.g. cetuximab) (5–8). Retrospective analyses, however, of ‘all-comer’ trials suggests mutational specificity in this regard: patients harboring K-RAS codon 13 mutations appear to benefit from cetuximab therapy, while those with codon 12 mutations were unresponsive (9–13). Moreover, progression-free survival on targeted therapies may also be codon-specific in non-small cell lung cancer (NSCLC) (14). Here, molecular modeling and reverse phase protein analysis pinpointed differential effector engagement and downstream signaling as potential mediators of mutation-specific therapeutic response (14). Together, these results suggest that distinct, codon-specific properties of RAS mutations have important clinical and biological implications.
Cancers display tissue-specific preferences for mutation of the RAS homologs (Table S1). In melanoma, N-RAS is by far the most frequently mutated RAS isoform and notably, 84% of these mutations localize to codon 61 versus only 7% to glutamine 12 (Table S1). A similar preference for codon 61 mutations is noted in thyroid cancer, but is not observed in other cancer types. Codon 12 and 13 mutations constitute more than 90% of K-RAS mutations observed in human colon, pancreatic, lung and ovarian cancers (Table S1 and (1)). Likewise, glycine 12 is the most common site of N-RAS mutation in acute myeloid leukemia (Table S1). The mechanistic basis for codon 61 selection in melanoma and thyroid cancer is unclear. Some have suggested that cytosine to thymidine transversions caused by ultraviolet (UV) light may explain the preference for certain mutations in melanoma, but the majority of codon 61 mutations do not exhibit a characteristic UV-damage signature (15). Alternatively, it is possible that codon mutation preferences reflect differences in oncogenic signaling.
Comparing the oncogenic potential of various RAS mutants is challenging for several reasons. RAS gene dosage clearly influences downstream signaling, and artifacts of RAS overexpression are well-described. Likewise, endomembrane localization is critical for physiologic RAS signal transduction (16), and may not be adequately recapitulated using exogenous protein expression. In addition, genetic alterations private to a given cell line or tumor sample could obscure distinct functions of individual RAS mutants. To circumvent these issues, we generated a knock-in allele (LSL-N-RasQ61R) allowing for the conditional, tissue-specific and somatic expression of N-RasQ61R under control of the endogenous promoter. We used this allele in combination with similarly designed knock-in N-RasG12D (17) and K-RasG12D (18) models to compare the transforming potential of each mutant when expressed at physiological levels in melanocytes.
Results
Generation and characterization of LSL-N-Ras alleles
To compare the ability of N-Ras mutants to promote melanoma formation, we employed three conditional knock-in alleles: LSL-N-RasG12D (17), LSL-K-RasG12D (18) and LSL-N-RasQ61R. Each allele contains a floxed transcriptional stop sequence followed by a single missense mutation in the endogenous Ras gene (G12D or Q61R, respectively; Fig. 1A). The codon 12 LSL-K-Ras and N-Ras alleles have been previously described (17, 18). We generated and confirmed a related LSL-N-RasQ61R allele using standard homologous recombination followed by Southern blot, PCR and genomic sequencing (Fig. S1A–D). To minimize strain-specific effects, all alleles were backcrossed more than 7 generations to C57Bl/6J in the presence of a conditional p16INK4a knockout allele (p16L; (19)) and a melanocyte-specific, 4-hydroxytamoxifen inducible CRE recombinase (Tyr-CRE-ERT2; (20)). Cohorts of all three alleles were born at normal Mendelian ratios (data not shown) and showed no defects in development or fertility.
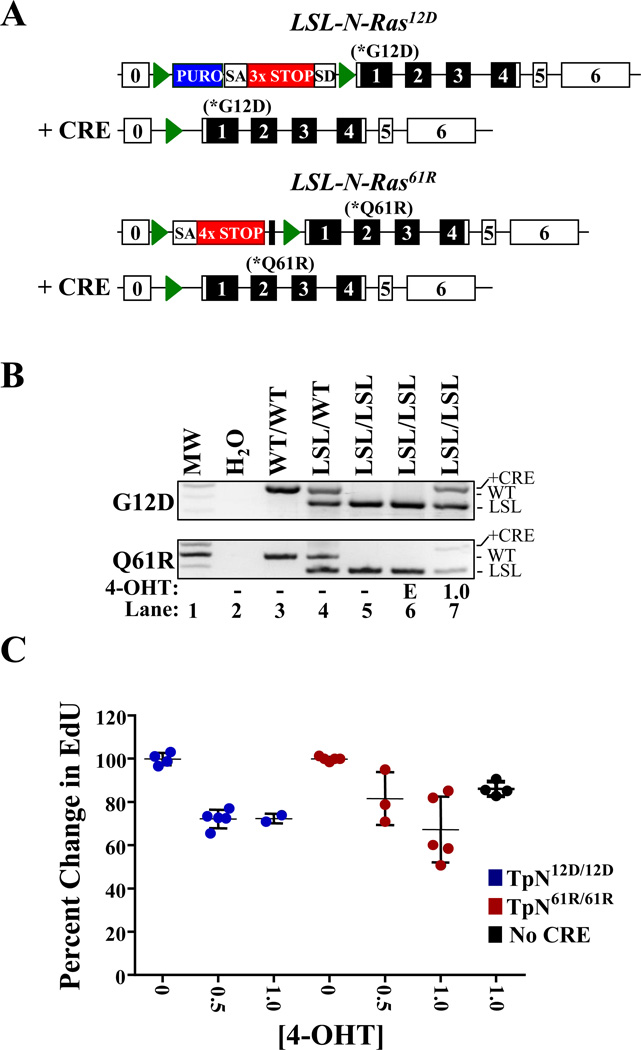
Figure 1. Activation of LSL-N-RasQ61R or LSL-N-RASG12D slows melanocyte proliferation.
(A) Diagrammatic representation of the LSL-N-RASG12D (17) and LSL-N-RasQ61R alleles in the presence or absence of CRE recombinase. Green triangles denote lox P sites and unnumbered black boxes represent FRT sites. Blue boxes indicate the location of a puromycin (PURO) resistance cassette. SA-splice acceptor; SD-splice donor; 3×STOP- transcription and translational stop sequence. (B) PCR genotyping of melanocytes treated for 6 days with ethanol vehicle (E; lane 7) or 1.0 µM 4-OHT (lane 7). DNA from mice of the indicated genotypes is included as a control (lanes 3–5). (C) Melanocytes cultured as in ‘B’ were incubated with EdU, a thymidine analog, for 16 hours and then analyzed by flow cytometry. EdU incorporation in vehicle treated cells was set to 100%. Each dot represents a biological replicate with the mean and standard error of the mean indicated by black lines.
Using primary melanocytes derived from syngeneic Tyr-CRE-ERT2 p16L/L LSL-N-RasG12D/G12D (TpN12D/12D) or LSL-N-RasQ61R/Q61R (TpN61R/61R) neonates, we verified the functionality of the LSL alleles. Melanocyte purities of >99% were confirmed using Tyrosinase-Related Protein 1 (TRP-1) as a marker for flow cytometry ((21); Fig. S2A). In culture, melanocytes were treated with ethanol vehicle or 4-hydroxytamoxifen (4-OHT) to induce CRE activity. CRE-dependent excision of the transcriptional stop element was verified by PCR (Fig. 1B), and resulted in the production of mutant N-Ras mRNA (Fig. S2B). Activation of either allele did not induce changes in melanocyte morphology or pigmentation (Fig. S2C), but induced a decrease in melanocyte proliferation as measured by EdU incorporation (Fig. 1C). Despite co-deletion of p16INK4a, physiological expression of either N-Ras mutant caused comparable anti-proliferative effects (Fig. 1C). Additionally, both TpN12D/12D and TpN61R/61R melanocytes failed to bypass senescence in the presence of 4-OHT, and the cells invariably ceased proliferating after 2–3 passages in culture (data not shown). Therefore, when expressed under the control of an endogenous promoter, neither N-RasG12D nor N-RasQ61R was capable of immortalizing p16INK4a-deficient melanocytes.
N-RAS-driven melanomagenesis is codon-specific
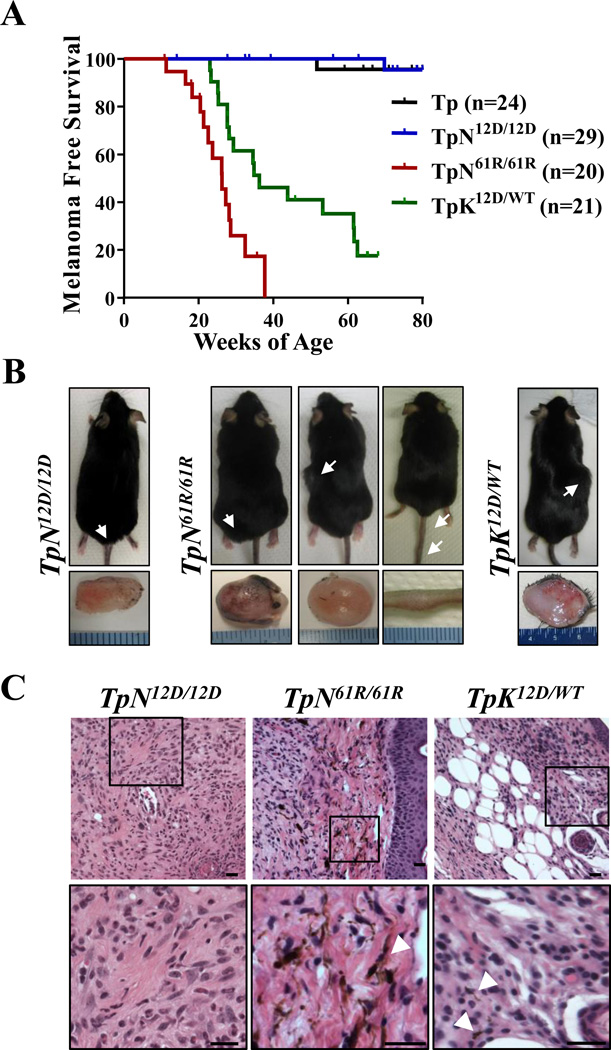
We next sought to examine the phenotypic effects of melanocyte-specific Ras mutations in vivo. Toward that end, we generated contemporaneous colonies of p16INK4a-deficient K-Ras or N-Ras mutant mice in a common genetic background. We elected to study N-Ras mutations in the homozygous state for two reasons: i) we noted no melanoma formation despite 80 weeks of monitoring in a large cohort of Tyr-CRE-ERT2 p16L/L LSL-N-rasG12D/WT (TpN12D/WT) animals (Fig. S3A), and ii) deep sequencing of nine N-RAS mutant human melanoma cell lines failed to detect the presence of a wild type allele, demonstrating consistent N-RAS loss of heterozygosity in vivo (22). Prior work from our group has demonstrated that melanocytic expression of K-RasG12D efficiently promotes melanoma in vivo in the setting of p16INK4a inactivation (19, 23). Herein, syngeneic Tyr-CRE-ERT2 p16L/L LSL-K-RasG12D/WT (TpK12D/WT), were maintained in a heterozygous state due to the homozygous lethality of K-Ras deletion (24). All mice in these cohorts were treated neonatally with 4-OHT (19) to induce melanocyte-specific expression of the desired Ras mutant and delete p16INK4a.
Initial examination of the skin, paws and tails of these mice revealed the presence of nevi and hyperpigmented regions on the paws and tails (Fig. S4A). The penetrance and severity of these phenotypes was allele-specific with TpK12D/WT animals having the most pronounced effect and the N-Ras codon 12 mutation producing the least (TpK12D/WT > TpN61R/61R > TpN12D/12D). To quantify nevi frequency, nevi presence was scored weekly for 10 weeks. As expected, nevi were rarely observed on Tyr-CRE-ERT2 p16L/L (Tp) mice, but consistently found on TpK12D/WT animals (Fig. S4B). Members of both the TpN12D/12D and TpN61R/61R cohorts had more nevi than control, Tp mice (p<0.001). However, the frequency with which N-RasQ61R triggered nevus formation was significantly higher than that observed in TpN12D/12D animals (p = 0.03). These data suggest that in p16INK4a-deficient melanocytes, physiological N-RasQ61R expression more efficiently promotes nevus formation than N-RasG12D mutation.
These established colonies of syngeneic TpN12D/12D, TpN61R/61R and TpK12D/WT mice were aged and serially assessed for melanoma formation. In accordance with previous data, TpK12D/WT animals developed tumors with high penetrance ((19); Fig. 2A, Table 1). Tumors were very rare in TpN12D/12D mice (1 tumor found in 29 mice observed for 80 weeks), whereas TpN61R/61R mice readily developed melanoma with high penetrance and a median latency of 26.3 weeks (Fig. 2A, B and Table 1). To explain this result, we considered the possibility that the LSL-N-RasQ61R and LSL-N-RasG12D alleles had different recombination efficiencies. Toward that end, we examined allelic recombination in primary melanocyte cultures and noted that if anything, the codon 61 allele recombined with lower efficiency than the codon 12 allele (Fig. S5A). We also considered the possibility that the recombined LSL-N-RasG12D allele was poorly expressed. Sequencing of cDNA from treated melanocytes confirmed 4-OHT-dependent expression of N-RasG12D mRNA (Fig. S2B). Lastly, functional validation of the LSL-N-RasG12D allele was accomplished by inducing allelic recombination in the hematopoietic lineage using an interferon-inducible Mx1-CRE driver. Expression of N-RasG12D in the hematopoietic compartment efficiently induced a myeloproliferative syndrome in accord with prior findings ((25) and data not shown). These data establish that when expressed at physiological levels in melanocytes, N-RasQ61R and K-RasG12D are inherently more transforming than N-RasG12D.
Figure 2. In vivo melanomagenesis is isoform and mutation-specific.
(A) Kaplan-Meier curve of melanoma-free survival. Animals were followed for a total of 80 weeks. Survival of each strain was compared pairwise to that of TpN61R/61R animals using a log-rank (Mantel-Cox) test. The following significant p-values were determined: Tp and TpN12D/12D - p<0.0001; TpK12D/WT- p<0.0027. (B) Representative images of the amelanotic tumors found throughout the skin of TpN12D/12D, TpN61R/61R and TpK12D/WT animals. White arrowheads indicate the tumor location. (C) Hematoxylin and eosin staining of representative tumors of the indicated genotypes. Black bars represent a length of 20µm. White arrowheads indicate areas of melanin deposition.
Table 1.
Comparative summary of melanomagenesis and macrometastasis in LSL-Ras-driven models.
| Genotype | Tumors/Treated Mice (%) |
Median Tumor Latency (wks) |
Macrometastasis |
|---|---|---|---|
| Tp | 1/24 (4.1%) | > 80 | None |
| TpN12D/12D | 1/29 (3.4%) | > 80 | None |
| TpN61R/61R | 14/20 (70%) | 26.3 | None |
| TpK12D/WT | 16/21 (76%) | 36.3 | None |
| TpLN61R/61R | 17/20 (85%) | 22.1 | 5/14 (36%) |
We performed additional analyses to determine if there were phenotypic differences in the tumors of TpN12D/12D, TpN61R/61R and TpK12D/WT mice. Similar growth rates were observed in tumors from TpN12D/12D, TpN61R/61R and TpK12D/WT mice, albeit only one tumor was found in the TpN12D/12D cohort (Fig. S5B). Moreover, melanomas from all three groups were histologically similar; containing both spindle-cell and desmoplastic cell types with no overt signs of macrometastatic spread (Fig. 2C, Table 1). In the TpN61R/61R tumors, we confirmed the presence of the recombined LSL-N-rasQ61R allele (Fig. S5C) and used quantitative real-time PCR to look for potential gene amplifications. Compared to TpK12D/WT tumors, in which endogenous N-Ras expression is unaltered, TpN61R/61R melanomas displayed no significant change in N-Ras mRNA levels, suggesting that the allele was not amplified during tumorigenesis (Fig. S5D). These data show that N-RasQ61R, N-RasG12D and K-RasG12D tumors, once established, are phenotypically similar, and suggests that oncogenic differences between the alleles is most pronounced during tumor initiation.
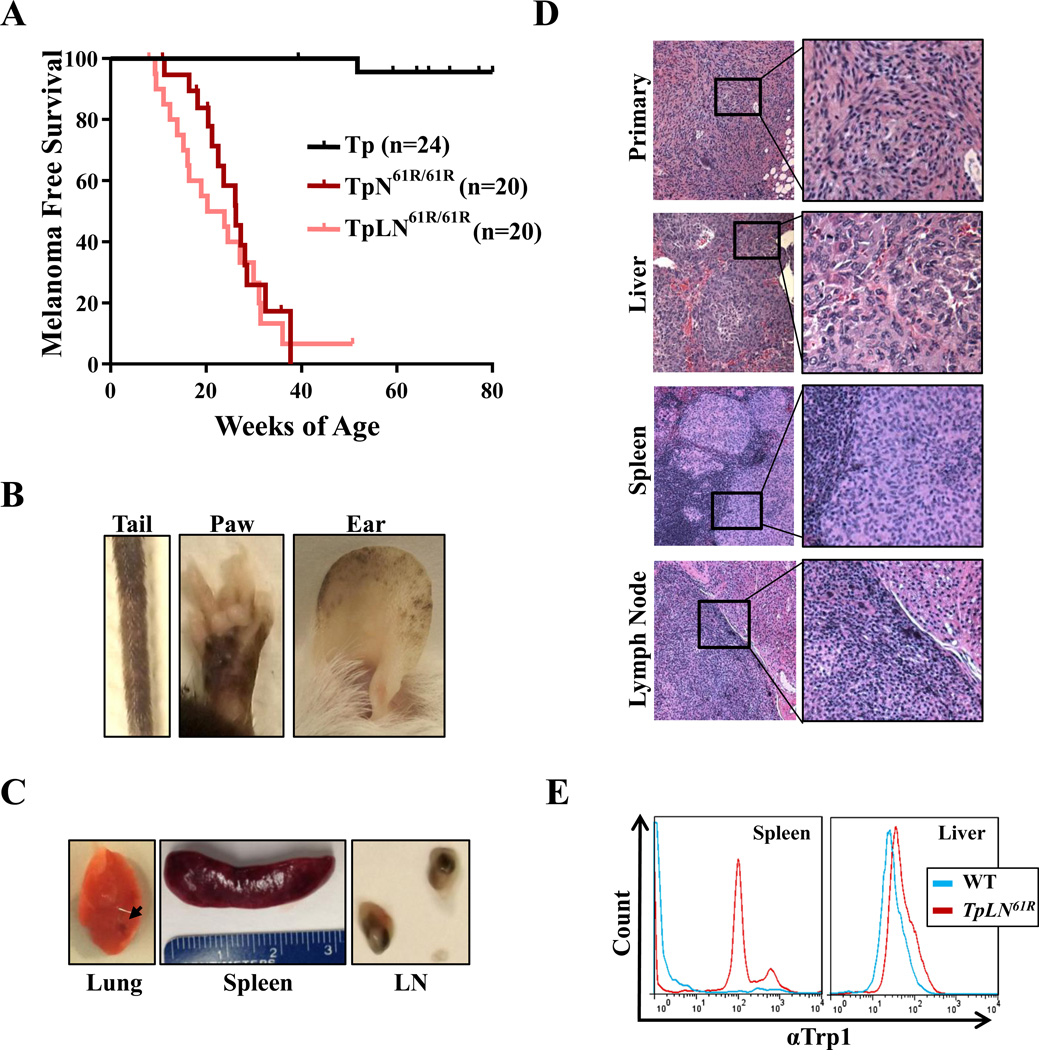
Lkb1 loss promotes melanoma metastasis in TpN61R/61R mice
Melanoma mortality is predominantly attributed to metastatic disease, however very few RAS-driven genetically engineered mouse models, including the TpN61R/61R model, show evidence of macrometastases (Tables 1 and (26)). We recently demonstrated that the loss of Liver Kinase 1 (Lkb1/Stk11) promotes widespread macrometastases in a K-Ras12D-driven mouse melanoma model (23). LKB1 is mutated in ~10% of malignant human melanomas (15, 27, 28) and functionally inactivated by oncogenic BRAF (22, 29, 30). To test whether Lkb1 loss promotes metastasis in TpN61R/61R tumors, we crossed these animals to a previously described conditional Lkb1 knockout allele (Lkb1f; (31)). TpLN61R/61R mice were treated neonatally or at adulthood with 4-OHT to stimulate melanocyte-specific deletion of both Lkb1 and p16INK4a and induce N-Ras61R expression. The onset of tumor formation in TpN61R/61R neonates was not affected by Lkb1 loss (Fig. 3A; Table 1); however, the propensity for distant metastasis was markedly affected. TpLN61R/61R animals, treated as adults or neonates, exhibited enhanced nevus formation relative to Lkb1-proficient counterparts (compare Fig. S4A to Fig. 3B). Upon sacrifice due to increasing tumor burden, many of TpLN61R/61R showed signs of metastasis. We performed thorough autopsies on 14 TpLN61R/61R mice, noting noting lymph node enlargement and other signs of macroscopic disease (Fig. 3C) in the majority of animals. The presence of visceral metastatic disease to the lung, spleen and/or liver was confirmed in 5 of these animals by histologic examination (36%; Fig. 3D, Table 1). Metastastases appeared similar in morphology and incidence to those observed in a prior K-Ras-driven, Lkb1−/− melanoma model (23). Flow cytometric analyses of TpLN61R/61R mice with primary melanomas showed the presence of infiltrating tumor cells expressing the melanocyte marker, Trp1 (Fig. 3E; Note that CD45+ cells were excluded from splenic analyses). Unlike cells from TpN61R/61R tumors, we were able to culture TpLN61R/61R melanoma cells in vitro. These cells lines exhibited varied morphologies, but were invariably Trp-1 positive (Fig. S6). Together, these findings establish TpLN61R/61R as a faithful murine model of metastatic, cutaneous melanoma driven by an endogenous, oncogenic N-RAS allele.
Figure 3. Lkb1 deficiency promotes metastasis in TpN61R/61R tumors.
(A) Kaplan-Meier curve comparing the melanoma-free survival of TpN61R/61R and TpLN61R/61Rmice treated neonatally with 4-OHT. A log-rank (Mantel-Cox) test revealed no significant difference in disease onset in these models (p>0.05). (B) Representative photographs showing severe melanocytic hyperproliferation in the ears, tails and paws of TpLN61R/61Rmice. (C) Images of macrometastases in the lung, spleen and lymph nodes (LN) of TpLN61R/61Rmice. (D) Representative hematoxylin and eosin stained TpLN61R/61Rtissues showing tumor invasion into the liver, spleen and lymph node. (E) Expression of TRP1 in the spleen and liver of TpLN61R mice with primary melanomas. The aqua line designates tissue from a wild type animal and the red line shows TRP1 expression in a TpLN61R/61Rmouse with observed macrometastases. For splenic tissues, CD45+ cells were excluded prior to analysis.
N-RASQ61R and N-RASG12D similarly bind melanogenic effector pathways
The distinct oncogenicity of H-, K- and N-RAS is often attributed to the isoform-specific, preferential engagement of downstream effector pathways (17, 32–35). We postulated that a similar mechanism might also drive codon-specific melanomagenesis. To this end, the interaction of purified K-RAS, N-RAS, N-RASG12D and N-RASQ61R with PI3K and RAF (i.e. the BRAF RBD) was examined in vitro. Each GTPase was first loaded with a fluorescent GTP analog (mant-GMPPNP). Next, purified PI3K or RAF-RBD was titrated into reactions containing a constant amount of GMPPNP-RAS protein at both 5 mM and 100 µM Mg2+ concentrations. Fluorescence quenching, caused by effector binding was monitored to determine the Kd for each protein-protein interaction. In these assays, N-RASQ61R bound both PI3K and the RAF-RBD with a lower affinity than either wild-type N-RAS or N-RASG12D (Table 2). However, the noted differences were small (<1-fold for PI3K; ~4-fold for the RAF-RBD) and unlikely to translate to an in vivo phenotype. Likewise, isothermal titration calorimetry experiments did not reveal significant differences in the RAF-RBD binding affinities of N-RASQ61R, N-RASG12D and wild-type N-RAS (Figure S8). These data indicate that distinct engagement of the oncogenic RAS effector pathways frequently targeted in melanoma (i.e. RAF and PI3K), is not the cause of codon-specific melanomagenesis.
Table 2.
Binding affinities of RAS-mant-GMPPNP to PI3Kγ and RAF-RBD.
| RAS Mutant |
mant-GMPPNP Binding | |
|---|---|---|
| KD to bRaf-RBD (µM) | KD to PI3K (µM) | |
| NRASWT | 0.07 ± 0.03 | 2.2 ± 0.4 |
| NRASG12D | 0.08 ± 0.04 | 1.8 ± 0.5 |
| NRASQ61R | 0.28 ± 0.04 | 2.7 ± 1.2 |
| KRASG12D | 0.06 ± 0.03 | 2.5 ± 0.8 |
Activation of MAPK and ERK is codon-independent in NRAS mutant melanomas
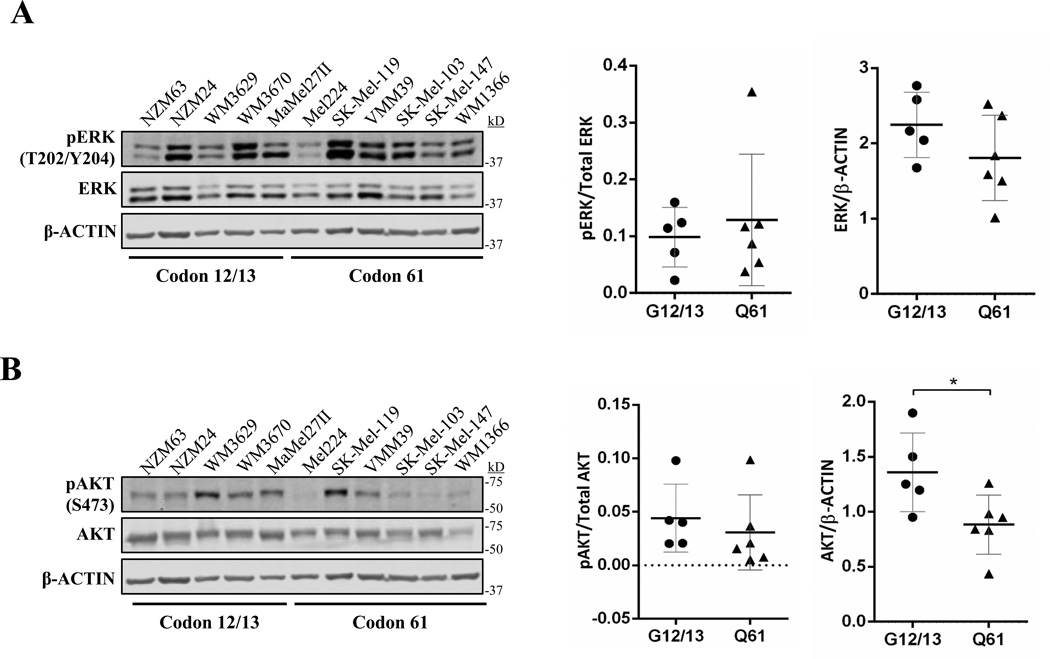
The distinct subcellular localizations of H-, K- and N-RAS are suggested to influence effector availability and contribute to isoform-specific RAS oncogenicity (17, 32–35). To determine whether the availability of specific effector pools in vivo contributes to codon-specific signaling, we examined MAPK and PI3K activation in a variety of human melanoma cell lines harboring a mutation in N-RAS codon 12, 13 or 61 (n=11). These cell lines exhibited variable levels of activated ERK and AKT that did not correlate with genotype (Fig. 4A and B). NRASG12/13-mutant cell lines exhibited a moderate increase (<2-fold) in total ERK and AKT expression; however, the significance of this observation is unclear (Fig. 4A and B, right) and this finding would not explain the increased transforming activity of codon 61 mutants. In addition, no difference was observed in the proliferation of N-RAS codon 12/13 and 61-mutant cell lines as measured by EdU incorporation (Fig. S9).
Figure 4. Activation of ERK and AKT in human melanoma cell lines is not codon-specific.
Shown are immunoblots for total and phosphorylated ERK (A), or AKT (B) in established human melanoma cell lines harboring the indicated NRAS mutations. Protein expression levels were quantified using LI-COR ImageStudio software. Each dot represents a single cell line. The mean is indicated by a line, and the standard error is shown as whiskers. * p<0.05
Due to the confounding potential of secondary mutations acquired in human melanoma cell lines, we also examined ERK and AKT activation in primary and immortalized melanocytes derived from the TpN12D/12D and TpN61R/61R models. Expression of each N-RAS allele was induced in vitro using CRE recombinase and verified by genomic PCR (Fig. S810A and B, top). Immortalized TpN61R/61R melanocytes were particularly resistant to sustained allelic recombination (Fig. S10B, top). Therefore, we analyzed these cells at an early time point following CRE induction (2 days). In both cases, we found that phospho-ERK and AKT levels were variable and codon-independent (Fig. S10A and B, bottom). These data, along with observations in human melanoma cell lines, demonstrate that melanomageneic and non-melanomageneic N-RAS mutants similarly activate the MAPK and PI3K pathways.
NRASQ61R exhibits distinct biochemical properties
Surprised by the finding that NRAS codon 12, 13 and 61 mutants appear equally to engage the oncogenic PI3K and MAPK effector pathways, we further investigated the biochemical properties that differentiate these two mutants. We observed during mant-GMPPNP-based binding assays, that the nucleotide exchange rate of NRASQ61R was significantly retarded compared to WT or NRASG12D (data not shown). To follow up on this observation, we measured the intrinsic and SOS-mediated nucleotide disassociation rates of purified NRAS, NRASG12D and NRASQ61R. To monitor exchange, each NRAS variant was pre-loaded with either mant-GMPPNP (mimicking the GTP bound state) or mant-GDP and then incubated with 1000-fold excess unlabeled nucleotide. The resulting change in fluorescence over time was used to monitor exchange rates. NRAS, NRASG12D and NRASQ61R showed a similar ability to exchange GDP (Table 3). However, the GMPPNP exchange rate of NRASQ61R was significantly slower than NRAS and NRASG12D (Table 3). This distinction became more pronounced when SOS was added to each reaction (Table 3). Specifically, SOS was unable to stimulate either GDP or GMPPNP exchange in the codon 61 variant (Table 3). These results indicate that NRASQ61R possesses a clear affinity for GTP that cannot be overcome by GEF interaction at the concentrations used in our assays.
Table 3.
Nucleotide binding affinities and intrinsic GTP hydrolysis rates of relevant NRAS isoforms.
| RAS Mutant |
GDP Off | GDP off + 1:2 SOScat | GMPPNP Off | GMPPNP Off + 1:2 SOScat | Intrinsic Hydrolysis |
|---|---|---|---|---|---|
| Rate (s−1 × 10−4) | Rate (s−1 × 10−4) | Rate (s−1 × 10−4) | Rate (s−1 × 10−4) | Rate (s−1 × 10−4) | |
| NRASWT | 1.5 ± 0.2 | 8.0 ± 0.7 | 1.7 ± 0.4 | 9.8 ± 0.9 | 4.6 ± 0.4 |
| NRASG12D | 1.3 ± 0.3 | 3.3 ± 0.4 | 1.6 ± 0.5 | 3.9 ± 0.6 | 2.3 ± 0.5 |
| NRASQ61R | 1.1 ± 0.2 | 1.3 ± 0.3 | 0.4 ± 0.2 | 0.5 ± 0.2 | 0.002 ± 0.03 |
These data, along with previous observations in other RAS isoforms (36), suggest a decreased intrinsic GTPase activity of NRASQ61R. Indeed, when directly assessed, the intrinsic GTP hydrolysis rate of NRASQ61R was much slower than NRASG12D or NRAS (1,150 and 2,300 times slower, respectively; See Table 3). These results, taken together, suggest that NRASQ61R has higher affinity for GTP relative to NRAS and NRASG12D. Therefore, we examined the relative stability of NRAS, NRASG12D and NRASQ61R using thermal unfolding measurements for both GDP- and GMPPCP-bound proteins. NRASQ61R remained stable until reaching 80 ± 5°C, whereas the wild type and NRASG12D proteins were destabilized at much lower temperatures (67 ± 3°C and 70 ± 3°C, respectfully; See Table S2). The thermostability of NRASQ61R was lower in the GDP-bound versus GMPPCP-bound state (74 ± 4°C versus 80 ± 5°C, See Table S2), suggesting that NRASQ61R adopts a conformation which stabilizes nucleotide binding, especially in the GTP-bound state. Together, our work reveals that NRASQ61R exhibits distinct nucleotide binding capacity, stability and GTPase resistance likely responsible for its exceptional melanomagenic properties.
Discussion
Our data explain a long-standing mystery in the field, demonstrating through biochemical and genetic analyses that the predominance of codon 61 mutants in human melanoma can be attributed to their distinct oncogenic properties. In our novel suite of mouse models, endogenous levels of NRasQ61R, but not NRasG12D were able to efficiently drive in vivo melanomagenesis (Fig. 2). Although prior work has linked isoform-specific RAS oncogenicity to differential effector binding (17, 32–35), we found that NRASQ61R and NRASG12D similarly engaged the PI3K and MAPK pathways (Table 2, Fig. 4, Fig. S8, Fig. S10). Our data suggest an alternative model for codon-specific oncogenicity, demonstrating that the melanomagenic NRASQ61R mutant possesses distinct biochemical properties not found in NRASG12D (Table 3).
Prior to this study, it was unclear whether the prevalence of NRAS codon 61 mutations in melanoma reflected a preferential pattern of mutagenesis or codon-specific differences in the biological function of each allele. We now show that NRASG12D does not efficiently drive cutaneous melanomagenesis (Fig. 2), and therefore, codon-specific RAS biology influences tumorigenic potential. Tissue- and temporal-specificity are also likely to affect oncogenic RAS activity. For instance, activation of the LSL-NRASG12D allele in hematopoietic stem cells and early melanocyte progenitors drives leukemogenesis and melanoma of the central nervous system ((25, 37, 38), data not shown). Moreover, it is possible that our experimental system, which relies upon p16INK4a loss to facilitate melanomagenesis, is biased to favor N-Ras codon 61 mutations. Patient studies have suggested exquisite cooperation between N-RAS codon 61 mutations and p16INK4a inactivation (39, 40), and N-RasQ61K transgenic mice appear to be ‘addicted’ to the activity of CDK4/6, which is enhanced by p16INK4a loss (41). Therefore, perhaps other cooperating oncogenic events commonly found in melanoma would more efficiently synergize with N-RasG12D in tumor formation. Nonetheless, p16INK4a loss is found in the majority of human melanomas, occurring through a variety of mechanisms (i.e. epigenetic silencing, mutation and deletion). Given the significance of this event in the clinical setting, we believe the choice of the p16INK4a-deficient genetic context is highly relevant to the human disease.
Widely efficacious and durable treatment options are not available for the 17–30% of patients with NRAS mutant, metastatic melanomas (1, 15, 42). With mounting data to suggest that genetically engineered mouse models of cancer more faithfully report therapeutic efficacy (43–45), a number of RAS-mutant melanoma models have been developed (See Table S3). Most of these models employ transgenic technologies that can alter the expression, subcellular compartmentalization, intracellular signaling and transforming potential of RAS (16). To this end, the activation of endogenous RAS oncogenes triggers minimal ERK and AKT activation in our system and others (Fig. S10 and (17, 18, 25)); yet, transgenic RAS alleles robustly stimulate these pathways (46, 47). Other murine melanoma models employ oncogenic RAS isoforms rarely observed in human melanomas (i.e. H-RAS and K-RAS mutants). As each RAS isoform can initiate a unique set of downstream signals, it is unclear to what degree these models faithfully recapitulate the oncogenic mechanisms found in human melanomas. For these reasons, we believe that the TpN61R and TpLN61R models will be extremely valuable for pre-clinical drug testing, especially given the metastatic nature of the Lkb1-deficient tumors.
NRASQ61R exhibits unique biochemical properties
In contrast to prior studies showing that preferential effector usage drives isoform-specific RAS oncogenesis (17, 32–35), we found that distinct biochemical properties inherent to each RAS mutant likely drives codon-specific melanomagenesis (Tables 2 and S2). Nearly 3 decades ago, Der and colleagues reported that HRAS codon 61 mutants, regardless of their transforming potential, exhibited a ~10-fold decrease in intrinsic GTP hydrolysis (48). Later, Donovan, Shannon and Bollag used predicted RAS-GTP levels to form the hypothesis that defects in intrinsic GTPase activity and nucleotide exchange dictate the transforming potential of individual RAS mutants (49). Our experimental results are consistent with this model, demonstrating that the melanomagenic NRASQ61R variant exhibits high affinity GTP binding, increased stability and reduced intrinsic GTPase activity when compared to NRASG12D (Tables 2 and S2).
These results suggest that NRASQ61R may more efficiently activate downstream effectors in vivo. However, the activation of ERK and AKT were similar in both primary melanocytes and human melanoma cultures harboring an NRAS codon 12 or 61 mutation (Figs. 4 and S10). Moreover, even through unbiased probes of the kinome using PhosphoScan technology (Cell Signaling), we were unable to detect consistent differences between NRasG12D and NRasQ61R signaling in primary melanocytes (data not shown). Due to feedback mechanisms within many pathways downstream of RAS, changes in signaling flux may not be readily apparent. Alternatively, enhanced cooperation with secondary oncogenic events during the initial stages of tumor development may be responsible for the initial selection of NRASQ61R mutants in melanoma. In line with this observation, RAF activation is a common event in both thyroid carcinomas and melanomas, the two major tumor types wherein NRAS codon 61 mutations predominate (1, 2). Mounting evidence supports the idea that the intensity of oncogenic RAS signaling can influence transformation potential (38). In addition, RAF binding in conjunction with unidentified allosteric regulators is speculated to further reduce the intrinsic GTPase activity of HRAS codon 61 mutants (36). These findings, along with our work, suggest that NRAS codon 61 mutants exhibit unique biochemical properties which promote the transformation of RAF-responsive tissues. Parsing the mechanism of this specific dependency will require the structural analyses of multiple NRAS mutants as well as comprehensive functional screens using endogenous RAS expression systems and relevant cell types. However, our data suggest that this work would be extremely valuable, identifying codon-specific tumor vulnerabilities for therapeutic targeting.
Methods
Murine Alleles and Husbandry
Animal work was conducted in accordance with protocols approved by the Institutional Care and Use Committee for animal research at the University of North Carolina and the Ohio State University. The Tyr-CRE-ERT2, p16L, Lkb1f, LSL-K-ras12D and LSL-N-Ras12D alleles have been previously described (17–20, 31). All animals in this study were backcrossed more than 7 generations to C57BL/6. The Lkb1f allele was initially established on an albino C57BL/6J background and then crossed into the TpN61R colony. Therefore, some of the mice in this cohort are albino. Analysis of tumor-free survival was conducted using GraphPad Prism software. To determine statistical significance, a Logrank (Mantel-Cox) test was performed for each experimental pairing.
Generation of the LSL-N-RasQ61R Allele
Standard homologous recombination procedures were used to insert a conditionally excisable transcriptional stop codon and point mutation into the endogenous N-Ras locus (See Fig. S1A). Founding mice were first crossed to C57Bl/6 FlpE mice (Jackson Labs #005703) to remove the neomycin resistance cassette. Southern blotting was performed on ES cells and founder DNA using standard procedures. PCR primers and cycling conditions used for genotyping LSL-N-RasQ61R were as follows: Q61R GENO2- 5'-GCAAGAGGCCCGGCAGTACCTA-3' (0.15 µM); Primer 1 – 5'-AGACGCGGAGACTTGGCGAGC-3' (0.15 µM); Primer 2 – 5'-GCTGGATCGTCAAGGCGCTTTTCC-3' (0.15 µM); Cycling - 95°C 15 min, 35 × [94°C 30s, 62°C 30s, 72°C 45s], 72°C 5 min. The resulting PCR products were 487 (wild type), 371 (LSL-N-rasQ16R), and 562 (LSL-N-rasQ16R+ CRE) base pairs in size.
Primary Melanocyte Culture
Skin was isolated from newborn pups and placed dermis side down in 0.25% trypsin, 0.1% EDTA for 3 hours at 37°C and 5% CO2. Using forceps, the epidermal layer was separated from the dermis. Using surgical scissors, epidermal cells were minced finely in phosphate buffered saline supplemented with 0.02% EDTA. To further dissociate the cell suspension, each sample was subjected to 2 rounds of program A on the GentleMACs dissociator (Miltenyi). The resulting samples were spun down and plated on rat tail collegen-coated dishes in melanocyte growth medium (Ham’s F10, 10 µg/mL insulin, 0.5 ng/mL bovine serum albumin, 5% fetal bovine serum, 1 µM ethanolamine, 1 µM phophoethanolamine, 10 nM sodium selenite, 20 nM TPA, 50 pM cholera toxin, 1× penicillin/streptomycin, 100 nM melanocyte stimulating hormone, and 0.05 mM di-butyril cyclicAMP). Media was changed every other day.
Induction of CRE Recombinase
For neonatal induction, pups were painted dorsally (postnatal days 2–4) with 25mg/mL 4-hydroxytamoxifen (4-OHT) dissolved in DMSO. Adult induction of CRE was performed as previously described (50). For cell culture studies in primary melanocytes, 4-OHT was dissolved in ethanol and added to the growth media. Cells were treated for 6 consecutive days replacing the media and 4-OHT every other day. In the immortalized TpN61R cultures, which were resistant to recombination, an adenoviral CRE recombinase was employed (Ad5-MCV-Cre-GFP, Baylor College of Medicine Vector Development Lab). Prior to proliferation or morphological assessment, melanocyte cultures were returned to untreated media for 3 days. Allelic recombination in tumors and primary cultures was confirmed by genomic PCR using the genotyping primer sets and conditions described for each allele.
Scoring of Murine Nevi
Nevi on a 4 cm2 dorsal area were counted by staff, blinded of the animal’s genotype, every week for 10 weeks. So as not to interfere with melanomagenesis, we chose not to depilate or shave these animals during the study. The presence of a single nevus at any time point was scored as a ‘1’. If no nevus was observed a ‘0’ was entered for that time point. The percentage of time nevus positive was calculated as: (sum of all 10 measurements/10)* 100. Each dot represents 1 animal with the mean indicated by a line. Comparisons between sample pairs were conducted using a student’s t-test.
Culture of Murine and Human Melanoma Cell Lines
The NZM24 and NZM63 cell lines were developed in the laboratory of Dr. Bruce C. Baguley (51) and obtained from the Cell Line Collection maintained by the Auckland Cancer Society Research Centre, Faculty of Medical and Health Sciences, The University of Auckland. The MaMel27II cell line was generated by Drs. G. Finlay, C. Posch and S. Ortiz (Mount Zion Cancer Research Center, San Francisco; (52)). WM3670, WM3629 and WM1366 cells were created in the Heryln lab and obtained from the Wistar Institute (53). VMM39 cells were the kind gift of C. Slingluff (U. Virginia)(54). The SK-Mel-119, SK-Mel-103 and SK-Mel-147 cell lines were generously provided by Dr. A. Houghton (Memorial Sloan-Kettering)(55). The Mel224 cell line was produced and supplied by J. Hansson (Karolinska Institutet). Tumor-derived murine cell lines form the TpLN61R/61R and TpK12D/WT models were generated, genotyped and maintained as previously described (23).
Authentication of Cell Lines
All cell lines were obtained from their original sources or authorized distributors and maintained as previously described (51–56). Cell lines from the Wistar institute (WM3670, WM3629 and WM1366) are routinely validated by short tandem repeat (STR) profiling using the AmpFISTR Identifiler PCR Amplification kit (Life Technologies) and were used for the experiments shown within 3 months of receipt. For primary cell lines where original STR data is not currently available, N-RAS mutations were validated by PCR followed by Sanger sequencing.
Flow Cytometry
Cell cycle analysis of primary melanocytes: After 10 days in culture, 4.4×105 primary melanocytes were seeded onto collagen coated 60mm dishes. A day later, the media was changed and cells were allowed to recover overnight. EdU was added to each culture at a concentration of 2.5ug/mL for a period of 16 hours. Cells were harvested and processed as described in the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen) using saponin for permeabilization. Cell cycle analysis of human melanoma cell lines: Human melanoma cell lines grown to 50–70% confluence were treated with 10 µM EdU for a period of 6 hours. Cells were harvested, processed, stained for EdU incorporation and analyzed on a Flowsight cytometer (Amnis). Trp1 Staining: To validate that our cultures contained a pure population of melanocytes and verify the identity of TpLN61R metastases, PEP1 antibody (αTrp1; (21)) was added at a 1:100 dilution to saponin permeabilized cells followed by incubation with an AlexaFluor488 conjugated anti-rabbit secondary (Molecular Probes, 1:1000 dilution). In the analysis of splenic tissues (Fig. 3E), co-staining with CD45 was first used to gate out any CD45+ cells.
In vitro protein purification
A vector encoding human N-RAS (1-172) was acquired from Addgene (vector #25256, NRAS-A). The G12D and Q61R mutations were each introduced into this vector using standard quick change mutagenesis. All proteins were expressed and purified from Rosetta 2 BL21(DE3) cells (Novagen, Madison, WI, USA). Briefly, cells were grown at 37°C in a shaking culture of Terrific Broth media supplemented with kanamycin. Once the cells reached an OD600 between 0.6–1.0, the culture was chilled in an ice bath (to ~20°C) and 0.1mM IPTG added. The cultures were then shaken for an additional 12–15 hours at 18°C. Cells were harvested by centrifugation and pellets stored at −80°C. N-RAS proteins were purified on Ni Sepharose6 columns following the manufacturer’s protocol (GE Life Sciences, Piscataway, NJ, USA). To further purify the proteins from contaminants, size-exclusion chromatography was performed (Superdex 75 10/300 GL; GE Life Sciences, Piscataway, NJ, USA). Purity exceeding 95% was confirmed by SDS-PAGE analysis.
The minimum catalytic domain of the human protein Son of Sevenless (SOScat) was expressed in the pQlinkH vector (Addgene) and purified as previously described (57). The E.coli codon-optimized RAS-binding domain of BRAF (amino acids 149–232) containing an N-terminal purification tag (MGHHHHHHSSGVDLGTENLYFQS) was synthesized by Genewiz (South Plainfield, NJ), sub-cloned into pET28a and expressed in BL21 (DE3) cells. The resulting BRAF-RBD was purified on Ni Sepharose6, cleaved overnight with TEV protease and then subjected to subtractive Ni column purification. The tag-less BRAF-RBD was further purified using size exclusion chromatography and verified to be > 95% pure by SDS-PAGE analysis. Purified PI3Kγ (amino acids 144-1102) was kindly provided by Genentech.
Loading of N-RAS with nucleotide derivatives
To observe nucleotide dissociation activity in the GTP-bound state, N-RAS was loaded with 2’−/−3’-O-(N’methylanthraniloyl) -guanosine-5'-[(β,γ)-imido]triphosphate (mantGMPPNP). N-RAS was exchanged out of excess MgCl2 into a buffer containing 20 mM HEPES, 50 mM NaCl, 125 mM (NH4)2SO4, 1 mM MgCl2 at pH 7.4. The concentration of N-RAS was adjusted to 100 µM and a 5-molar excess of mantGMPPNP was added. Alkaline phosphatase conjugated to sepharose beads was added to 1:10 volume and EDTA was added to 1 mM to increase the nucleotide exchange rate. The protein mixture was incubated at 4°C until all unlabeled nucleotide was converted to guanosine. The alkaline phosphatase beads were removed by centrifugation and 10 mM MgCl2 was added to induce nucleotide binding. The concentration of mantGMPPNP-bound RAS was determined by measuring the absorbance of the mant fluorophore (ε350 = 5700 M−1cm−1). For assays requiring unlabeled N-RAS in the GTP-bound state, N-RAS·GMPPCP was prepared in an identical manner.
For assays observing N-RAS nucleotide dissociation activity in the GDP-bound state, the protein was loaded with mantGDP (2’−/−3’-O-(N’methylanthraniloyl) –guanosine diphosphate) as previously described (58). N-RAS was exchanged into a buffer containing 20 mM HEPES, 125 mM (NH4)2SO4, 100 µM EDTA, 200 µM DTPA at pH 8.0 to remove excess MgCl2 and nucleotide. A 5-molar excess of mantGDP was added to the protein and the mixture was incubated for 30 min at 37°C to increase the nucleotide exchange rate. MgCl2 was then added to 20 mM and the protein was placed at 4°C for 2 hours to induce binding, after which N-RAS was exchanged into a buffer containing 20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 100 µM DTPA, pH 7.4 for storage.
Quantitative Real-time PCR
RNA from melanocytes or melanomas was isolated using the Qiagen All-Prep Kit. Equal amounts of RNA were subjected to reverse transcription using a PrimeScript First Strand cDNA Synthesis kit (Takara). Real-time PCR was run in triplicate and a standard curve of known plasmid concentration was included on every plate (ABI Taqman primer set #Mm00477878). Molecules of N-RAS/µg total RNA were calculated assuming a cDNA conversion efficiency of 100%.
Determination of N-RAS binding to BRAF-RBD and PI3Kγ
For quantitative binding to BRAF-RBD and PI3Kγ (amino acids 144-1102), N-RAS·mantGMPPNP was incubated with different concentrations of the desired effectors in a buffer containing 50 mM HEPES, 50 mM NaCl, 5 mM MgCl2 at pH 7.4. Nucleotide dissociation was initiated by the addition of 1000-molar excess of unlabeled nucleotide and the rate of dissociation was determined by monitoring the change in fluorescence of the mantGMPPNP loaded protein (excitation and emission wavelengths of 365 and 435 nm, respectively) using a Spectramax M2 plate reader (59). Each nucleotide dissociation curve was fit to a one-phase single exponential to determine kobs. The dissociation rates were plotted against the effector concentrations and fit to the equation:
Y: Observed rate, A: Maximum rate, B: Minimum rate, P1: Concentration of protein 1 (cell), P2: Concentration of protein 2 (ligand/effector), which was used to determine the N-RAS·mantGMPPNP : effector binding constant (KD) (60).
Nucleotide exchange in the absence and presence of SOScat
Nucleotide dissociation rates were measured using a PerkinElmer LS50B fluorimeter as previously reported (58). N-RAS was loaded with mant-labeled nucleotide and added to a final concentration of 1 µM in 1 mL of exchange buffer (20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 100 µM DTPA, pH 7.4). A 1000-fold excess of unlabeled nucleotide was added to initiate the dissociation reaction and the rate of nucleotide dissociation was measured by monitoring the change in fluorescence (λex = 365 nm, λem = 435 nm). For reactions that showed dissociation rates too slow to reach completion within the time frame of the assay, 25 mM EDTA was added to induce complete nucleotide dissociation. For GEF-facilitated nucleotide exchange, the minimum catalytic domain of son of sevenless (SOScat) was added to a 1:2 RAS-to-SOScat ratio (chosen to limit RAS binding to the allosteric site in SOScat). The fluorescence data was normalized and fit to one-phase exponentials to determine rates.
Determination of intrinsic GTP hydrolysis activity of N-RAS
To measure GTP hydrolysis rates, the phosphate sensor FLIPPi (Addgene, Cambridge, MA, USA) was used for the reaction, which binds to free phosphate in solution (61). Due to the nature of the assay, all trace phosphate was removed from buffers prior to use to minimize background fluorescence. N-RAS was exchanged into a buffer containing 20 mM HEPES, 20 mM (NH4)2SO4, 1 mM EDTA, 1 mM inosine, pH 8.0. N-RAS was incubated with GTP and passed through a PD-10 column to remove trace Mg2+ (62). N-RAS was diluted to 10 µM and combined with 10 µM FLIPPi, and 2 mM MgCl2 was added to initiate the hydrolysis reaction. A phosphate standard curve was used to convert the raw fluorescence output to a measurement of [GTP] hydrolyzed.
Thermal Melting Analysis by Circular Dichroism (CD) Spectroscopy
A Jasco J-815 CD Spectrometer was used for CD measurements. N-RAS was exchanged into a buffer containing 10 mM KH2PO4/K2HPO4 at pH 7.45 and diluted to 15 µM. MgCl2 and guanine nucleotide (GDP or GMPPCP) was added to a final concentration of 500 µM and 80 µM respectively, immediately before analysis. A thermal melt scan from 20° to 90°C was performed to determine the temperature (Tm) at which half of the protein is unfolded.
RAS Mutational Analysis in Human Cancers
Data were downloaded from COSMIC (release v64, March 26, 2013)(1). Cell lines and cultured cells were removed from the analysis to prevent inclusion of events secondary to culture. Remaining events were limited to observations of somatic missense mutations in codons 12, 13 or 61 of HRAS, NRAS or KRAS. Multiple subtypes and histologies were then summed to give information for the displayed tumor types.
Supplementary Material
Significance.
This work explains the curious predominance in human melanoma of mutations of codon 61 of N-RAS over other oncogenic N-RAS mutations. Using conditional ‘knock-in’ mouse models, we show that physiological expression of N-RASQ61R, but not N-RASG12D, drives melanoma formation.
Acknowledgements
We thank Wayne Joseph, Drs. Graeme Finlay, Christian Posch, Susana Ortiz, Vincent Hearing, Meenhard Herlyn, Altaf Wani, Kevin Haigis and Tyler Jacks for reagents, equipment and/or animals, Drs. Ruben Carrasco, Yuri Fedoriw and Arlin Rogers for advice, Dr. Nabeel Bardeesy for critical reading of the manuscript, and the UNC Structural Bioinformatics Core Facility (Brenda Temple), Animal Histopathology Core, Biostatistics Core (Dominic Moore), Protein Expression and Purification Core (Mischa Machius), Macromolecular Interactions Facility (Ashutosh Tripathy), and Mouse Phase I Unit (Luke Hunter) for technical assistance. Purified PI3Kγ was generously provided by Genentech.
Financial Support: This work was supported by NIH R00AG036817 (C.E.B.), NIH R01CA163896 (N.E.S.), NIH U01CA141576 (N.E.S.), American Cancer Society IRG-67-003-50 (C.E.B.), NIH T32ES007126 (C.E.B), NIH T32CA009156 (G.A.S.) and The UNC University Cancer Research Fund (UCRF).
Abbreviations List
- 4-OHT
4-hydroxytamoxifen
- AKT
v-akt murine thymoma viral oncogene
- BRAF
v-raf murine sarcoma viral oncogene homolog B
- CDK4/6
cyclin dependent kinase 4/6
- DMSO
dimethyl sulfoxide
- DTPA
diethylene triamine pentaacetic acid
- EDTA
Ethylenediaminetetraacetic acid
- EdU
5-ethynyl-2’-deoxyuridine
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinases
- FLIPPi
fluorescence resonance energy transfer (FRET) sensors for phosphate (P(i))
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GMPPNP
guanosine-5'-[(β,γ)-imido]triphosphate
- GMPPCP
guanosine-5'-[(β,γ)-methyleno]triphosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- H-RAS
Harvey rat sarcoma viral oncogene homolog
- K-RAS
Kirsten-rat sarcoma viral oncogene homolog
- LKB1/STK11
liver kinase B1/serine threonine kinase 11
- MAPK
mitogen-activated protein kinase
- N-RAS
neuroblastoma RAS viral oncogene homolog
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- RAF
rapidly accelerated fibrosarcoma
- SOS
Son of Sevenless
- TEV
tobacco etch virus protease cleavage site
- TPA
12-O-Tetradecanoylphorbol-13-Acetate
- TRP-1
Tyrosinase-Related Protein 1
Footnotes
Conflict of Interest Disclosure: The authors listed above have no financial or non-financial interests to disclose in relation to this work.
References
- 1.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- 4.Frech M, Darden TA, Pedersen LG, Foley CK, Charifson PS, Anderson MW, et al. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 1994;33:3237–3244. doi: 10.1021/bi00177a014. [DOI] [PubMed] [Google Scholar]
- 5.Garm Spindler KL, Pallisgaard N, Rasmussen AA, Lindebjerg J, Andersen RF, Cruger D, et al. The importance of KRAS mutations and EGF61A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2009;20:879–884. doi: 10.1093/annonc/mdn712. [DOI] [PubMed] [Google Scholar]
- 6.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer research. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 9.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Ye Y, Sun H, Shi G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer chemotherapy and pharmacology. 2013;71:265–272. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 11.Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p.G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: A systematic review and meta-analysis. Cancer. 2012 doi: 10.1002/cncr.27804. [DOI] [PubMed] [Google Scholar]
- 12.Modest DP, Jung A, Moosmann N, Laubender RP, Giessen C, Schulz C, et al. The influence of KRAS and BRAF mutations on the efficacy of cetuximab-based first-line therapy of metastatic colorectal cancer: an analysis of the AIO KRK-0104-trial. International journal of cancer Journal international du cancer. 2012;131:980–986. doi: 10.1002/ijc.26467. [DOI] [PubMed] [Google Scholar]
- 13.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA : the journal of the American Medical Association. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 14.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. Journal of the National Cancer Institute. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omerovic J, Prior IA. Compartmentalized signalling: Ras proteins and signalling nanoclusters. The FEBS journal. 2009;276:1817–1825. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- 17.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature genetics. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 19.Monahan KB, Rozenberg GI, Krishnamurthy J, Johnson SM, Liu W, Bradford MK, et al. Somatic p16(INK4a) loss accelerates melanomagenesis. Oncogene. 2010;29:5809–5817. doi: 10.1038/onc.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–267. doi: 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- 21.Hearing VJ. Antibodies specific to melanocyte-specific proteins available from the Hearing Laboratory. Pigment Cell & Melanoma Research. 2009;22:1. [Google Scholar]
- 22.Jeck WR, Parker J, Carson CC, Shields JM, Sambade MJ, Peters EC, et al. Targeted next generation sequencing identifies clinically actionable mutations in patients with melanoma. Pigment Cell Melanoma Res. 2014 doi: 10.1111/pcmr.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer cell. 2012;21:751–764. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes & development. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117:2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damsky WE, Jr., Bosenberg M. Mouse melanoma models and cell lines. Pigment Cell Melanoma Res. 2010;23:853–859. doi: 10.1111/j.1755-148X.2010.00777.x. [DOI] [PubMed] [Google Scholar]
- 27.Rowan A, Bataille V, MacKie R, Healy E, Bicknell D, Bodmer W, et al. Somatic mutations in the Peutz-Jeghers (LKB1/STKII) gene in sporadic malignant melanomas. The Journal of investigative dermatology. 1999;112:509–511. doi: 10.1046/j.1523-1747.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 28.Guldberg P, thor Straten P, Ahrenkiel V, Seremet T, Kirkin AF, Zeuthen J. Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene. 1999;18:1777–1780. doi: 10.1038/sj.onc.1202486. [DOI] [PubMed] [Google Scholar]
- 29.Esteve-Puig R, Canals F, Colome N, Merlino G, Recio JA. Uncoupling of the LKB1-AMPKalpha energy sensor pathway by growth factors and oncogenic BRAF. PloS one. 2009;4:e4771. doi: 10.1371/journal.pone.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Molecular cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, et al. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. The Journal of biological chemistry. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 33.Voice JK, Klemke RL, Le A, Jackson JH. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. The Journal of biological chemistry. 1999;274:17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 34.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. The Journal of biological chemistry. 2001;276:15609–15615. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 35.Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Molecular and cellular biology. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buhrman G, Wink G, Mattos C. Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure. 2007;15:1618–1629. doi: 10.1016/j.str.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen M, Kusters-Vandevelde HV, Viros A, Groenen PJ, Sanchez-Laorden B, Gilhuis JH, et al. Primary melanoma of the CNS in children is driven by congenital expression of oncogenic NRAS in melanocytes. Cancer discovery. 2013;3:458–469. doi: 10.1158/2159-8290.CD-12-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Haigis KM, Firestone AJ, McNerney ME, Li Q, Davis E, et al. Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation. Cancer discovery. 2013;3:993–1001. doi: 10.1158/2159-8290.CD-13-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonsson A, Tuominen R, Grafstrom E, Hansson J, Egyhazi S. High frequency of p16(INK4A) promoter methylation in NRAS-mutated cutaneous melanoma. The Journal of investigative dermatology. 2010;130:2809–2817. doi: 10.1038/jid.2010.216. [DOI] [PubMed] [Google Scholar]
- 40.Eskandarpour M, Hashemi J, Kanter L, Ringborg U, Platz A, Hansson J. Frequency of UV-inducible NRAS mutations in melanomas of patients with germline CDKN2A mutations. Journal of the National Cancer Institute. 2003;95:790–798. doi: 10.1093/jnci/95.11.790. [DOI] [PubMed] [Google Scholar]
- 41.Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nature medicine. 2012;18:1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim WY, Sharpless NE. Drug efficacy testing in mice. Current topics in microbiology and immunology. 2012;355:19–38. doi: 10.1007/82_2011_160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Combest AJ, Roberts PJ, Dillon PM, Sandison K, Hanna SK, Ross C, et al. Genetically engineered cancer models, but not xenografts, faithfully predict anticancer drug exposure in melanoma tumors. The oncologist. 2012;17:1303–1316. doi: 10.1634/theoncologist.2012-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, et al. Predicting drug responsiveness in human cancers using genetically engineered mice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:4889–4899. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer research. 2005;65:4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 47.VanBrocklin MW, Robinson JP, Lastwika KJ, Khoury JD, Holmen SL. Targeted delivery of NRASQ61R and Cre-recombinase to post-natal melanocytes induces melanoma in Ink4a/Arflox/lox mice. Pigment Cell Melanoma Res. 2010;23:531–541. doi: 10.1111/j.1755-148X.2010.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 49.Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochimica et biophysica acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- 50.Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan PF, Shimamura T, et al. HIF1alpha and HIF2alpha independently activate SRC to promote melanoma metastases. The Journal of clinical investigation. 2013;123:2078–2093. doi: 10.1172/JCI66715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stones CJ, Kim JE, Joseph WR, Leung E, Marshall ES, Finlay GJ, et al. Comparison of responses of human melanoma cell lines to MEK and BRAF inhibitors. Frontiers in genetics. 2013;4:66. doi: 10.3389/fgene.2013.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4015–4020. doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28:85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molhoek KR, Brautigan DL, Slingluff CL., Jr. Synergistic inhibition of human melanoma proliferation by combination treatment with B-Raf inhibitor BAY43-9006 and mTOR inhibitor Rapamycin. Journal of translational medicine. 2005;3:39. doi: 10.1186/1479-5876-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houghton AN, Albino AP, Cordon-Cardo C, Davis LJ, Eisinger M. Cell surface antigens of human melanocytes and melanoma. Expression of adenosine deaminase binding protein is extinguished with melanocyte transformation. The Journal of experimental medicine. 1988;167:197–212. doi: 10.1084/jem.167.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer research. 2007;67:1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 57.Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Lenzen C, Cool RH, Wittinghofer A. Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods in enzymology. 1995;255:95–109. doi: 10.1016/s0076-6879(95)55012-7. [DOI] [PubMed] [Google Scholar]
- 59.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. The Journal of biological chemistry. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 60.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. The Journal of biological chemistry. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 61.Gu H, Lalonde S, Okumoto S, Looger LL, Scharff-Poulsen AM, Grossman AR, et al. A novel analytical method for in vivo phosphate tracking. FEBS letters. 2006;580:5885–5893. doi: 10.1016/j.febslet.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shutes A, Der CJ. Real-time in vitro measurement of intrinsic and Ras GAP-mediated GTP hydrolysis. Methods in enzymology. 2006;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.