Abstract
Purpose
Asparaginase is a standard and critical component in the therapy of childhood acute lymphoblastic leukemia (ALL), but it is also associated with several toxicities.
Experimental design
We recently reported the results of an association study between asparaginase pathway genes and event free survival (EFS) in childhood ALL patients. The same polymorphisms were interrogated here in relation to allergies, pancreatitis and thrombotic events following treatment with E.coli asparaginase.
Results
Among patients of discovery group, allergies and pancreatitis were more frequent in individuals who are homozygous for the triple repeat allele (3R) of asparagine synthetase ASNS gene, resulting in remarkably higher risk of these toxicities associated with 3R3R genotype (OR for allergies =14.6, 95% CI= 3.6–58.7, p<0.0005 and OR for pancreatitis = 8.6, 95% CI= 2.0–37.3, p=0.01). In contrast, the ASNS haplotype *1 harbouring double repeat (2R) allele had protective effect against these adverse reactions (p≤0.01). The same haplotype was previously reported to confer reduction in EFS. The risk effect of 3R3R genotype was not replicated in validation cohort, whereas the protective effect of haplotype *1 against allergies was maintained (p≤0.002). Analysis with additional polymorphisms in ASNS locus in lymphoblastoid cell lines showed that haplotype *1 is diversified in several subtypes of which one was associated with reduced in vitro sensitivity to asparaginase (rs10486009p=0.01) possibly explaining an association seen in clinical setting.
Conclusions
This finding might have implication for treatment individualization in ALL and other cancers employing asparagine depletion strategies.
Keywords: asparaginase, polymorphisms, pharmacogenetics, childhood leukemia, treatment, adverse reactions
Introduction
Over the past four decades, treatment of childhood acute lymphoblastic leukemia (ALL) has improved importantly such that ~ 80–85% of patients are cured with current therapy regimen. Up to 20% of patients experience treatment failure whereas treatment-related toxicities are often life-threatening and are the primary cause of interruption or discontinuation of chemotherapy. Asparaginase (ASNase) is a standard component in the childhood ALL treatment (1). It is required by all cells for survival and is normally produced by the enzyme asparagine synthetase (ASNS). Malignant lymphoblasts are thought to have low ASNS levels and thus depend on extracellular sources of asparagine for their rapid growth. Depletion of asparagine by ASNase selectively kills leukemia cells by decreasing protein biosynthesis (2). Associations between success of ALL treatment and ASNase dose intensity or formulation have been reported in several clinical studies (3–5). E.coli derived enzymes is more potent, it is associated with higher efficacy, but also with higher toxicity (5–7). Side effects related to ASNase treatment include allergic reactions that occur in 20–40% patients and require change of drug formulation; Two most serious and most frequent dose-limiting asparaginase-related toxicities are pancreatitis and thrombotic events reported in up to 18% and 5% of ALL patients, respectively (8, 9). Pancreatitis usually develops after the first few doses of asparaginase suggesting that it may occur as a result of an underlying predisposition rather than as a cumulative drug effect (9).
We recently analyzed relationship between event free survival (EFS) in childhood ALL patients and genes in ASNase pathway (10), which were selected based on differential expression between asparaginase resistant and sensitive cells (11–13). We showed that promoter variant of transcriptional factor ATF5 involved in ASNS regulation, is associated with higher promoter activity and confers higher risk of ALL relapse in patients who received E.coli ASNase (10). Association with lower EFS has been also found with tandem repeat (14 in ASNS gene and with resulting haplotype (arbitrarily named haplotype *1) (10). Here we report the analysis of the same set of polymorphisms in ASNS ATF5 and ASS1 arginosuccinate synthase 1) in relation to ASNase-related acute complications (allergies, pancreatitis and thrombotic events) in two independent childhood ALL cohorts.
Patients and methods
Study population and endpoints in the analysis
The study population consisted of 285 Caucasian children (98% of French-Canadian origin) diagnosed with ALL at the Hospital Sainte-Justine, (HSJ, Montreal, Quebec, Qc, Canada) between January 1989 and July 2005 (QcALL cohort or test group) who received E.coli asparaginase as a part of Dana-Farber Cancer Institute ALL Consortium protocols DFCI 87-01, 91-01, 95-01 or 00-01 (Table 1) (5, 6, 10, 15). Details of asparaginase administration across these treatment protocols are described elsewhere (10, 16). The information on asparaginase-related toxicity was assessed by retrospective chart review. Pancreatitis was defined as an elevation in the serum amylase level >3 times normal associated with clinical signs and symptoms consistent with the diagnosis (9). Pancreatitis cases were classified by duration of symptoms as severe or mild/moderate (16). Hypersensitivity reactions to asparaginase were characterized by local manifestations at the injection site as well as systemic manifestations (erythema, swelling, urticaria, rash, pruritus, tachypnea, and wheezing) (17). Thrombosis was identified by clinical symptoms and confirmed by radiological imaging based on institutional guidelines (18).
Table 1.
Baseline characteristics of ALL patients in the test (QcALL) and validation (DFCI) cohort
| No of subjects and frequency (%) | ||||
|---|---|---|---|---|
| Characteristic | QcALL (n=285) | DFCI (n=248) | ||
| Sex | ||||
| Female | 130 | (45.6) | 114 | (46.0) |
| Male | 155 | (54.4) | 134 | (54.0) |
| Age, y | ||||
| <10 | 224 | (78.6) | 203 | (81.9) |
| ≥10 | 61 | (21.4) | 45 | (18.1) |
| WBC, ×109/L | ||||
| <50 | 242 | (84.9) | 202 | (81.5) |
| >50 | 43 | (15.1) | 46 | (18.5) |
| Cell type | ||||
| B | 267 | (93.6) | 227 | (91.5) |
| T | 18 | (6.4) | 21 | (8.5) |
| Risk groups | ||||
| Standard | 137 | (48.1) | 153 | (61.7) |
| High | 148 | (51.9) | 95 | (38.3) |
| Treatment protocol | ||||
| 87-01 | 20 | (7.0) | ||
| 91-01 | 57 | (20.0) | ||
| 95-01 | 92 | (32.3) | 73 | (29.4) |
| 00-01 | 116 | (40.7) | 175 | (70.6) |
Protocol distribution is different between QcALL and DFCI group (p=0.001), whereas the remaining characteristics do not differ significantly. WBC, white blood cell count.
Previously obtained genotypes in asparaginase pathway genes were used for the analysis, as described in Rousseau et al (10), including 8, 2 and 4 SNPs in ATF5, ASNS and ASS1 genes, respectively (Supplemental Table 1). The estimates of linkage disequilibrium (LD), and haplotype phase was obtained by PHASE software, version 2.0 (19). Association of genotypes/haplotypes with presence of each ASNase related toxicity was assessed by chi-square test. Adjustment for multiple testing (including all polymorphisms and all toxicities analyzed) was estimated by false discovery rate (FDR) (10). Analyses of haplotypes within significantly associated gene were not further corrected. For significant associations, genotypes/haplotypes were grouped in two categories and the genotype-associated risk was expressed as odds ratio (OR) with 95% confidence interval (CI). A validation set of Caucasian patients called the Dana-Farber Cancer Institute (DFCI) group (Table 1) was composed of a 248 patients who received E.coli ASNase within DFCI 95-01 and 00-01 ALL treatment protocol in remaining (without HSJ) consortium institutions (5, 6, 16).
Cellular proliferation assay
In vitro sensitivity to asparaginase was assessed in lymphoblastoid cell lines (LCLs) from 89 individuals of Northern and Western Europe (CEU), as described by Chen et al. (17) The drug concentration resulting in 50% inhibition of cell growth (IC50) during 48h incubations time was estimated using several E.coli asparaginase concentrations ranging from 0.01–10 IU and the GraphPad software by fitting sigmoid dose-response curves. Obtained values were correlated to genotypes using Mann-Whitney or Kruskal-Wallis test.
Informed consents were obtained from parents or guardians before enrolment into the study. The study was approved by institution ethics committees.
Results
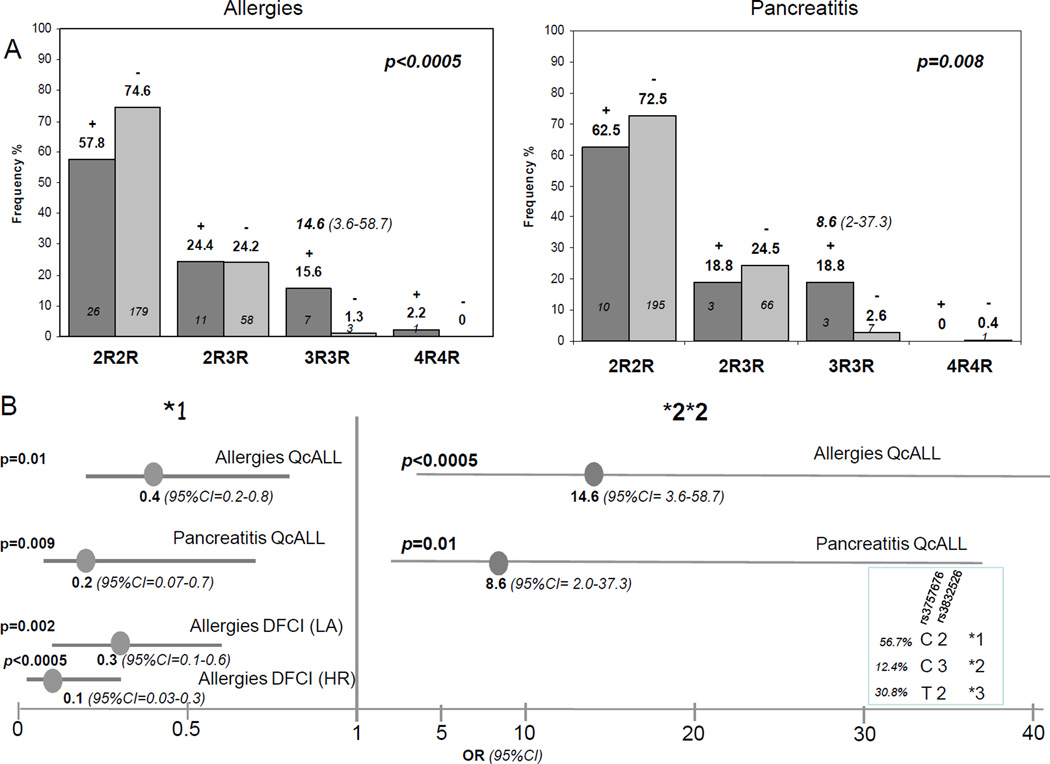
Allergies, pancreatitis and thrombotic events occurred in discovery group (QcALL) with the frequency of 15.8%, 5.6% and 3.5%, respectively. Pancreatitis was in most cases severe (in 13 out of 16 cases) and systemic allergies also occurred more frequently (in 37 out of 45 subjects with allergic reactions). Analysis between these toxicities and SNPs in ASNS, ATF5 and ASS1 genes revealed an association of tandem repeat polymorphism rs3832526 in ASNS gene with both pancreatitis and allergies (p=0.008 and p<0.0005, respectively, Figure 1A). These complications were more frequent among patients that were homozygous for the triple repeat allele (3R) resulting in 8–14 fold risk elevation (OR for allergies =14.6, 95% CI= 3.6–58.7, and OR for pancreatitis = 8.6, 95% CI= 2.0–37.3). The association with allergy remained significant with FDR of lower than 1%, whereas association with pancreatitis remained significant only with FDR of 16%. We further analyzed ASNS haplotypes composed of the tandem repeat polymorphism and promoter C-181T substitution rs3757676). Two haplotypes were associated with allergies and pancreatitis; Homozygosity for haplotype *2, uniquely tagged by 3R allele conferred higher risk of these toxicities (high sensitivity haplotype), whereas haplotype *1 defined by C-181 and 2R alleles had protective effect (low-sensitivity haplotype, OR for allergies =0.4, 95% CI= 0.2–0.8, and OR for pancreatitis = 0.2, 95% CI= 0.07–0.7, p≤0.01, Figure 1B). In our previous analysis the low-sensitivity haplotype *1 conferred reduction in EFS in QcALL cohort (10). We further performed the analyses of ASNS gene in replication (DFCI) cohort. Pancreatitis and allergies occurred in this group with the frequency of 8.5% and 23%, similar to the frequencies reported for the 00–01 clinical trial (16). Distribution of severe/moderate pancreatitis and systemic/local allergies differed from discovery group, 33.3% pancreatic cases had moderate form and among patients with allergies, 50.1% had local manifestation. The risk effect of 3R3R genotype (or haplotype *2 was not seen, whereas protective effect of haplotype *1 against allergies was maintained, particularly against local allergic manifestation and in patients assigned to high risk group (p=0.002 and p<0.0005, respectively, Figure 1B).
Figure 1. Asparaginase-related acute complications in childhood ALL in relation to tandem repeat polymorphism and resulting haplotypes in asparaginase synthase (ASNS) gene.
A. Tandem repeat polymorphism in relation to allergies and pancreatitis in ALL patients of discovery cohort. The frequency of individuals with genotypes of tandem repeat polymorphism in patients with (+, dark gray bars) and without (−, light gray bars) allergies and pancreatitis (left and right side panel, respectively). Numbers of individuals represented by each bar, p value for the difference across genotype groups and risk associated with 3R3R genotype (odds ratio, OR, with 95% confidence interval, CI) are indicated on each plot.
B. Risk of allergies and pancreatitis in relation to ASNS haplotypes. Linear display of risk (OR with 95% CI) associated with haplotype *1 and *2 in discovery (QcALL) and replication group (DFCI) based on C-181T substitution (rs3757676) and tandem repeat polymorphism (rs3832526). Protective effect of haplotype *1 in DFCI group was more apparent against local allergies (LA) and in high risk (HR) group. Sequence and frequency of 3 major haplotypes are indicated in the box on the right-bottom side.
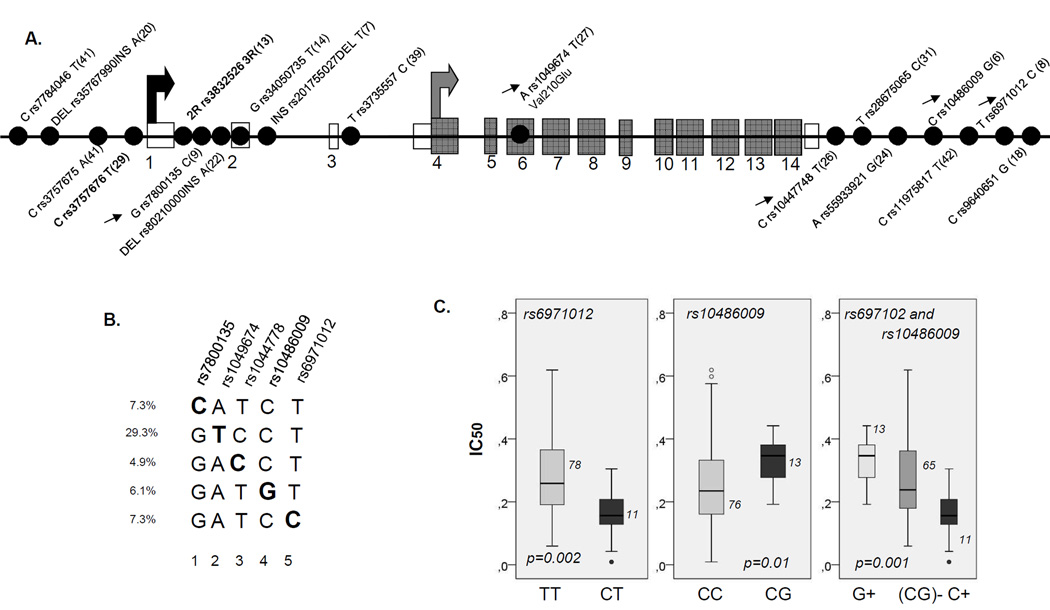
To further understand the variability of ASNS gene, we retrieved an information on all SNPs in coding and regulatory regions with minor allele frequency higher than 5%, that might have been identified (20) since our first analysis of this gene (10). We also included SNPs within 5kb upstream and downstream the from gene boundaries. Sixteen additional tag SNPs were analyzed, which were all except one (Val210Glu) located beyond the coding region (Figure 2A). Given high number of resulting haplotypes and low frequency of adverse events, detailed stratified analysis would have limited power in patients. We analyzed instead whether polymorphisms defining particular haplotype subtypes might have functional role as estimated by in vitro sensitivity assay in lymphoblastoid CEU cell lines. The protective *1 haplotype was diversified in 5 subtypes (defined by SNPs at 5 different positions, Figure 2B). Two polymorphisms (rs10486009 and rs6971012, positions 4 and 5 in Figure 2B) correlated with in vitro sensitivity to ASNase, (p=0.01 and p=0.002, respectively, Figure 2C). The TT genotype of rs6971012 had lower sensitivity, which was further reduced by the G allele of rs10486009 (Figure 2C, p=0.001) suggesting that it may contribute to protective effect of haplotype *1 observed in clinical setting. In contrast, only one haplotype defined by 3R allele remained after addition of other ASNS polymorphisms and it did not affect in vitro sensitivity to ASNase.
Figure 2. ASNS polymorphisms, haplotype *1 sub-classification and cellular proliferation assay.
A. Linear display of SNPs in ASNS locus. Linear display refers to all SNPs with minor allele frequency (MAF) > 5% located in coding regions and within upstream and downstream gene boundaries of ~5kb. Exonic and coding sequences are represented by open and gray boxes, respectively, and SNPs are represented by gray dots. Rs number and MAF are indicated next to each polymorphism. The black and gray arrows indicate the transcription and translation start site, respectively, estimated according to reference sequence NM_183356.3. Polymorphisms analyzed in patients are indicated in bold character. Arrows next to the polymorphisms indicate those depicted in Figure 2B.
B. Haplotype *1 subtypes. Diversification of ASNS haplotype *1 on several subtypes, based on the identified polymorphic positions (positions 1 to 5 indicated by respective rs numbers). Haplotype resolution is performed using all SNPs presented in Figure 2A; only those that are relevant for haplotype *1 diversification are presented.
C. In vitro sensitivity to asparaginase in relation to rs6971012 and rs10486009. Box plots representing ASNase IC50 in CEU lymphoblastoid cell lines with and without C allele of rs6971012with and without G allele of rs10486009 and with combined rs6971012 and rs10486009 genotypes (G+, C+ or none of these alleles, (CG)-) are presented in left, middle and right panel, respectively. The number of individuals represented by each bar and the p value obtained by Mann-Whitney or Kruskal-Wallis test is indicated on the plot.
Discussion
Differences in susceptibility to asparaginase have been attributed to variable levels of ASNS expression in number of studies: ASNS levels in leukemia cell lines, patients lymphoblasts and surrounding mesenchyme cells suggested that elevated ASNS levels may counteract the ASNase effect and underlie the resistance to treatment (11, 21, 22). Lower ASNS expression might then be expected to mediate higher sensitivity to treatment and possibly higher frequency of ASNase-related complications. We found that the 3R3R genotype of tandem repeat polymorphism correlated more frequently with pancreatitis and allergies in discovery group. Tandem repeat polymorphism is located in intron 1, but upstream from translation initiation site and was reported to act as an enhancer element (14). The 3R allele increased ASNS promoter activity in embryonic kidney cell line (14). We did not observe relationship between 3R3R genotype with mRNA levels (10), or in this study, with in vitro sensitivity to ASNase. The effect of 3R allele on therapeutic responses to ASNase is also ambiguous. Recent study reported that 3R allele can affect early response to ALL treatment, as defined by the number of leukemic blasts following one ASNase dose (23). We did not find an association of 3R3R genotype with reduced EFS (10) and in this study we did not replicate an association of 3R3R genotype with adverse reactions of ASNase. Other polymorphisms/haplotypes in ASNS gene cannot explain this discrepancy; there was only one haplotype defined by 3R allele when additional polymorphisms were included in the analysis. The differences can nevertheless be due to low frequency of 3R3R genotype, different distribution of severe/moderate and systemic/local allergies, or different geographical location of discovery and replication group. Other genes beyond those studied here, as well as disease and treatment characteristics might also play a role. For example, it has been shown that leukemic cells carrying TEL/AML1 fusion gene are more sensitive to treatment with ASNase compared to other subtypes of ALL (24); several polymorphisms of aspartate metabolic pathway have been associated with ASNase sensitivity in vitro using ALL cells and LCL cell lines (25); top ranking SNPs for ASNase -related allergies have been identified in the gene coding for glutamate receptor in genome-wide association study (17). Distribution of treatment protocols differed between discovery and replication group (p=0.001, Table 1) and patients might have received different ASNase doses. An association with pancreatitis and allergies with 3R3R genotype was nevertheless maintained in the discovery group when analysis was limited to patients treated with DFCI 95-01 and 00-01 protocols (p ≤ 0.02, not shown). Other baseline characteristics between two groups did not differ significantly (Table 1). Further and larger studies are needed to confirm the role of 3R3R genotype and to explore whether this finding is applicable to other protocols and other populations.
The results obtained for haplotype *1, harbouring 2R allele, seems more consistent and correlated with lower frequency of allergies in both discovery and replication patient cohort. This is in agreement with previously reported association of the same haplotype with reduced EFS (10), suggesting its lower sensitivity in response to ASNase treatment. Analysis of additional polymorphisms in ASNS locus revealed diversification or haplotype *1 in several subtypes; one of them (defined by minor allele of rs10486009) seems particularly interesting because it was associated with reduced sensitivity to ASNase in vitro, possibly explaining lower sensitivity of haplotype *1 seen in clinical setting. Our finding might be as well of interest for the treatment of other cancers since asparagine depletion strategies using ASNS inhibitors and asparaginase have been suggested in pancreatic and ovarian malignancies (26, 27). Tumor specific up-regulation of ASNS was also reported in castration-resistant prostate cancer and correlated with the progression to a therapy-resistant disease state (28).
In conclusion, we reported an association of 3R3R genotype of ASNS gene with a higher frequency of ASNase–related adverse reactions. The association was not seen in the replication group suggesting limited study power or possible modulating role of other genes and/or disease and treatment features or patient origin. Haplotype harboring 2R allele seems to have protective role against ASNase allergies in both discovery and replication patient set. Extension of the analysis to additional polymorphisms and cellular proliferation assay in response to ASNase treatment, identified variants possibly explaining lower sensitivity of this haplotype observed in clinical setting.
Supplementary Material
Statement of translational relevance.
Acute lymphoblastic leukemia (ALL) is the most frequent malignancy of childhood. The treatment of pediatric ALL has greatly improved in the past four decades due to the introduction of effective combination risk-adapted therapies. However, therapy resistance in a significant number of children is still a major obstacle to successful treatment. Intensive treatment has also significant short-term side effects and long-term consequences. Identification of genetic component underlying this variability, would allow traditional treatment to be complemented by genotype-based drug dose adjustment. Asparaginase is a critical component of ALL treatment. We recently analyzed genes of asparaginase action pathway in relation to the risk of relapse in ALL. Here we investigated whether the same genes may affect asparaginase-related toxicities and found that particular polymorphisms and haplotypes in asparagine synthetase gene may affect these adverse reactions in ALL patients. The study provides a new insight into the pharmacogenetics of asparaginase-related treatment complications in ALL.
Acknowledgement
We thank all patients and their parents who consented to participate in genetics studies related to leukemia. Canadian Institutes of Health Research, Leukemia Lymphoma Society of Canada, Charles Bruneau Foundation, and Centre d'excellence en Oncologie pédiatrique et en soins palliatifs supported this study. Dana-Farber Cancer Institute ALL treatment protocols are supported by the National Cancer Institute/NIH grant P01 CA 68484.
Grant Support
Dana-Farber Cancer Institute ALL treatment protocols are supported by the National Cancer Institute/NIH grant 5 P01CA068484.
References
- 1.Silverman LB, Declerck L, Gelber RD, Dalton VK, Asselin BL, Barr RD, et al. Results of Dana-Farber Cancer Institute Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1981–1995) Leukemia. 2000;14:2247–2256. doi: 10.1038/sj.leu.2401980. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz B, Madras BK, Meister A, Old LJ, Boyes EA, Stockert E. Asparagine synthetase activity of mouse leukemias. Science. 1968;160:533–535. doi: 10.1126/science.160.3827.533. [DOI] [PubMed] [Google Scholar]
- 3.Pession A, Valsecchi MG, Masera G, Kamps WA, Magyarosy E, Rizzari C, et al. Long-term results of a randomized trial on extended use of high dose L-asparaginase for standard risk childhood acute lymphoblastic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:7161–7167. doi: 10.1200/JCO.2005.11.411. [DOI] [PubMed] [Google Scholar]
- 4.Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Dana-Farber Consortium Protocol 91-01. Blood. 2001;97:1211–1218. doi: 10.1182/blood.v97.5.1211. [DOI] [PubMed] [Google Scholar]
- 5.Moghrabi A, Levy DE, Asselin B, Barr R, Clavell L, Hurwitz C, et al. Results of the Dana-Farber Cancer Institute ALL Consortium Protocol 95-01 for children with acute lymphoblastic leukemia. Blood. 2007;109:896–904. doi: 10.1182/blood-2006-06-027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman LB, Stevenson KE, O~Brien JE, Asselin BL, Barr RD, Clavell L, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000) Leukemia. 2010;24:320–334. doi: 10.1038/leu.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duval M, Suciu S, Ferster A, Rialland X, Nelken B, Lutz P, et al. Comparison of Escherichia coli-asparaginase with Erwinia-asparaginase in the treatment of childhood lymphoid malignancies: results of a randomized European Organisation for Research and Treatment of Cancer-Children~s Leukemia Group phase 3 trial. Blood. 2002;99:2734–2739. doi: 10.1182/blood.v99.8.2734. [DOI] [PubMed] [Google Scholar]
- 8.Verma N, Kumar K, Kaur G, Anand S. L-asparaginase: a promising chemotherapeutic agent. Crit Rev Biotechnol. 2007;27:45–62. doi: 10.1080/07388550601173926. [DOI] [PubMed] [Google Scholar]
- 9.Kearney SL, Dahlberg SE, Levy DE, Voss SD, Sallan SE, Silverman LB. Clinical course and outcome in children with acute lymphoblastic leukemia and asparaginase-associated pancreatitis. Pediatr Blood Cancer. 2009;53:162–167. doi: 10.1002/pbc.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rousseau J, Gagne V, Labuda M, Beaubois C, Sinnett D, Laverdiere C, et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood. 2011;118:5883–5890. doi: 10.1182/blood-2011-05-355560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stams WA, den Boer ML, Holleman A, Appel IM, Beverloo HB, van Wering ER, et al. Asparagine synthetase expression is linked with L-asparaginase resistance in TEL-AML1-negative but not TEL-AML1-positive pediatric acute lymphoblastic leukemia. Blood. 2005;105:4223–4225. doi: 10.1182/blood-2004-10-3892. [DOI] [PubMed] [Google Scholar]
- 12.Estes DA, Lovato DM, Khawaja HM, Winter SS, Larson RS. Genetic alterations determine chemotherapy resistance in childhood T-ALL: modelling in stage-specific cell lines and correlation with diagnostic patient samples. British journal of haematology. 2007;139:20–30. doi: 10.1111/j.1365-2141.2007.06763.x. [DOI] [PubMed] [Google Scholar]
- 13.Fine BM, Kaspers GJ, Ho M, Loonen AH, Boxer LM. A genome-wide view of the in vitro response to l-asparaginase in acute lymphoblastic leukemia. Cancer research. 2005;65:291–299. [PubMed] [Google Scholar]
- 14.Akagi T, Yin D, Kawamata N, Bartram CR, Hofmann WK, Song JH, et al. Functional analysis of a novel DNA polymorphism of a tandem repeated sequence in the asparagine synthetase gene in acute lymphoblastic leukemia cells. Leukemia research. 2009;33:991–996. doi: 10.1016/j.leukres.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Dana-Farber Consortium Protocol 91-01. Blood. 2001;97:1211–1218. doi: 10.1182/blood.v97.5.1211. [DOI] [PubMed] [Google Scholar]
- 16.Vrooman LM, Stevenson KE, Supko JG, O~Brien J, Dahlberg SE, Asselin BL, et al. Postinduction dexamethasone and individualized dosing of Escherichia Coli L-asparaginase each improve outcome of children and adolescents with newly diagnosed acute lymphoblastic leukemia: results from a randomized study--Dana-Farber Cancer Institute ALL Consortium Protocol 00-01. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1202–1210. doi: 10.1200/JCO.2012.43.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SH, Pei D, Yang W, Cheng C, Jeha S, Cox NJ, et al. Genetic variations in GRIA1 on chromosome 5q33 related to asparaginase hypersensitivity. Clinical pharmacology and therapeutics. 2010;88:191–196. doi: 10.1038/clpt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grace RF, Dahlberg SE, Neuberg D, Sallan SE, Connors JM, Neufeld EJ, et al. The frequency and management of asparaginase-related thrombosis in paediatric and adult patients with acute lymphoblastic leukaemia treated on Dana-Farber Cancer Institute consortium protocols. British journal of haematology. 2011;152:452–459. doi: 10.1111/j.1365-2141.2010.08524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchanan CC, Torstenson ES, Bush WS, Ritchie MD. A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. Journal of the American Medical Informatics Association : JAMIA. 2012;19:289–294. doi: 10.1136/amiajnl-2011-000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aslanian AM, Fletcher BS, Kilberg MS. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J. 2001;357:321–328. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwamoto S, Mihara K, Downing JR, Pui CH, Campana D. Mesenchymal cells regulate the response of acute lymphoblastic leukemia cells to asparaginase. The Journal of clinical investigation. 2007;117:1049–1057. doi: 10.1172/JCI30235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastorczak A, Fendler W, Zalewska-Szewczyk B, Gorniak P, Lejman M, Trelinska J, et al. Asparagine synthetase (ASNS) gene polymorphism is associated with the outcome of childhood acute lymphoblastic leukemia by affecting early response to treatment. Leukemia research. 2014;38:180–183. doi: 10.1016/j.leukres.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Krejci O, Starkova J, Otova B, Madzo J, Kalinova M, Hrusak O, et al. Upregulation of asparagine synthetase fails to avert cell cycle arrest induced by L-asparaginase in TEL/AML1-positive leukaemic cells. Leukemia. 2004;18:434–441. doi: 10.1038/sj.leu.2403259. [DOI] [PubMed] [Google Scholar]
- 25.Chen SH, Yang W, Fan Y, Stocco G, Crews KR, Yang JJ, et al. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2011;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, et al. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer research. 2007;67:3345–3355. doi: 10.1158/0008-5472.CAN-06-2519. [DOI] [PubMed] [Google Scholar]
- 27.Lorenzi PL, Reinhold WC, Rudelius M, Gunsior M, Shankavaram U, Bussey KJ, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Molecular cancer therapeutics. 2006;5:2613–2623. doi: 10.1158/1535-7163.MCT-06-0447. [DOI] [PubMed] [Google Scholar]
- 28.Sircar K, Huang H, Hu L, Cogdell D, Dhillon J, Tzelepi V, et al. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. The American journal of pathology. 2012;180:895–903. doi: 10.1016/j.ajpath.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.