SUMMARY
Genes are packaged into nucleosomal arrays, each nucleosome typically having two copies of histones H2A, H2B, H3, and H4. Histones have distinct post-translational modifications, variant isoforms, and dynamics. Whether each histone copy within a nucleosome has distinct properties, particularly in relation to the direction of transcription, is unknown. Here we use ChlP-exo to resolve the organization of individual histones on a genomic scale. We detect widespread subnucleosomal structures in dynamic chromatin, including what appear to be half nucleosomes consisting of one copy of each histone. We also detect interactions of H3 tails with linker DNA between nucleosomes, which may be negatively regulated by methylation of H3K36. Histone variant H2A.Z is enriched on the promoter-distal half of the +1 nucleosome, whereas H2BK123 ubiquitylation and H3K9 acetylation are enriched on the promoter-proximal half in a transcription-linked manner. Subnucleosome asymmetries might serve as molecular beacons that guide transcription.
Keywords: Chromatin structure, nucleosomes, histone modifications, genomics
INTRODUCTION
Nearly every gene in a eukaryotic nucleus is packaged into chromatin by an array of nucleosomes (Jiang and Pugh, 2009; Rando and Ahmad, 2007; Segal and Widom, 2009). How these genic arrays are structured in relation to transcription is only partly understood. The first nucleosome in each array typically resides at a canonical distance from the transcription start site (TSS) and at the edge of a 5’ nucleosome-free promoter region (NFR). A subset of quiescent genes, typically regulated by the SAGA complex, may have nucleosomes over their promoters which are lost or depleted upon gene activation. The +1 nucleosome is the gateway to transcription as it is the first nucleosome encountered by the transcription machinery. Arrays continue into gene bodies, having nucleosome repeat lengths (NRL) of ~165 bp in budding yeast. This regularity dissipates towards the middle of genes.
In the wake of DNA replication nucleosomes are assembled by chaperones that first escort dimers of histones H3/H4 into tetramer intermediates on ~60 bp of DNA, which are then rapidly flanked by two sets of H2A/H2B dimers that together wrap ~147 bp of DNA ~1.65 times around the octamer core (Luger et al., 2012). This basic two-step assembly process has been a tenet in chromatin biology for over 25 years (Kornberg and Lorch, 1999). However, early studies hinted at alternative pathways that produce subnucleosomal particles (Weintraub et al., 1975; Weintraub et al., 1976). Regardless, nucleosomal and subnucleosomal structures with respect to the organization of individual histones and their chromatin context have not been defined on a genome-wide scale.
A nucleosome has two-fold symmetry of histone organization, and thus might have a symmetrical distribution of histones, variants and modifications about its dyad axis. However, RNA polymerase II engages the NFR-proximal face of a nucleosome differently than it engages its distal back-end, as it transcribes a gene. Either as a cause or a consequence of an asymmetric polymerase-nucleosome relationship, the levels of histones, variants and their modifications might be asymmetrically distributed within specific nucleosomes.
The ChlP-exo assay locates formaldehyde-induced protein-DNA crosslinks along a genome at very high resolution in vivo (Rhee and Pugh, 2011, 2012b). Here we apply ChlP-exo to the budding yeast four core histones, histone variant H2A.Z, the linker histone HI, and the transcription-linked histone modifications H3K4me3, H3K36me3, H3K79me2/3, H3K9ac, and H2BK123ub. With this we examine nucleosome substructure and symmetry at the ~60,000 nucleosome-occupied sites in the budding yeast genome. Our study suggests a surprising model of intra- and inter-nucleosomal histone interplay that may reflect a variety of subnucleosomal structures and their dynamics. A substantial fraction of all genes display differential histone occupancy on one half of their nucleosomes versus the other. Nucleosomes at specific positions within arrays have an asymmetric organization of transcription-linked histone variants and modifications. Together, these finding paint a strikingly detailed and unexpected view of subnucleosomal structure in vivo and its relationship with the direction of transcription.
RESULTS
H3 Tails Engage Linkers
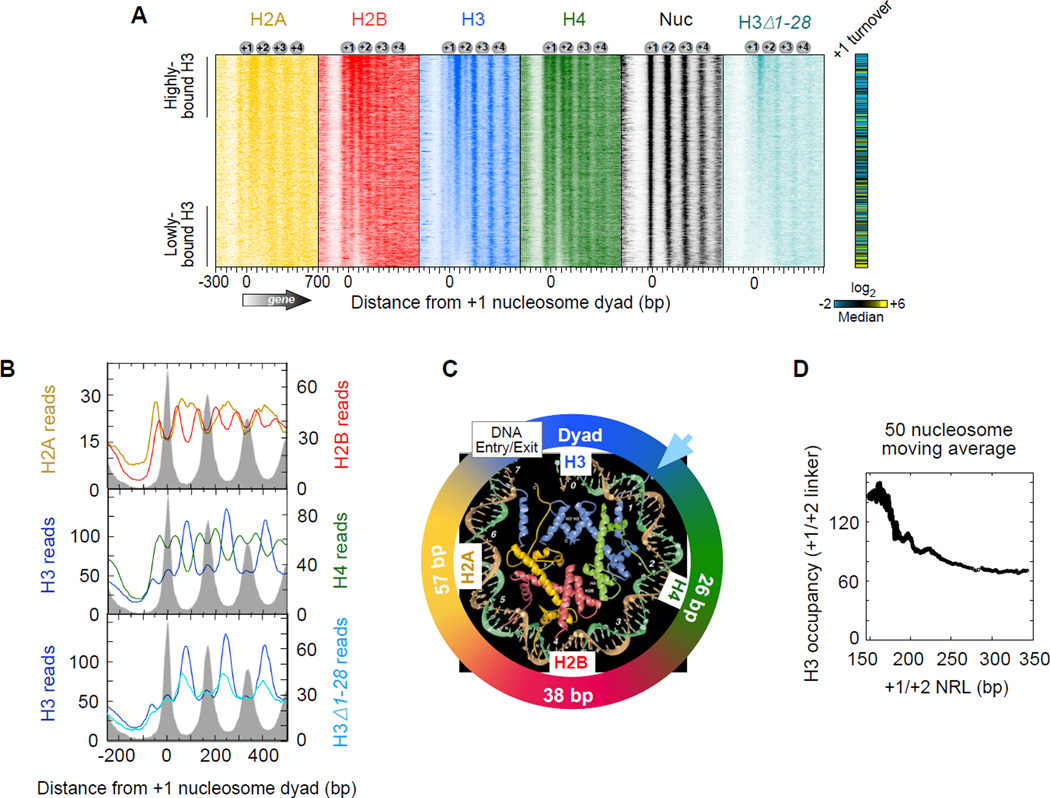
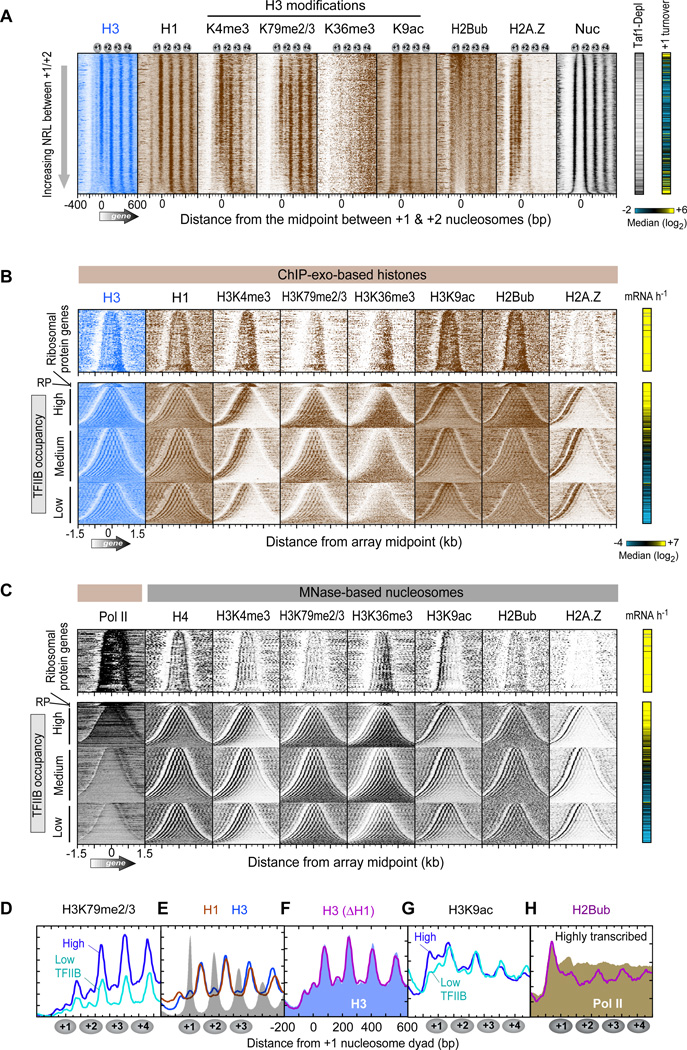
Figure 1A displays the distribution of ChlP-exo crosslinking points (exonuclease stop sites) for each of the four core histones around the 5’ end of genes. None of the histones substantially crosslinked in NFRs, as expected of their general nucleosome-free status, although SAGA-regulated (Tafl-depleted) genes tended to have higher histone occupancy in promoter regions (data not shown). H2B and H4 each displayed two regions of crosslinking for each nucleosome (two vertical stripes for each “Nuc” stripe in Figure 1A and averaged in Figure 1B). Their crosslinks corresponded to the genomic locations expected from the crystal structure of the nucleosome core particle that is centered on the genomic coordinates of nucleosome midpoints (dyads), as defined by MNase digestion (Figure 1C). H4 and H2B were about 26 and 38 bp from the dyad, respectively. H2A crosslinked broadly near linker DNA, which is consistent with in vitro studies (Shukla et al., 2011; Usachenko et al., 1994). This broad crosslinking represents two adjacent H2A from two adjacent nucleosomes. Sorting arrays by the length of the linker DNA between the +1 and +2 nucleosomes resolved H2A into separate peaks (Figure S1). For brevity, we focused subsequent analyses on the +1 and +2 nucleosome positions, although equivalent conclusions can be drawn at other resolvable genic positions.
Figure 1. Subnucleosomal Detection of Histones across Yeast Genes.
(A) Histone occupancy (color intensity) as detected by ChlP-exo was plotted relative to +1 nucleosome dyads (“Nuc”, defined by MNase) at annotated mRNA genes (4,738 rows; values reported in Table S3). Rows were sorted by H3 occupancy in the linker between the +1 and +2 nucleosomes (Table S2). Histone turnover rate in the +1 nucleosome region is from (Dion et al., 2007), where yellow, black, and blue represent high, medium, and low turnover, respectively.
(B) Composite (average) plot of panel A. Gray fill indicates nucleosome midpoint distribution defined by MNase (tags plotted relative to consensus).
(C) Regions of histone crosslinking projected onto the crystal structure of one half of the nucleosomal core particle (Luger et al., 1997). Colored segments of the circle denote regions of crosslinking, centered at the indicated distance from the nucleosome dyad. Arrowhead denotes where the H3 tail emerges from between the DNA gyres.
(D) Relationship between NRL and H3 occupancy levels between the +1 and +2 nucleosomes (Table S2). Data were plotted as a 50-nucleosome moving average for all NRL >145 bp (minimum size of a nucleosome). Data were not background-subtracted.
See also Figure S1.
Surprisingly, H3 crosslinking peaked in linker regions rather than at nucleosome dyads where the bulk of H3 resides (Figure 1A,B). A minor H3 peak was detected at the dyad. In the nucleosome crystal structure, the base of the H3 N-terminal tail emerges from the nucleosome core near the linker DNA (Figure 1C). To experimentally test whether H3 tails were responsible for linker crosslinking, ChlP-exo was performed in a strain where most, but not all, of the H3 tail was deleted (Δl–28) (Morgan et al., 1991). H3-linker crosslinking was diminished in this strain (Figure 1A,B), suggesting that H3 tails are in close proximity to linker DNA within genic nucleosome arrays. This conclusion is further supported by in vitro reconstitution experiments (Zheng et al., 2005). The overall H3 pattern was not grossly altered in the H3Δ1–28 strain (Figure 1A,B), indicating that amino acids 1–28 of H3 tails lack a predominant or nonredundant role in organizing nucleosomes within arrays. The remaining eight amino acids of the tail (residues 29–36) could nevertheless be involved.
Remarkably, H3 crosslinks were largely absent from the edges of NFRs adjacent to +1 nucleosomes (which might be thought of as very long linkers, Figure 1A,B). When arrays were sorted by +1/+2 linker-length, H3 tail-linker crosslinking diminished at linkers >~30 bp in length (i.e., where the nucleosome repeat length or NRL exceeds ~180 bp in Figures 1D and S1). Genes at both extremes of this linker-length distribution tended to have more dynamic nucleosomes and be of the Tafl-depleted (and thus SAGA/TATA/stress-regulated) class (right-most panels in Figure S1), which is consistent with the greater plasticity and inducibility of these genes (Huisinga and Pugh, 2004; Tirosh and Barkai, 2008). Thus, nucleosome dynamics, +1/+2 linker-length, diminished H3 interactions at the +1/+2 linker, and gene inducibility appear to be linked. This may reflect an ability to mobilize (e.g., re-position or dissociate) a noncanonically positioned +1 nucleosome, which would occur adjacent to long linkers. Indeed, chromatin remodelers may access nucleosome via adjacent long linkers (Ranjan et al., 2013).
In principle, since transcription and genic arrays have directionality, H3 tail-linker interactions might arise predominantly from H3 in the adjacent upstream nucleosome or H3 in the adjacent downstream nucleosome, resulting in asymmetric substructures. Alternatively, H3 in both flanking nucleosomes might contributed similarly. We reasoned that H3 tail-linker occupancy levels should correlate most with the occupancy level of the nearest neighboring nucleosome or H4 subunit, since H3 normally interacts with H4 within a nucleosome. H3-linker occupancy correlated similarly with both flanking H4 occupancy levels (blue arrow in Figure 2A, Rproximal = 0.39 vs. Rdistal = 0.31) and also with total flanking nucleosomal H4 levels (i.e., both H4 copies within a flanking nucleosome, Figure S2A–C). Similar trends were evident with H3 (Δl–28) (Figure S2D, Rproximal = 0.37 vs. Rdistal = 0.30). We conclude that H3-linker interactions are largely derived from both flanking nucleosomes, although the correlations suggest there may be slightly more interactions arising from the more upstream nucleosome.
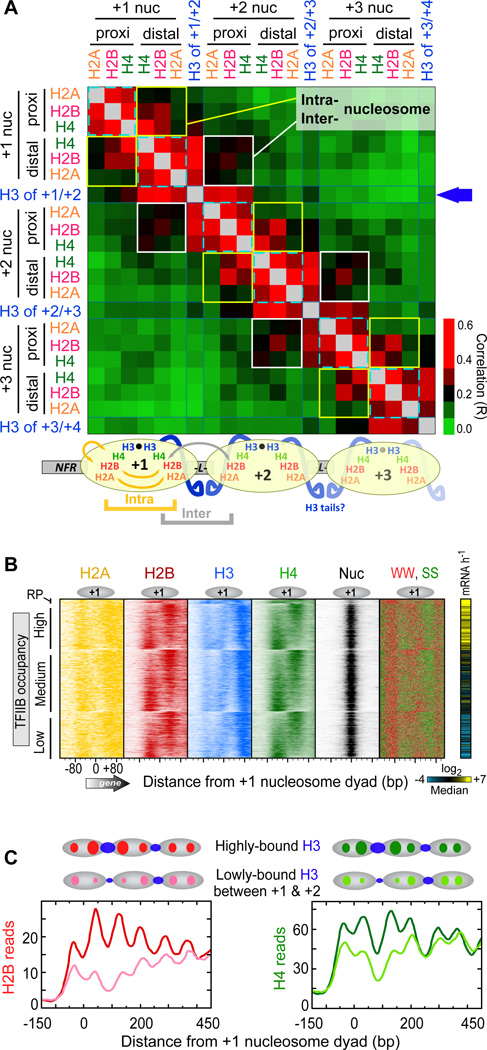
Figure 2. Asymmetry in Histone Occupancy.
(A) Heat map of correlation coefficients (R) for all pair-wise combinations of NFR-proximal and NFR-distal histone occupancies at +1, +2, and +3 nucleosomes (Table S2). To simplify the patterning, correlations above 0.4 were coded with the same red color. The types of intra- and inter-nucleosomal correlative interactions that the data suggest are illustrated below the heat map.
(B) Histone distribution relative to the +1 nucleosome of mRNA genes (4,738 rows) orientated by TSS, and sorted by H4 NFR-distal versus NFR-proximal ratios (Table S4). Rows were grouped by ribosomal protein (RP, n=128) genes, then by high (n=l,381), medium (n=l,852), and low (n=l,377) TFIIB occupancy in the promoter region (Rhee and Pugh, 2012b). Also shown is the distribution of SS (green) and WW (red) dinucleotides (IUPAC: W=A/T, S=C/G; 4 bp bin). The right panel shows transcription frequency (Holstege et al., 1998), where yellow, blue, black represent high, medium, and low rates, respectively.
(C) Composite distribution of histones separated out by the most highly- versus the most lowly-occupied H3 in the +1/+2 linker (respectively shown as dark and light colored traces, using 30* percentile cutoffs; n=l,455 for each, derived from Table S3). Gray ovals demarcate nucleosome intervals.
See also Figure S2.
Evidence for Subnucleosomal Structures
Since the histone octamer is a fundamental unit of chromatin, we expected histone occupancies on the NFR-proximal half of a nucleosome to correlate with histone occupancies on its NFR-distal half. Indeed, occupancies did correlate between the two halves (yellow boxed areas in Figure 2A, Rave = 0.22). However, the correlations were surprisingly modest when compared to stronger cross-correlations of the core histones located on the same half of each nucleosome (cyan dashed areas, Rave = 0.45). Non-adjacent combinations were uncorrelated (Rave = 0.08). Although the H3 that was crosslinking in the linker was not included in this assessment, it too behaved similarly. These relationships were also evident in plots of individual arrays (Figure 1A) and in composite plots (see Figure 2C). In general, H4-H4 cross-nucleosomal correlations were higher than for H2A-H2A and H2B-H2B, which may reflect a tighter cross-nucleosomal linkage between H4. We interpret these correlations to reflect differential histone occupancy (or DNA crosslinking) on one half of a canonical genic nucleosome compared to the other, subject to the controls and caveats described below.
Figure 2B displays the distinct histone occupancy levels on the two halves of canonical +1 nucleosomes, where all datasets were grouped by promoter activity (TFIIB occupancy) of the associated gene, then sorted based on H4 distal/proximal ratios. Thus, where H4 occupancy was higher on the NFR-distal half of +1, the occupancy of the other histones was also higher on that half. A reciprocal relationship existed on the NFR-proximal half. These findings suggest that the histones on one half of a genic nucleosome may be more coordinated in their DNA occupancy than they are across the two halves of the same nucleosome.
About 50% of all analyzed nucleosomes showed a >2-fold differential of H2B occupancy on one half versus the other, whereas only about 12% showed the same differential with H4. These represent arbitrary thresholds, as there is a continuum of differential occupancy but subject to nonbiological biases in detection, which we addressed below. Thus, differential occupancy is detected between two halves of a nucleosome in a substantial number of cases. The more pronounced differential seen with H2B suggests that additional differential occupancy may exist between H3/H4 and H2A/H2B on the same half of a nucleosome. The low cross-nucleosome correlations relative to the same-side correlations at nucleosome positions +1, +2, and +3 (Figure 2A) indicates that these relationships exist broadly across multiple genic nucleosome positions.
SS-richness (where S denotes G or C) in DNA promotes nucleosome assembly (Kaplan et al., 2009; Mavrich et al., 2008; Tillo and Hughes, 2009). We therefore examined the distribution of SS dinucleotides at +1 nucleosomes and found them to partially reflect differential proximal versus distal histone occupancy (Figure 2B). This suggests that an imbalance of SS (or GC content) on one half of a nucleosome or the other may contribute to differential histone occupancy, much as it contributes to overall nucleosome occupancy. While this also raises the question as to whether any sequence specificity in formaldehyde crosslinking or sample processing is responsible for the histone ChlP-exo patterning, our analyses of this issue in Figure S3A,B suggest otherwise.
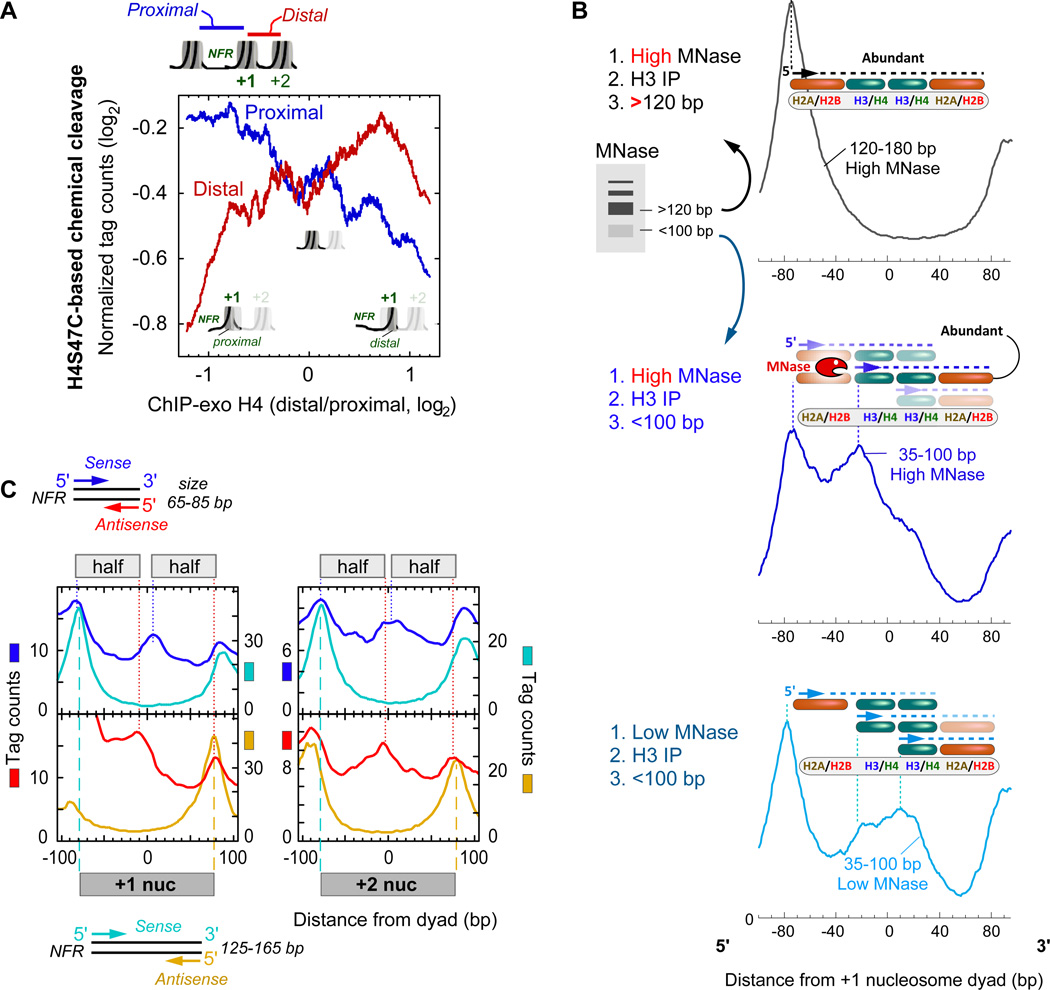
We sought independent evidence for differential distal versus proximal histone occupancy using an assay that did not involve formaldehyde or ChlP-exo. Nucleosome organization has been mapped genome-wide at high precision using an engineered cysteine at residue 47 in H4 to catalyze hydroxyl-radical cleavages in DNA near nucleosome dyads (Brogaard et al., 2012). This separates proximal from distal nucleosomal DNA. As shown in Figure 3A, H4 distal/proximal occupancy ratios for the +1 nucleosome, as determined by ChlP-exo, positively correlated with levels of distal H4S47C-cleaved fragments and negatively correlated with proximal fragments. As described in the Extended Experimental Procedures, these correlations and the abrupt changes in the trends at the extremes of the ratios are predicted outcomes of differential proximal versus distal histone occupancy.
Figure 3. Evidence for Differential Proximal versus Distal Histone Occupancy using Chemical Cleavage and MNase.
(A) Evidence for differential distal versus proximal histone occupancy in H4S47C-mediated chemical cleavage data (Brogaard et al., 2012). Average levels of DNA fragments (tag counts) released on the NFR-proximal (blue) or NFR-distal (red) side of 4,738 genic +1 nucleosomes are plotted as a function of H4 distal/proximal occupancy ratio at +1 nucleosomes. Data were smoothed using a 500 value moving average. See Table S5 for data processing.
(B) Cleavage at canonical nucleosomal dyads using low MNase activity. Chromatin was treated with either high or low MNase activity, H3-immunoprecipitated, then size-selected in the indicated range. The average distribution of unshifted tag 5’ ends around +1 nucleosomes (n=4,738) was orientated from left to right in the 5’−3’ direction (regardless of strand). Note that plotting strands separately (sense versus antisense with respect to the direction of transcription) produces essentially identical plots within the relevant +1 region, when plotted in the 5’ to 3’ direction (not shown). Tag counts are normalized and thus their vertical scales are not comparable between traces. Above each trace are illustrated interpretations of the peaks. The interpretations were constrained by the experimental design so that properly sized fragments spanned H3 and included at least two dimer sets of histones. See Table S5 for data processing.
(C) Composite distribution of MNase cleavage sites (paired-end tag 5’ ends) reported by (Henikoff et al., 2011), plotted relative to the +1 and +2 nucleosome midpoint (Table S2) of annotated mRNA genes (n=4,738) with respect to TSS orientation. Full nucleosome (125–165 bp, cyan/orange) and subnucleosome (65–85 bp, blue/red) were computationally size-selected and their occupancy (tag counts) plotted. Cyan/blue vertical lines indicate peak 5’ ends on the sense strand, and red/orange vertical line indicates peak 5’ ends on the antisense strand of paired-end reads. Data show DNA solubilized with low MNase activity (2.5-min digestion).
See also Figure S3.
We next used MNase ChlP-seq as a third independent probe of subnucleosomal structure. MNase probes nucleosomal particles in their native state. MNase at low activity preferentially cleave histone-free DNA, rather than DNA within the nucleosome core. Since we were probing for subnucleosomal particles, we size-selected for library inserts in the 35–100 bp range, rather than the normal range of 120–180 bp for full nucleosomes. In this population, we detected enhanced cleavages (5’ ends) starting where H3/H4 interfaces with H2A/H2B and extending through where the canonical dyad resides (Figure 3B, where the bottom trace plateaus from −30 to +30 bp from the dyad). We interpret this cleavage to reflect enhanced DNA accessibility internal to and on one side of what otherwise would be a full nucleosome. Enhanced cleavage at the expected nucleosome edge (~75 bp from the dyad) was also detected with these small fragments. Both sets of cleavages are consistent with subnucleosomal structures consisting of hexasomes (nucleosome that lack one H2A/H2B dimer) and half-nucleosomes.
For comparison, Figure 3B also plots the distribution of subnucleosomal fragments from standard high MNase digestion (blue middle trace). This level of MNase is expected to partially nibble in from the edges of full nucleosomes up to the junction between H2A/H2B and H3/H4, due to the loose association of DNA in that region. Since full nucleosomes are quite abundant, these cleavages are expected to dominate the distribution of small fragments, thereby obscuring the presence of subnucleosomal fragments. As a control, large DNA fragments (120–180 bp) have their predominant cleavages occuring at the expected edges of full nucleosomes. Together these results are consistent with MNase detecting hexasomal and half-nucleosomal structures.
We next sought to confirm our MNase experiments using a fourth assay, paired-end MNase-seq data of (Henikoff et al., 2011). This assay did not involve formaldehyde or ChlP. As shown in Figure 3C, cleavages were again detected in the dyad region amongst the population of small DNA fragments. Based on the mechanism by which MNase cleaves DNA, we suggest that in these instances the DNA is lifted off of one half or quarter of the histone core where it is accessible to MNase, or alternatively results from differential distal versus proximal histone occupancy. This implies that at least some nucleosomes at the 5’ end of genes (other regions not excluded) have at least one half (demarcated by the dyad) that is intact and the other half or quarter that is disassembled.
The concept of alternative nucleosomal substructures was first described by Weintraub et al in 1975: “A basic unit of chromosome structure is a tetramer containing all 4 histones” (Weintraub et al., 1975; Weintraub et al., 1976), where “half-nucleosomes” were reconstituted in vitro with pure histones and DNA. “Unfolded” nucleosomes have been isolated from cells, whereby the normally inaccessible H3-H3 dyad interface was found to be accessible to external mercury probes (Chen et al., 1991). Other subnucleosomal structures, including hexasomes have been suggested (Annunziato, 2005; Black and Cleveland, 2011; Zlatanova et al., 2009). More recently, hemisomes have been suggested for centromeric nucleosomes (Dalal et al., 2007; Krassovsky et al., 2012), and have been reconstituted in vitro using centromeric H3 (Cse4) and canonical H3 (Furuyama et al., 2013). Our ChlP-exo datasets provide little insight into centromeric nucleosome structures, in that the four core histones were not detected at centromeric locations perhaps due to inefficient crosslinking or extraction (Figure S3C). However, adjacent, non-centromeric subnucleosomal structures were detected. Partial-nucleosomes appear enriched at two positions to the right of centromeres, although full nucleosomes also were nearby. To the left of the centromeres, full nucleosomes predominated.
Histone Coordination between Nucleosomes
Less H3 crosslinking within the +1/+2 linker was accompanied by a parallel decrease in flanking histone occupancy (i.e., the NFR-distal half of +1 and NFR-proximal half of +2, Figure 2C, also evident in Figure 1A). Histone occupancies on the two flanks of a linker were as correlated on average as they were across two halves of the same nucleosome (Figure 2A, white versus yellow boxed areas, Rave. = 0.20 versus 0.22, respectively). These averaged values were essentially the same at all nucleosome positions, where examined (+1, +2, +3). Thus, there appears to be coordination in histone occupancy between two halves (or parts thereof) of adjacent nucleosomes towards a shared linker. In a slight contrast, H4 was more correlated within a nucleosome than between nucleosome. Speculatively, this might be due to the presence of H3/H4 tetramers in addition to other substructures.
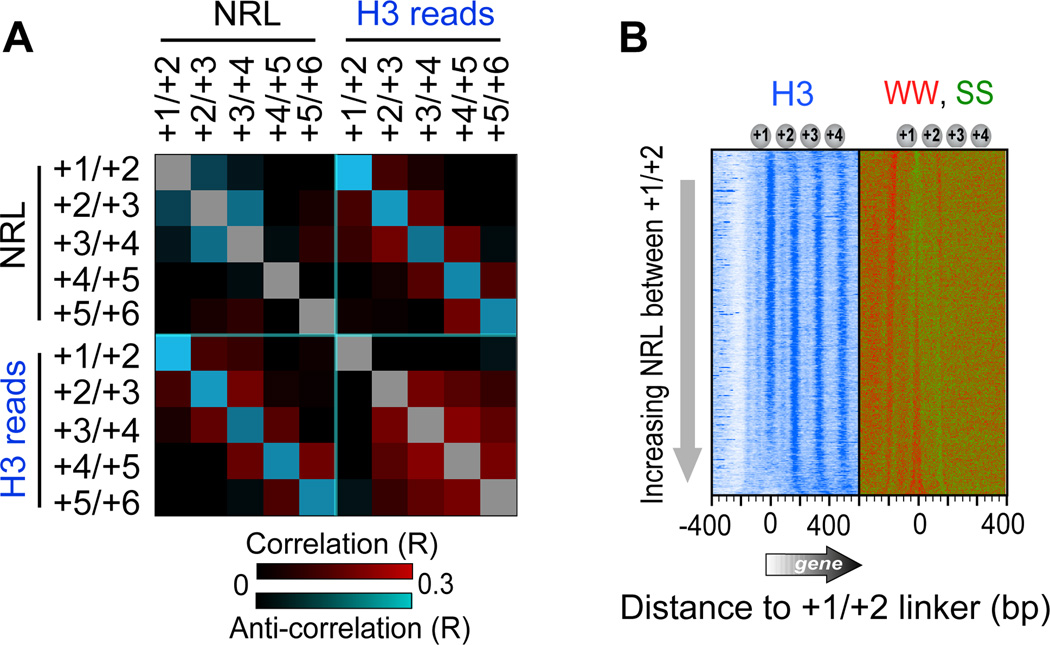
NRLs (distances between MNase-defined nucleosome midpoints) anti-correlated with flanking NRLs, and also anti-correlated with underlying H3 tail-linker occupancy levels (Figures 4A and S1). Thus, despite generally uniform positioning imposed by chromatin remodelers (Zhang et al., 2011b), certain nucleosome neighbors gravitate towards each other (illustrated in Figure 7, bottom panel). Longer linkers (NRL) arise on their other flank, and this is coupled to histone depletion of the nucleosomal halves that abut these long linkers. This is consistent with longer linkers having higher histone exchange rates (Figures 1A and S1) and diminished H3 tail interactions. Since linker length is measured using full nucleosomes (regardless of histone occupancy), their measurement should not be influenced by histone depletion. Remarkably, WW enrichment (where W denotes A or T) “painted” the linker-length landscape (Figure 4B, WW, SS panel). Thus, linker WW nucleotides may promote deviations from uniform positioning established by remodelers, and in doing so enhance occupancy dynamics of adjacent histones.
Figure 4. Sequence-based Localized Nucleosome Positioning.
(A) Heat map representing all pair-wise correlations (R) between NRLs and H3-linker occupancy levels at nucleosomes +1 through +6. Cyan and red represent negative and positive correlations, respectively. Calculations were based on nucleosomes at 3,194 genes longer than 1 kb (Table S2). NRL reports dyad-to-dyad distances in MNase-based maps (Zhang et al., 2011b). Similar observations were made with positions determined by ChlP-exo (not shown).
(B) Occupancy level of H3 ChlP-exo and SS/WW dinucleotide frequencies, relative to the midpoint between the +1 and +2 nucleosomes (Table S6), sorted by NRL between the +1 and +2 nucleosomes (Table S2).
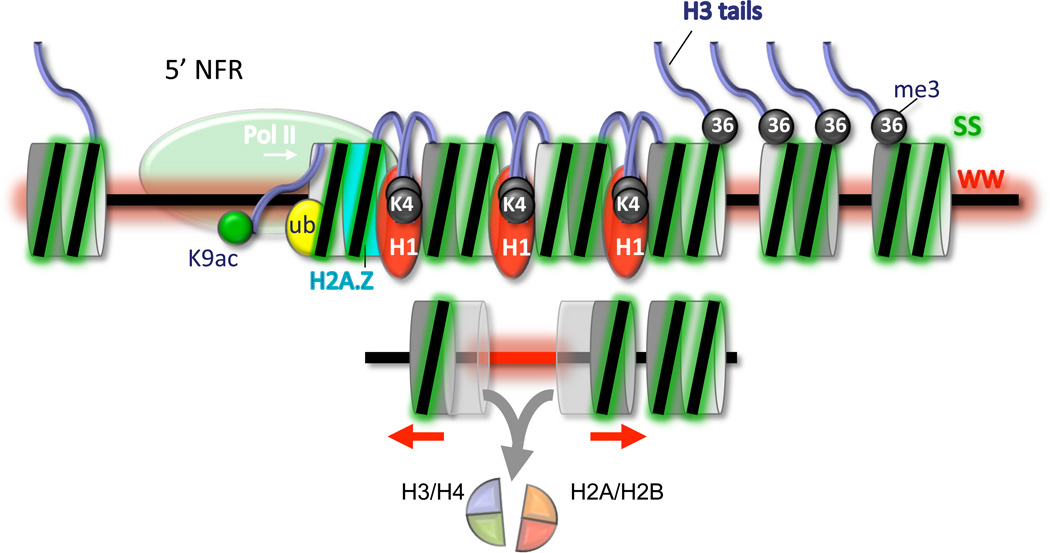
Figure 7. Composite Model of Nucleosomal Arrays at Genes.
The nucleosome-free promoter region (5’ NFR) is depicted with RNA polymerase II. Within arrays, the two sides of a nucleosome are depicted as two-toned gray disks, fused to form nucleosomes. In the upper panel, histone marks, variants, and WW/SS dinucleotide enrichment are depicted by different colors. “K9ac”, “ub”, “K4”, and “36” denote H3K9ac, H2Bub, H3K4me3, and H3K36me3, respectively. These features are placed at their predominant locations within arrays. The lower panel illustrates longer WW-rich linkers (red) that are linked to histone destabilization on the linker-flanking half of each nucleosome (symbolized by transparency). The red arrows indicate that those (sub)nucleosomes are closer than normal to their adjacent nucleosomes.
H3K36me3 Negatively Regulates Linker Interactions
H3 tails are methylated (me) and acetylated (ac) in a genome-wide location-specific manner so as to potentially regulate nucleosomal arrays and transcription. We examined whether H3 modifications alter H3-linker interactions by conducting ChlP-exo after immunoprecipitation with histone modification-specific antibodies. H3K4me3 and H3K79me2/3 were enriched at their previously published array positions (i.e., nucleosome positions +1, +2, +3 for H3K4me3 and at all positions for H3K79me2/3) in a transcription-linked manner (Figure 5A–D). In addition, linker crosslinking patterns were consistent with expectations from MNase-based maps. Therefore, these marks had no overt effect on H3-linker interactions.
Figure 5. Subnucleosomal Organization of Histone Marks and Variants.
(A) Occupancy levels of histones and their marks relative to the +1/+2 linker, and sorted by linker length (n=4,738 genes; Table S7). The right panels demarcate Tafl-depleted genes (black lines) (Rhee and Pugh, 2012b) and histone turnover rate of the +1 nucleosome region (yellow indicates “hot” nucleosomes) (Dion et al., 2007).
(B) Same as panel A except that entire genic arrays are shown (5’ to 3’ from left to right). Arrays are sorted by array length and grouped by ribosomal protein (RP) genes (including an expanded vertical view), and the remaining by TFIIB occupancy (Rhee and Pugh, 2012b). The right panel shows transcription frequency (Holstege et al., 1998).
(C) Similar as panel B, except measured by MNase ChlP-seq from previous studies (Albert et al., 2007; Batta et al., 2011; Zhang et al., 2011a; Zhang et al., 2011b). Pol II denotes RNA polymerase II measured by ChlP-exo.
(D–H) Composite distribution of the indicated histones or marks relative to the +1 nucleosome midpoint at genes having TFIIB promoter occupancy in the top and bottom 30% (Rhee and Pugh, 2012b). E–F, are for all genes. F, H3 in a wild type strain is shown as a blue fill, and H3 in an hholΔ strain as a magenta trace. H, H2Bub (magenta) and Pol II (brown fill) are for genes having the top 30% of TFIIB occupancy.
See also Figure S4.
In contrast, H3K36me3 had a markedly less coherent ChlP-exo pattern (Figure 5A,B and Figure S4A), despite recapitulating the known array asymmetry (i.e., depletion at the 5’ end of genes). This was surprising because MNase-based maps of nucleosome cores having H3K36me3 display very robust array patterning (Figure 5C) (Zhang et al., 2011a). K36 is located at the base of the H3 tail where it emerges between the DNA gyres of the nucleosome core (Luger et al., 1997). The diffuse pattern of crosslinking associated with H3K36me3 suggests that while H3K36me3 nucleosome cores are well-positioned in genic arrays (based on MNase maps), this mark is inhibitory to H3 tail-linker crosslinking. Conceivably, K36me3 might alter the trajectory of the H3 tail as it emerges from the DNA gyres, as reported for tail mutants (Ferreira et al., 2007), or bind histone modifying/remodelling enzymes such as the Rpd3S histone deacetylase complex or the ISW1 complex (Carrozza et al., 2005; Keogh et al., 2005) with the result of blocking H3 tail-linker interactions. A more trivial explanation may be that K36me3 renders K36 less reactive to formaldehyde, if indeed K36 is the major point of crosslinking. To experimentally test this, we performed ChlP-exo on H3 containing alanine instead of lysine at position 36 (K36A mutant). This mutant H3 appeared to crosslink normally to linker DNA (Figure S4B), indicating that K36 is not the predominant H3 crosslink to linker DNA, although its methylation alters the potential of the tail to crosslink to linkers.
We examined linker histone HI and found its crosslinking pattern to be almost identical to that of H3 (Figure 5A,B,E). To test whether HI regulates H3-linker interactions we deleted HI (hholΔ strain). However, we observed no effect on H3-linker interactions (Figure 5F), nor any effects on nucleosome organization. Thus HI does not play a widespread or nonredundant role in organizing nucleosomal arrays in Saccharomyces.
Asymmetry of H3K9ac, H2Bub, and H2A.Z
Nucleosomal arrays that encompass genes have asymmetry, as a whole, with respect to transcription-linked histone modifications, being distinct at the 5’ ends of genes compared to internal and 3’ locations (Henikoff, 2008; Rando and Ahmad, 2007). This asymmetry is an integral part of the transcription cycle. Since RNA polymerase II makes distinct approaches to the proximal versus distal sides of these nucleosomes, we examined whether this might be reflected in asymmetric deposition of histone marks and variants. Remarkably, at highly but not lowly transcribed genes we found H3K9ac to be enriched primarily on the NFR-proximal half of the +1 nucleosome (Figure 5B,G). This is the half of the +1 nucleosome where H3 crosslinking was almost nonexistent in the general population. Indeed, a similar transcription-linked enrichment at the NFR-proximal side of +1 was not observed for H3 or for other transcription-linked H3 marks. This is consistent with the notion that the K9ac mark is transient, and that most H3 is unacetylated at H3K9 even at highly transcribed genes. Thus, H3K9ac rather than transcription per se, may be more directly linked to H3 tail contacts with the edge of NFRs.
H2BK123 is located within the core C-terminal a-helix of H2B, and its ubiquitylation (ub) is linked to the transcription cycle (Batta et al., 2011; Fleming et al., 2008; Pavri et al., 2006). Like H3K9ac, H2Bub was highly enriched on the NFR-proximal half of the +1 nucleosome in a transcription-linked manner (Figure 5B,H). In light of this and the prior observation that loss of ubiquitylation results in accumulation of RNA polymerase II at promoters (Batta et al., 2011), we speculate that H2B ubiquitylation at the NFR-proximal half of the +1 nucleosome facilitates the movement of RNA polymerase II into gene bodies. Further into gene bodies, crosslinking of additional H2Bub occurred in linkers, and was contributed by both flanking H2B (Figure S4C), as seen with H3. Its basis is currently unclear, although preferential crosslinking of ubiquitylated H2B to H3 could generate such a pattern.
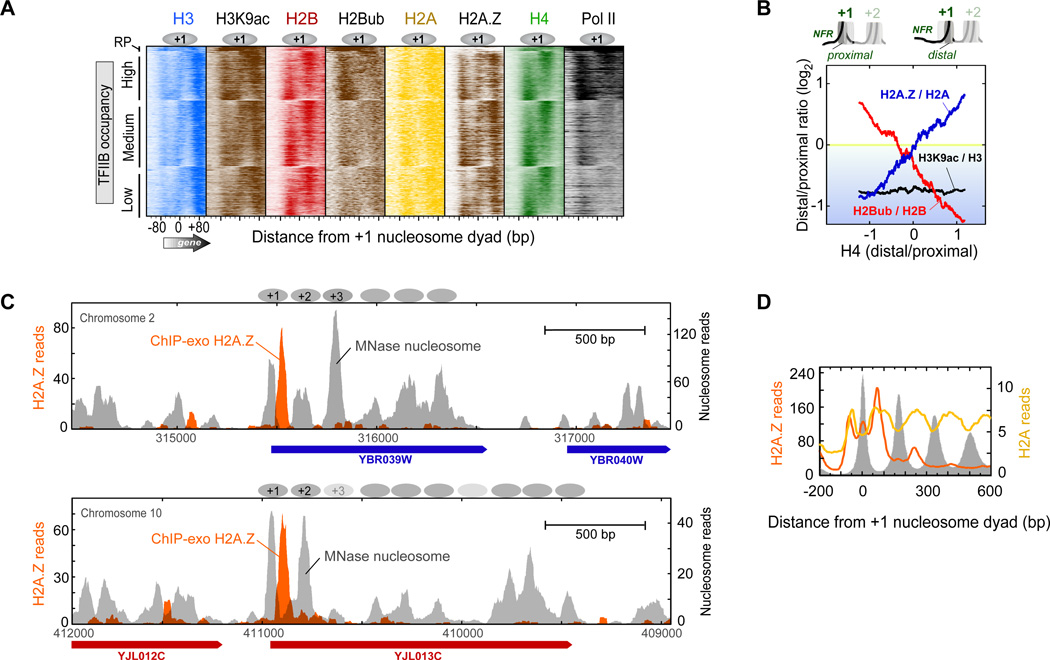
The apparent nucleosome asymmetry of histone marks could be an indirect consequence of differential histone occupancy (Figure 6A). To address this possibility, we normalized the level of each mark to the underlying occupancy level of the relevant histone (e.g., H3K9ac/H3). We then calculated the log2 occupancy ratio at the distal half of the +1 nucleosome to its proximal half, and plotted it as a function of H4 distal/proximal ratio. Accordingly, H3K9ac retained its overall preference for the NFR-proximal half of +1, as indicated by the black trace in Figure 6B being below zero and having zero slope. One caveat is that H3 and H3K9ac on the distal half was not positionally resolved from the proximal half of the +2 nucleosome.
Figure 6. Asymmetric Density of Histone Marks and Variant H2A.Z.
(A) Proximal vs. distal occupancy of histone marks. Data were plotted as described in Figure 2B.
(B) Shown is the distal/proximal occupancy ratio of the indicated histone marks or variant at +1 nucleosomes (n=4,738), after normalizing to (dividing by) the relevant underlying core histone occupancy (intervals designated in Table S2). Ratios were log2 transformed, sorted by H4 distal/proximal ratio (x-axis), and smoothed using a 500-nucleosome moving average.
(C) Example of asymmetrically placed H2A.Z at the NFR-distal half of the +1 nucleosome at specific loci. Orange filled plot shows the distribution of H2A.Z crosslinking sites (raw sequencing tags) measured by ChlP-exo. Gray filled plot shows nucleosome midpoints detected by MNaseChlP-seq.
(D) Composite distribution H2A.Z (orange trace) and H2A (yellow trace) detected by ChlP-exo, relative to the +1 nucleosome midpoint at all mRNA genes. Gray fill indicates nucleosome midpoint distribution detected by MNase ChlP-seq.
When normalized to H2B, ubiquitylated H2B remained biased towards the NFR-proximal half of the +1 nucleosome (red trace being generally below zero in Figure 6B). In addition, it displayed a tendency towards occupying the half that was most depleted of histones (negative slope of the trace). These effects were accentuated at highly transcribed genes (not shown). As depletion likely reflects dynamics, we infer that higher ubiquitylation density reflects nascent histone assembly occurring on the depleted side, which is what H2B ubiquitylation is thought to promote.
As expected, histone variant H2A.Z was highly enriched at the +1 nucleosome position (Figure 5A,B) (Albert et al., 2007), and was asymmetrically placed towards the NFR-distal half of the +1 nucleosome (Figure 5A and Figure 6C,D). Compared to H2A, H2A.Z was preferentially enriched where H4 was enriched (positive H2A.Z/H2A slope in Figure 6B), thereby linking the presence of H2A.Z to stably occupied histones. An exception was at long +1/+2 linkers, where the trend was reversed. This is consistent with the SWR1/SWR-C complex requiring long DNA to deposit H2A.Z into nucleosomes (Ranjan et al., 2013; Yen et al., 2013). We conclude that the transcription machinery generally encounters H2A.Z on the distal half of the +1 nucleosome. Such heterotypic nucleosomes are intrinsically unstable (Bonisch and Hake, 2012), and thus might facilitate the passage of RNA polymerase II (Weber et al., 2014).
DISCUSSION
Subnucleosomal Structures Suggest Mechanisms for Nucleosome Dynamics
The results presented here provide insight into potential mechanisms of nucleosome instability at the 5’ ends of genes that differs from the canonical view. Collectively, the data show that histone occupancies on one half of a nucleosome dyad are more strongly coordinated with each other than histone occupancy across the two sides of the dyad (Figure 7, bottom panel). The effect is stronger for H2A/H2B than for H3/H4, leading us to surmise that both hexasomes (2 copies of H3/H4 and one copy of H2A/H2B) and half-nucleosomes (one copy of H2A/H2B/H3/H4) exist in vivo, in addition to the more abundant standard nucleosomes. This observation is consistent with early views of in vivo nucleosomes (Weintraub et al., 1975; Weintraub et al., 1976) and with biochemical studies that reconstitute such half-nucleosomes on DNA (Furuyama et al., 2013). Such partial nucleosomes may not be present or evident in typical in vitro reconstitution studies, possibly due to missing factors (e.g., chaperones and/or DNA sequence) and/or the use of assays that are unsuitable for their detection in a subpopulation.
In support of the physiological importance of subnucleosomal structures, nucleosome positions that were associated with noncanonical linker lengths, which are linked to differential distal vs proximal histone occupancies, had distinct properties. They tended to have higher histone turnover, and were associated with Tafl-depleted/SAGA-regulated genes. Differential histone occupancy was also associated with distinctive densities of histone marks. Thus, regardless of its structural basis, differential proximal versus distal histone occupancy is associated with distinct functional properties compared to all other nucleosomes at the same relative position. Differential occupancy was not correlated with transcription frequency of the underlying mRNA gene, which indicates that it is not necessarily linked to transcription. Whether it is linked to noncoding transcription or other types of genomic regulation remains to be determined.
In addition to previously published works, evidence for half-nucleosomes in this study come from several different assays: 1) correlated distal/proximal occupancy ratios of all four core histones, measured by the ChlP-exo assay performed on in vivo crosslinked chromatin, 2) differential occupancy measured by H4S47C-based chemical cleavage of noncrosslinked chromatin; 3) MNase cleavage at dyads of native crosslinked chromatin; 4) MNase cleavage at dyads using noncrosslinked chromatin and paired-end sequencing from a different lab; and 5) an enrichment of DNA sequences (GC) that favor nucleosome formation on the side where histone occupancy was the highest. A number of control analyses indicated that DNA sequence-based biases in either crosslinking or DNA sequencing was not a trivial explanation for differential occupancy. However, we cannot unequivocally rule this possibility out.
Our conclusions on half-nucleosomes run counter to the current view that nucleosome assembly/disassembly proceeds through an H3/H4 tetramer (dimer of dimers) intermediate. Conceivably, however, nucleosome assembly/disassembly might proceed by multiple pathways. For example, ab initio nucleosome reassembly in the wake of DNA or RNA polymerase might proceed via the classical pathway of chaperone-assisted assembly of H3/H4 tetramers (dimer of dimers) followed by chaperone-assisted assembly of H2A/H2B dimers. However, chromatin that is intrinsically dynamic in the absence of a passing polymerase might exchange histones via hexasome and half-nucleosome intermediates. In this way, a hexasome might lose either a single H2A/H2B dimer or a single H3/H4 dimer. Chaperones, chromatin remodelers, DNA/RNA polymerases, histone variants/modifications, and the underlying DNA sequence could influence pathway selection in response to environmental cues.
Having more dynamic nucleosomes, for example near the 5’ ends of genes, might alter accessibility to transcription factor binding sites or transcriptional start sites, which includes creating alternative coding and noncoding transcription start sites. The class of genes that are particularly associated with subnucleosomal structures tend to also have abnormal +1/+2 linker lengths. These genes tend to be SAGA-regulated and TATA-containing and thus subject to a wide range of regulation.
Chromatin remodelers play a predominate role in organizing nucleosomes into uniform arrays. However, the work presented here and elsewhere suggest that the underlying DNA sequence may help maintain a prescribed organizational state at certain genes that differs somewhat from the canonical pattern that remodelers offer (Kaplan et al., 2009; Mavrich et al., 2008; Tillo and Hughes, 2009). These genes tend to have dynamic chromatin, and this fluidity may allow the underlying DNA sequence to reposition certain nucleosomes away from remodeler-imposed spacing uniformity. We observed coordinated histone occupancy between two halves of adjacent nucleosomes that share a common linker. Nucleosome repositioning may alter this relationship resulting in altered nucleosome stability.
Nucleosome splitting at the dyad, as one possible interpretation presented here, might explain the confounding biophysical observation that mechanical disruption of nucleosomes with optical traps produce a large cooperative tension transition that cannot readily be explained by gradual unwrapping of DNA from the histone octamer surface (Brower-Toland et al., 2002). Instead the abrupt transition may reflect the splitting of a nucleosome into two halves.
Nucleosomes are Asymmetric with Respect to Transcription
Our findings suggest that RNA polymerase II first encounters a unique and asymmetric +1 nucleosome (Figure 7). On the proximal face, which encounters polymerase first, these asymmetric features include H3 tail interactions with the edge of the NFR that are coupled to H3K9 acetylation. Further into gene bodies, H3 tails engage linkers, without dependence on H3K9ac, but their purpose remains unclear. Loss of most (but not all) of the H3 tail has surprisingly little impact on nucleosome organization, as does loss of histone HI. Methylation of H3 lysines 4 and 79 (with the former on the H3 tail) has little apparent impact on H3 tail-linker interactions. However, H3K36 methylation substantially delocalizes the crosslinking of H3 tails, such that it loses substantial specificity for linkers. K36 methylation might alter the trajectory of the H3 tail away from linkers, or the H3K36 modification might bind known regulatory factors such as Rpd3S or ISW1 complexes (Carrozza et al., 2005; Smolle et al., 2012), thereby preventing crosslinking.
Back at the +1 nucleosome, selective placement of H2B ubiquitylation on the NFR-proximal face may be important for allowing RNA polymerase II passage as well as promoting nucleosome reassembly in the wake of transcription. The presence of H2A.Z on the distal face of the +1 nucleosome may further facilitate the passage of polymerase, through destabilization of the promoter-proximal half of the nucleosome. This subnucleosomal asymmetry as well as overall array asymmetry may be applicable to multicellular eukaryotes including humans, because histones, nucleosome organization, and modifications are highly conserved across species.
EXPERIMENTAL PROCEDURES
ChlP-exo assay
Saccharomyces strains (listed in Table S1) were grown in rich media and subjected to formaldehyde crosslinking, then processed through the ChlP-exo assay (Rhee and Pugh, 2012a), using either SOLiD or lllumina adaptors. Briefly, cells were disrupted and chromatin pellets were isolated, and then solubilized and fragmented by sonication. Fragmented chromatin was then subject to immunoprecipitation using magnetic bead conjugated antibodies directed against either TAP tagged histones or directly against histone or their modifications. After washing the beads to remove unbound proteins and DNA, and while still on the beads the immunoprecipitates were polished, A-tailed, and ligated to an appropriate sequencing library adaptor. Samples were then subjected to lambda exonuclease digestion, which processively removes nucleotides from 5’ ends of double stranded DNA until blocked by a protein-DNA crosslink induced by formaldehyde treatment. The result is single-stranded DNA, which was then eluted from the magnetic beads, and converted to double-stranded DNA by primer annealing and extension. A second sequencing adaptor was then ligated to exonuclease treated ends, then PCR amplified, gel purified, and sequenced.
MNase-seq
Figure 3B experiments involved MNase-seq, where crosslinked chromatin was treated with 240 units of MNase for either 5 or 25 minutes (low versus high), the subjected to H3 immunoprecipitation, lllumina library construction, gel size selection, and deep sequencing. Libraries were sequenced by an Applied Biosystems 5500×1 SOLiD System, lllumina HiSeq2000, or lllumina NextSeq500. Sequencing tags were mapped to the reference yeast genome obtained from Saccharomyces Genome Database (R55-10-Nov-2006).
Data Analysis
Occupancy levels (tag counts) for various histone positions were typically plotted relative to genomic reference points of MNase-derived nucleosome dyads (listed in Table S2), and summed within the intervals specified in Table S2. Where occupancy correlations between datasets are reported, the Excel function “correl” was used. Tables S3–S7 present the underlying values and calculations used in the figures. Array midpoints represent the coordinate located half-way between the dyad of the +1 nucleosome and the dyad of the terminal genic nucleosome. See Extended Experimental Procedures for detailed experimental procedures, analyses methods, and rationale.
Supplementary Material
Highlights.
Histone H3 tails have regulated interactions with nucleosomal linkers
Histone occupancy is semi-independent across nucleosomal halves
Histone occupancy is semi-interdependent across adjacent nucleosomes
Subnucleosomal histone marks/variants are asymmetrical in the direction of transcription
ACKNOWLEDGEMENTS
We thank Yunfei Li, Rohit Reja, and William Lai for bioinformatic support, Matthew Rossi for sharing unpublished ChIP input DNA data, and members of the Pugh laboratory, the Penn State Center for Eukaryotic Gene Regulation, and Philipp Korber for valuable discussions. We are grateful to Bongsoo Park for computational assistance. National Institutes of Health grant HG004160 supported this work. BFP has a financial interest in Peconic, LLC, which utilizes the ChlP-exo technology implemented in this study and could potentially benefit from the outcomes of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Sequencing data are available at NCBI Sequence Read Archive under accession number SRA059355.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, ten tables, and extended experimental procedures.
REFERENCES
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes & development. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen TA, Smith MM, Le SY, Sternglanz R, Allfrey VG. Nucleosome fractionation by mercury affinity chromatography. Contrasting distribution of transcriptionally active DNA sequences and acetylated histones in nucleosome fractions of wild-type yeast cells and cells expressing a histone H3 gene altered to encode a cysteine 110 residue. J Biol Chem. 1991;266:6489–6498. [PubMed] [Google Scholar]
- Dalai Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Molecular and cellular biology. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Molecular cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Codomo CA, Henikoff S. Reconstitution of hemisomes on budding yeast centromeric DNA. Nucleic Acids Res. 2013;41:5769–5783. doi: 10.1093/nar/gkt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, andHenikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature reviews Genetics. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Molecular cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nature reviews Genetics. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci U S A. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes LP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome research. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Molecular and cellular biology. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. ChlP-exo Method for Identifying Genomic Location of DNA-Binding Proteins with Near-Single-Nucleotide Accuracy. Curr Protoc Mol Biol. 2012a;Chapter 21 doi: 10.1002/0471142727.mb2124s100. Unit21 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012b;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla MS, Syed SH, Goutte-Gattat D, Richard JL, Montel F, Hamiche A, Travers A, Faivre-Moskalenko C, Bednar J, Hayes JJ, et al. The docking domain of histone H2A is required for HI binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 2011;39:2559–2570. doi: 10.1093/nar/gkq1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Iswl and Chdl maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome research. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usachenko SI, Bavykin SG, Gavin IM, Bradbury EM. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc Natl Acad Sci U S A. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, andHenikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Molecular cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Palter K, Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975;6:85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Worcel A, Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976;9:409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome research. 201la;21:875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 201lb;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.