Abstract
Disease relapse remains the major clinical challenge in treating T cell acute lymphoblastic leukemia (T-ALL), particularly those with PTEN loss. We hypothesized that leukemia-initiating cells (LICs) are responsible for T-ALL development and treatment relapse. In this study, we used a genetically engineered mouse model of Pten−/− T-ALL with defined blast and LIC-enriched cell populations to demonstrate that LICs are responsible for therapeutic resistance. Unlike acute and chronic myelogenous leukemia, LICs in T-ALL were actively cycling, were distinct biologically and responded differently to targeted therapies in comparison to their differentiated blast cell progeny. Notably, we found that T-ALL LICs could be eliminated by co-targeting the deregulated pathways driven by phosphoinositide 3-kinase (PI3K) and Myc, which are altered commonly in human T-ALL and are associated with LIC formation. Our findings define critical events that may be targeted to eliminate LICs in T-ALL as a new strategy to treat the most aggressive relapsed forms of this disease.
Keywords: PTEN, Myc, leukemia-initiating cells, targeted therapy, therapeutic resistance
Introduction
T cell acute lymphoblastic leukemia (T-ALL) is a common hematological malignancy that is associated with a significant risk of disease relapse and poor prognosis (1). Activating mutations in NOTCH1 are present in approximately half of T cell leukemia patients (2), and deletion or mutations of the PTEN tumor suppressor gene have been reported in 8-63% of pediatric T-ALL patients (3-6) and are correlated with poor prognosis (7,8). Furthermore, PTEN is frequently inactivated non-genetically in primary T-ALL cells (9) and constitutive activation of the NOTCH signaling pathway down-regulates PTEN expression (6,10), suggesting that PTEN and its controlled PI3K/AKT/mTOR pathway are critical for the etiology of human T-ALL.
Individual cells within tumor masses exhibit notable heterogeneity in their functional and pathological properties. Recent evidence supports the model that the growth of many types of cancers, including leukemia, is sustained by a subset of cells, termed cancer stem cells (CSCs), that are responsible for the initiation and propagation of the disease and show ability to initiate cancer in xenografts or genetically engineered mouse models (11-14). CSCs are characterized by their self-renewal capacity and ability to generate all cell types that comprise the cancer, and are functionally distinct from the bulk of tumor cells that lack the ability to initiate tumor growth. A consequence of this model is that therapies that only eliminate the bulk of the tumor cells without targeting CSCs will result in re-emergence of disease (15).
In hematopoietic malignancies, frequent relapse following conventional chemotherapies suggests that leukemia-initiating cells (LICs) are spared by the treatment, which may be attributed to properties shared with normal hematopoietic stem cells such as maintenance of a quiescent state (16-18). In acute myelogenous leukemia (AML), most LICs isolated from patient samples are quiescent(18) and protected from chemotherapeutic agents (19,20). Maintenance of a quiescent state has also been associated with LICs in chronic myelogenous leukemia (CML) and resistance to tyrosine kinase inhibitors (21,22). However, the identity and cell cycle status of LICs in T-ALL has not been explored to date. In order to understand the cellular and molecular basis of T-ALL relapse and develop more effective therapies, there is a need to characterize T-ALL LICs using functional assays and determine the relationship between LICs and disease relapse in models that mimic human T-ALL pathogenesis.
We previously generated a VE-Cadherin-Cre+;Ptenloxp/loxp;RosafloxedSTOP-LacZ (Pten null) T-ALL model to investigate the molecular mechanisms underlying Pten inactivation-mediated leukemogenesis. In this model, disease is initiated by the conditional deletion of Pten in the fetal liver hematopoietic stem cells and animals develop T-ALL with 100% penetrance in the absence of activating Notch1 mutations (23). In addition to Pten deletion, at least two subsequent key events associated with human leukemogenesis have been identified, namely β-catenin activation and a T cell receptor α–c-Myc (Tcrα/δ-c-Myc) translocation, which lead to the transformation of T progenitor cells to self-renewable LICs enriched in the CD3+c-kitmidLin− subpopulation (23). Therefore the Pten null T-ALL model, with functionally defined LIC-enriched (CD3+c-kitmid) and blast (CD3+c-kit-) populations, provides a valuable platform to assess the effects of small molecule inhibitors on T-ALL development and to investigate the mechanisms underlying resistance to therapies. Here, we demonstrate that leukemia-initiating cells in Pten null T-ALL are actively cycling and targetable using combination therapy directed against the deregulated PI3K pathway and Myc.
Materials and Methods
Mice and transplantation assays
PtenloxP/loxP;VE-Cadherin-Cre+;Rosa26 floxedSTOP-LacZ+ 129/BALB/c mixed background mice were generated as previously described (23). NOD-SCID-IL2Rγ−/− (NSG) mice were obtained from UCLA’s Defined Flora Mouse Facility. 104 Pten null T-ALL BM or splenocytes harvested from primary Pten null T-ALL mice were transplanted by tail vein injection into non-irradiated NSG recipients as previously described (23) to generate cohorts of mice with T-ALL to perform analyses. To evaluate leukemia-initiating cell activity after drug treatment, bone marrow was harvested from 7 or 14 day treated mice and transplanted into secondary NSG recipients at 103−105 cell doses. All animal experiments were approved by the UCLA Animal Research Committee and conducted according to relevant regulatory standards.
Rapamycin, JQ1, and VX-680 in vivo treatment
Mice were administrated the following single agents i.p. daily for 7 or 14 days: 4 mg/kg rapamycin (LC Laboratories), 50 mg/kg JQ1 (Bradner Laboratory, Dana-Farber Cancer Institute), or 75 mg/kg VX-680 (LC Laboratories). For combination treatments, mice were treated daily for 7 or 14 days with 4 mg/kg rapamycin and 50 mg/kg JQ1 or 4 mg/kg rapamycin and 40 mg/kg VX-680. At indicated time points, BM and spleen were harvested for pathological and flow cytometric analysis.
Cell lines
Pten null T-ALL cells were derived from primary thymocytes harvested from a Pten null mouse with T-ALL and maintained in DMEM (Life Technologies) supplemented with 20% fetal bovine serum (FBS) (Omega Scientific), 10 ng/mL Interleukin-2 (IL-2) and 10 ng/mL Interleukin-7 (IL-7) (both Peprotech), 10 mM HEPES, non-essential amino acids (NEAA), sodium pyruvate, glutamine, penicillin and streptomycin (all Life Technologies), and 2-mercaptoethanol (βME) (Sigma). Human T-ALL lines were generously provided by Drs. G. Cheng at UCLA (Jurkat), A. Ferrando at Columbia University (MOLT-3, MOLT-4, and T cell lymphoma line CUTLL1), C. Radu at UCLA (CCRF-CEM), and X. Chen at the University of Maryland (MOLT-16). No additional cell line authentication was performed. Jurkat, CCRF-CEM, MOLT-3, and MOLT-4 cells were maintained in RPMI (Life Technologies) supplemented with 10% FBS, glutamine, penicillin and streptomycin. MOLT-16 and CUTLL1 cells were maintained as the other human cell lines but supplemented with 20% FBS.
Proliferation and viability assays
Cell viability was assessed by measuring the conversion of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Life Technologies) to formazan or by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Cells were seeded 24 hours before drug addition. After 48 hour incubation with drug or appropriate vehicle (DMSO or ethanol), MTT or CellTiter 96 AQueous One Solution Assay reagents were added as specified in reagent protocols and absorbance was read using a BioRad Benchmark microplate spectrophotometer.
Statistical analysis
The difference between experimental groups was compared using Student's t test analysis. Survival in animal experiments was represented with Kaplan-Meier plots (Graphpad Prism). IC50 was calculated using Graphpad Prism.
Results
Leukemia-Initiating Cells are Responsible for Therapeutic Resistance in Pten null T-ALL
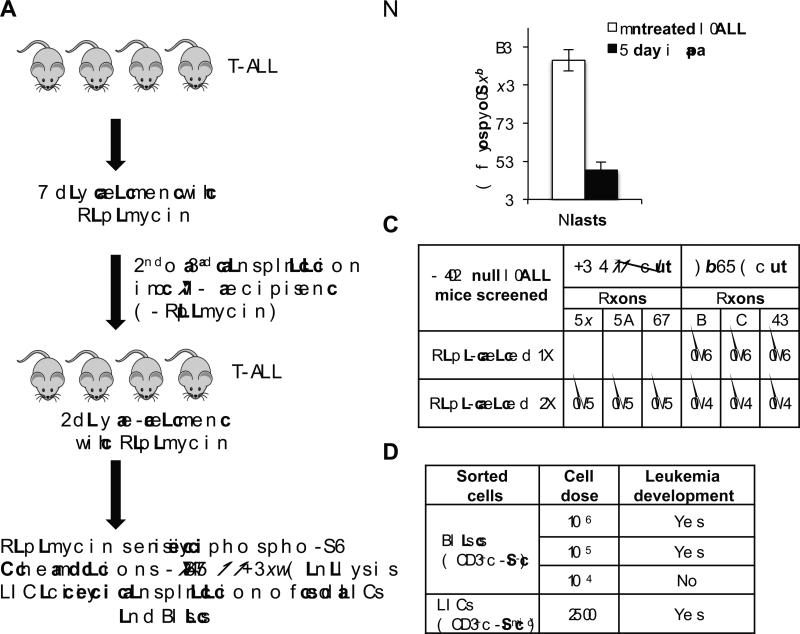
We previously found that rapamycin, a specific TORC1 inhibitor, abrogated LIC formation by preventing the Tcrα/δ-c-Myc translocation, a critical secondary event for Pten null T-ALL development in this model (24). However, when treatment began at the acute T-ALL stage, rapamycin extended survival but all Pten null animals eventually succumbed to T-ALL (24). To understand the cellular and molecular mechanisms underlying relapse, we treated Pten null TALL mice with rapamycin for 7 days, harvested bone marrow cells, and transplanted these cells into secondary and tertiary NOD-SCID-IL2Rγ−/− (NSG) recipients in the absence of drug. We then retreated the transplant recipients with rapamycin to test whether rapamycin-treated T-ALL remained sensitive to the drug or acquired resistance mutations (Fig. 1A). Rapamycin significantly reduced the percentage of phospho-S6+ cells, a surrogate marker for mTOR activity, in secondary and tertiary recipients from greater than 70% to 15% after 2 days of retreatment, suggesting that the majority of T-ALL cells remained sensitive to rapamycin (Fig. 1B). In addition, we did not detect “hotspot” mutations in Notch1 or Fbxw7, common alterations found in human T-ALL, after treatment (Fig. 1C). Importantly only LICs, not blasts, harvested from tertiary re-treated animals could efficiently initiate T-ALL upon transplantation (Fig. 1D). Altogether these data suggest that resistance to rapamycin is due to failure to eliminate LICs and not mutations in the mTOR or NOTCH pathways.
Figure 1.
Leukemia-initiating cells are responsible for therapeutic resistance in Pten null T-ALL. A, schematic diagram of strategy to investigate mechanisms underlying resistance to rapamycin. Pten null mice with T-ALL were treated for 7 days with rapamycin, and then bone marrow was harvested and transplanted into secondary and tertiary recipients in the absence of rapamycin. These recipients rapidly developed T-ALL and were re-treated for 2 days with rapamycin. After re-treatment, hematopoietic cells were harvested from Pten null T-ALL animals and analyzed for levels of phospho-S6, mutations in Notch1 and Fbxw7, and leukemia-initiating cell activity in blast and LIC compartments. B, quantitation of phospho-S6+ cells in Pten null TALL blast cells in bone marrow harvested from mice after 2 days of rapamycin treatment in vivo (4 mg/kg daily) using intracellular flow cytometric analysis. C, summary of mutational analysis of Notch1 exons 26, 27 and 34 and Fbxw7 exons 8, 9, and 10 using genomic DNA from splenocytes harvested from T-ALL mice after single or double rapamycin treatment. Ratios indicate number of T-ALL mice with mutations (Mut) over total number T-ALL mice screened. D, summary of transplantation assay with sorted fractions. Blast and LIC sorted populations were transplanted into NSG recipients at indicated doses and leukemia development was evaluated in recipients. Data in B are represented as mean ± SEM.
Co-targeting Deregulated PI3K and Myc Efficiently Eliminates Pten null LICs
The deregulated c-Myc oncogene, either via Tcrα/δ-c-Myc translocation or through its overexpression, is essential for LIC formation and leukemia development in the Pten null T-ALL model (24) and associated with LICs in NOTCH1-induced T-ALL (25). Therefore we hypothesized that combination therapy that co-targets the PI3K pathway and deregulated c-Myc might eliminate LIC and T-ALL. The bromodomain and extraterminal (BET) subfamily of bromodomain proteins (BRD2, BRD3, and BRD4) have emerged as potent regulators of MYC expression in various tumor types, and the bromodomain inhibitor JQ1 has been demonstrated to down-regulate MYC transcription, reduce expression of MYC target genes, and show anti-leukemic effects in in vitro and in vivo models (26-30).
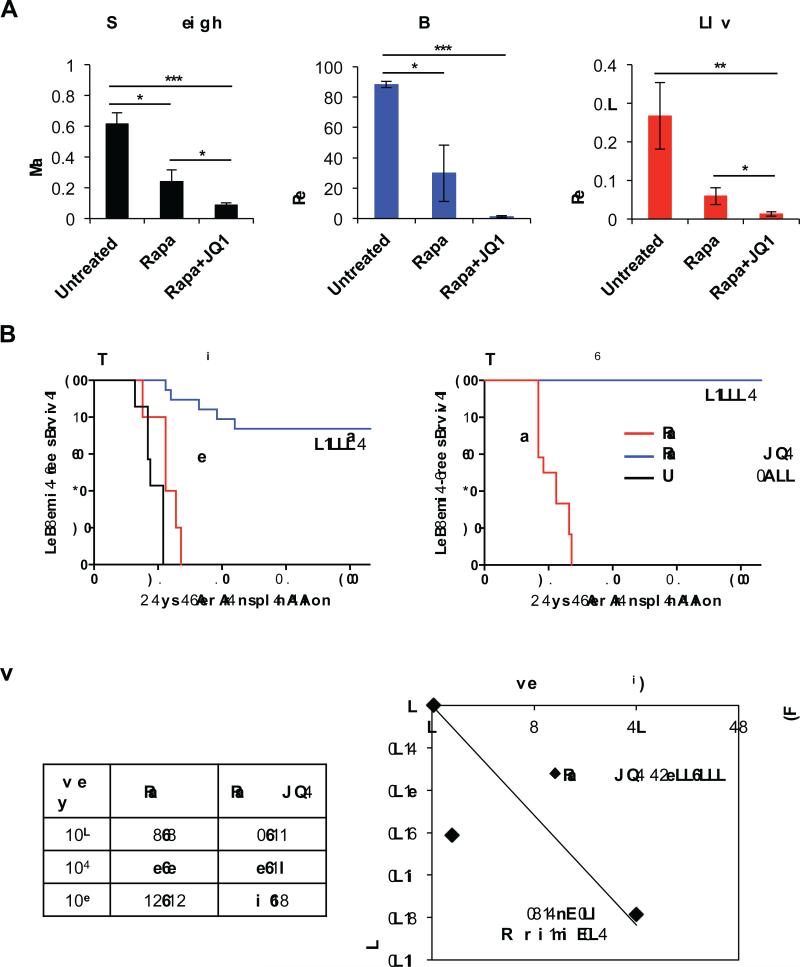
To test whether JQ1 can be used in combination with rapamycin for therapeutic utility, we transplanted leukemic cells from primary Pten null T-ALL into NSG recipients to generate a cohort of Pten null T-ALL animals to perform parallel analyses of treatment with agents, alone or in combination (Supplementary Fig. S1A). To determine the specific responses of blasts and LICs, we quantified the percentages of Lin−;CD3+;c-kit− blast and Lin−;CD3+;c-kitmid LIC-enriched subpopulations according to our previous studies (23,24) in the untreated and treated animals. Phospho-S6 levels in blasts were significantly decreased after 7 days of rapamycin treatment in vivo, demonstrating inhibition of mTOR by rapamycin in this compartment (Supplementary Fig. S1B). JQ1 significantly enhanced the effects of rapamycin in reducing splenomegaly and almost completely eliminated leukemic blasts after 7 days of daily in vivo treatment (Fig. 2A; left two panels). Importantly, the percentage of LICs in the bone marrow was markedly reduced with the combination treatment (Fig. 2A; right panel). JQ1 single agent treatment, however, did not effectively eliminate blast cells in vivo, as JQ1 single treated animals appeared weak during treatment and showed splenomegaly upon sacrifice (data not shown). To assess the functional significance of the reduction in LICs with combination treatment, we performed limiting dilution transplantation analyses using 103−105 bone marrow cells harvested from 7 day treated animals (Supplementary Fig. S1A). While all animals receiving a 103 cell dose of rapamycin treated cells succumbed to T-ALL by approximately 30 days, none of the animals receiving the same dose of dual treated cells developed any sign of illness over more than 100 days (Fig. 2B) and no leukemia cells could be detected in the euthanized animals using sensitive genomic PCR analysis (Supplementary Fig. S1C). Our regression analysis showed that combination treatment reduced the frequency of the LIC to ~1/200,000 (Fig. 2C), indicating that eliminating LIC number and activity are directly associated with this remarkable therapeutic response.
Figure 2.
Rapamycin and JQ1 combination treatment reduces Pten null T-ALL disease burden and LICs. A, measurement of splenic mass and quantitation of blast and LIC populations in bone marrow of Pten null T-ALL mice by flow cytometric analysis after 7 day treatment in vivo. B, Kaplan-Meier survival curve representing morbidity of NSG recipients transplanted with 104 and 103 cell doses of treated bone marrow (BM) cells. C, fraction of secondary recipients that developed leukemia when transplanted with limiting dilutions of rapamycin or rapamycin and JQ1 combination treated donor cells, and logarithmic plot and limiting dilution analysis to calculate LIC frequency after rapamycin and JQ1 combination treatment. LIC frequency after 7 day rapamycin and JQ1 combination treatment is ~1/200,000. Data in A are represented as mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
T-ALL May be Stratified and Treated According to its Molecular and Genetic Defects
The robust response of our Pten null T-ALL model to combination treatment of rapamycin and JQ1 prompted us to investigate the molecular mechanisms underlying T-ALL elimination and to test whether similar combination treatment can be applied to human T-ALL. For this, we generated a Pten null T-ALL in vitro culture system to circumvent the limitation of the in vivo model in conducting cellular, biochemical, and pathway analyses. By optimizing culture conditions that allow for the survival and expansion of primary leukemia cells harvested from Pten null T-ALL mice, we established Pten null T-ALL cell lines that require critical T cell cytokines IL-2 and IL-7 for growth, are CD3+;TCRβ+;LacZ+, express high levels of c-Myc and retain leukemia-initiating activity in vivo upon transplantation (Schubbert et al. manuscript in preparation).
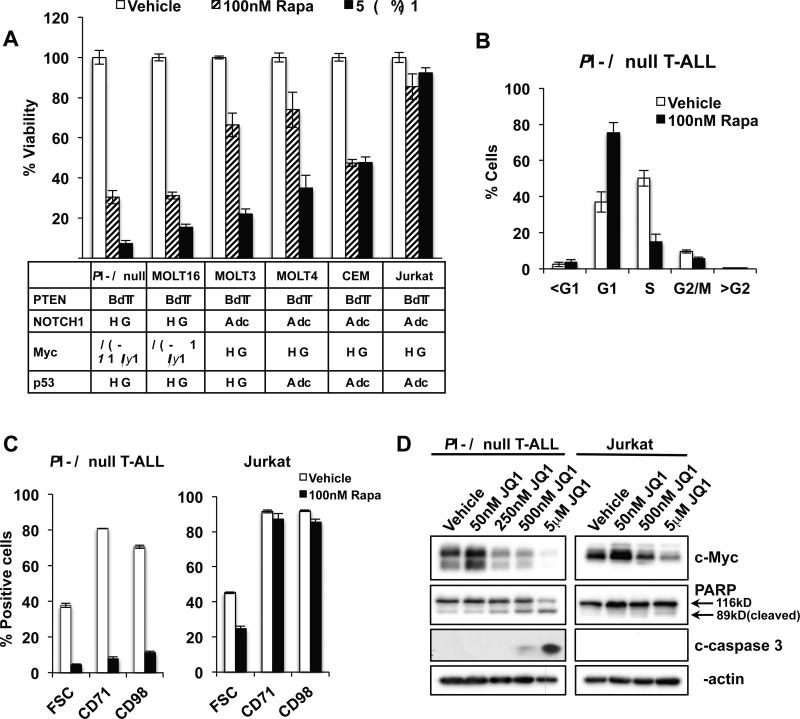
To assess the therapeutic utility of rapamycin and JQ1 in human T-ALLs, we first investigated the effect of these drugs on the growth and viability of our newly developed Pten null T-ALL line in comparison to several human PTEN null T-ALL lines. Interestingly, we found that MOLT-16 cells, which harbor similar genetic alterations (PTEN null with a Tcrα-c-Myc translocation, lacking p53 mutations and the activated, intracellular form of NOTCH1) (31) as our Pten null TALL model, are very sensitive to both rapamycin and JQ1 (Fig. 3A and Supplementary Fig. S2A and S3A). Other human PTEN null T-ALL lines, including those with NOTCH1 and p53 mutations, are less sensitive to rapamycin and JQ1, with Jurkat cells being most resistant (Fig. 3A and Supplementary Fig. S2A and S3A). Further analysis of Pten null T-ALL cells revealed that rapamycin caused accumulation of cells in G1 and a rapid and robust decrease in cell size and down-regulation of the surface expression of the transferrin receptor CD71 and amino acid transporter CD98 (Fig. 3B, 3C, and Supplementary Fig. S2B). mTOR activity regulates nutrient transporter levels in Pten null T-ALL (24) and the control between mTOR activity and nutrient sensing and uptake may play a critical role for cell survival, proliferation, and differentiation (32). Jurkat cells were less sensitive to reduction in cell size by rapamycin and levels of nutrient receptor expression remained high after treatment (Fig. 3C). Similarly, JQ1 significantly down-regulated c-Myc protein levels and induced apoptosis-associated cleavage of PARP and caspase 3 in Pten null T-ALL while only moderately reduced c-Myc levels were observed in Jurkat cells in the absence of apoptosis (Fig. 3D and Supplementary Fig. S3B). Interestingly, MOLT-16 cells were markedly more sensitive to JQ1-mediated induction of apoptosis compared to other human T-ALL lines tested (Supplementary Fig. S3B and S3C). The induction of apoptosis in Pten null T-ALL and MOLT-16 in response to JQ1 occurred following marked G1 arrest (Supplementary Fig. S3D) and was not associated with upregulation of p53 (Supplementary Fig. S3C).
Figure 3.
The effects of rapamycin and JQ1 on T-ALL correlate with their associated genetic alterations. A, viability of T-ALL lines after rapamycin or JQ1 treatment for 48 hours measured by an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Table below indicates mutational status of T-ALL lines. B, quantitation of cell cycle stage using propidium iodide and flow cytometric analysis after 24 hour treatment with 100 nM rapamycin in Pten null T-ALL. C, cell size and surface expression of CD71 and CD98 as measured by flow cytometry in Pten null T-ALL and Jurkat cells in response to 100 nM rapamycin at 24 hours. D, immunoblot for c-Myc, PARP, and cleaved caspase 3 (c-caspase 3) in Pten null T-ALL and Jurkat cells after 24 hour JQ1 treatment at indicated doses. Data in A through C are represented as mean ± SEM.
We investigated the efficacy of rapamycin and JQ1 treatment in 4 human pediatric T-ALL samples using in vitro growth and viability assays. Interestingly all 4 human T-ALL samples showed sensitivity to rapamycin but were less sensitive to JQ1 (Supplementary Fig. S4A and S4B). No PTEN mutations were found in any of the human samples (Supplementary Table 1), but phospho-AKT levels were similar or greater than PTEN null Jurkat cells and at least 2 fold greater than CUTLL1 cells, a PTEN-positive human T cell lymphoma line (33) (Supplementary Fig. S5A and S5B). The sensitivity of the human T-ALL samples to rapamycin is consistent with the notion that PI3K/AKT pathway status, which may result from both genetic and non-genetic regulatory mechanisms, may provide a biomarker for predicting patient response to inhibitors of PI3K or downstream targets (9,34). Levels of c-Myc in the primary human T-ALL samples were low in comparison to Jurkat cells (Supplementary Fig. S5A and S5B), which may in part explain the weak response to JQ1.
LICs and Leukemic Blasts are Biologically Distinct and Respond Differently to Targeted Therapies
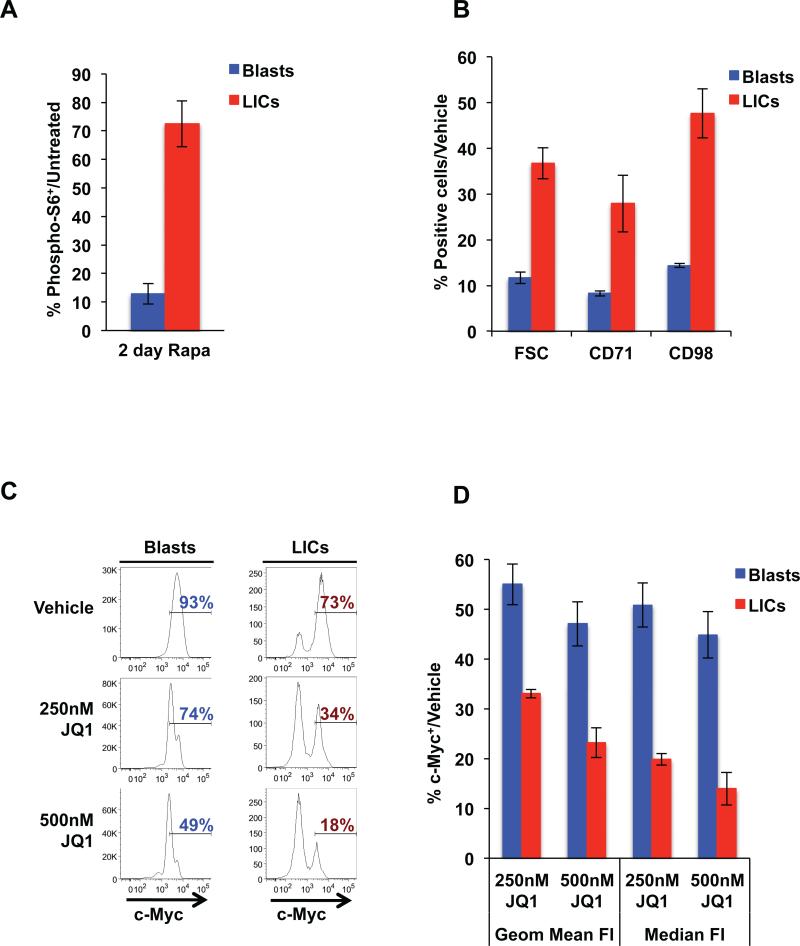
The remarkable responses of blast and LIC populations to rapamycin and JQ1 combination treatment in Pten null T-ALL in vivo prompted us to examine the specific effects of these agents in these distinct compartments. We hypothesized that the failure of rapamycin as a single agent to eliminate Pten null LICs in vivo may be due to the differential cellular and biochemical responses of LICs and blasts to rapamycin. Indeed, we found that a 2-day treatment with rapamycin in vivo greatly reduced levels of phospho-S6 in the blast cell compartment while levels of phospho-S6 remained high in LICs (Fig. 4A). Furthermore, we found that Pten null LICs are markedly less sensitive to the effects of rapamycin on reducing cell size and nutrient receptor levels in comparison to blast cells (Fig. 4B). These data suggest that failure to inhibit mTOR and effectively down-regulate nutrient receptors in the LIC compartment may underlie the mechanism of rapamycin resistance.
Figure 4.
Pten null LICs and leukemic blasts respond differently to targeted therapies. A, quantitation of phospho-S6+ cells in blast and LIC compartments by flow cytometric analysis of bone marrow harvested after 2 day rapamycin treatment in vivo. The percentage of phospho-S6+ in blasts and LICs is shown relative to untreated levels in respective compartments. B, quantitation of cell size and surface expression of CD71 and CD98 in blast and LIC compartments as measured by flow cytometry in Pten null T-ALL in response to 100 nM rapamycin at 24 hours. C, representative fluorescence-activated cell sorting (FACS) histogram plots showing the frequency of c-Myc+ cells in blast and LIC populations in Pten null T-ALL after 24 hour JQ1 treatment in vitro. D, quantitation of geometric mean and median fluorescence intensities (FI) of c-Myc using flow cytometric analysis after 24 hour JQ1 treatment in vitro. Data in A, B, and D are represented as mean ± SEM.
Because Myc is associated with LIC activity in both our model and NOTCH1-driven T-ALL, we evaluated the effect of JQ1 on Myc levels in the blasts and LIC-enriched compartments. While JQ1 reduced levels of c-Myc in both blast and LIC populations, we found that LICs are significantly more sensitive to JQ1-mediated down-regulation of c-Myc than their blast progenies (Fig. 4C; 18% vs. 49% c-Myc+ in LICs and blasts, respectively, after 500 nM JQ1 treatment). Quantitative analysis revealed that LICs show 2-3 fold greater reduction in c-Myc levels after JQ1 treatment compared to blast cells (Fig. 4D). The enhanced sensitivity of Pten null LICs to c-Myc down-regulation by JQ1 may be particularly valuable to efficiently target aberrant c-Myc levels in Pten null LICs and eliminate these cells. Taken together, these data demonstrate that LIC and blast subpopulations are not only functionally distinct but respond differently to targeted treatments.
LICs in Pten null T-ALL Are Actively Cycling
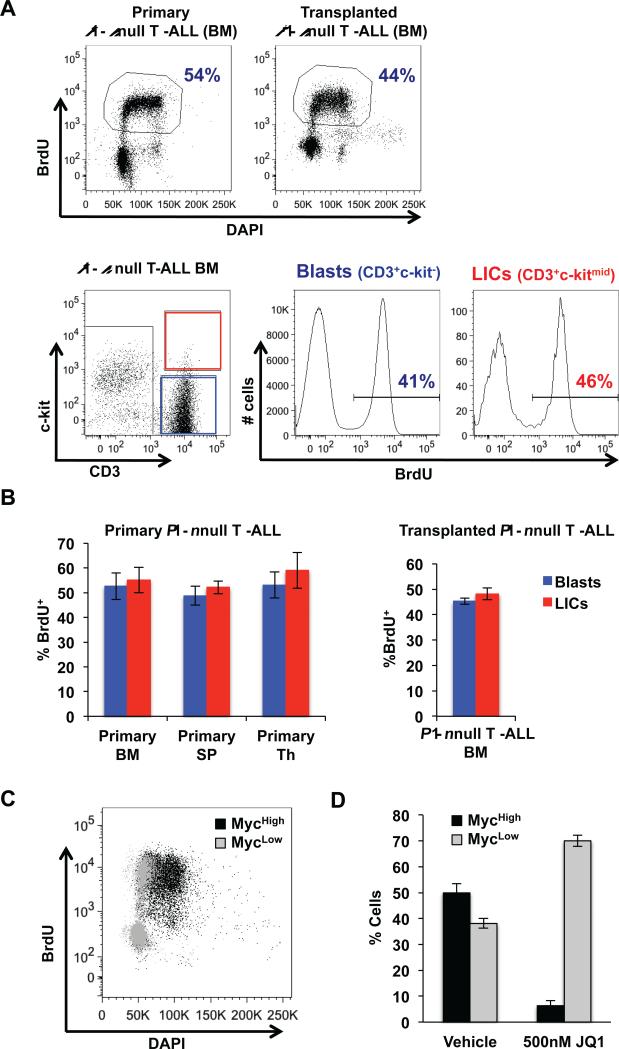
The proto-oncogene c-Myc is a key regulator of cell growth and cell cycle progression (35). Since Pten null LICs over-express c-Myc, we evaluated the cell cycle status of LICs. Intriguingly, when we measured BrdU incorporation in the bone marrow of primary and transplanted Pten null T-ALL in vivo, we found that a large portion of LICs were actively cycling (~50% BrdU+ after 1 hour pulse labeling) similar to blast cells (Fig. 5A and quantified in 5B). This observation contrasts with studies of LICs in other hematopoietic malignancies, including AML and CML, in which the LICs reside mostly in a quiescent state (13,19,21), however is similar to the increased levels of proliferation and self-renewal reported for NrasG12D mutant hematopoietic stem cells (36,37).
Figure 5.
Leukemia-initiating cells in Pten null T-ALL are actively cycling. A, percentage BrdU incorporation using flow cytometric analysis in primary and transplanted Pten null T-ALL mice and representative histogram plots of blast and LIC cell fractions of bone marrow (BM) harvested from transplanted Pten null T-ALL mice after 1 hour labeling in vivo. B, quantitation of BrdU incorporation by flow cytometric analysis in BM, spleen (SP), and thymus (Th) harvested from primary Pten null leukemic mice (left) and BM harvested from transplanted Pten null T-ALL mice (right) after 1 hour labeling in vivo. C, representative FACS plot showing overlay of BrdU incorporation in population with high c-Myc levels (Mychigh) and population with low c-Myc levels (Myclow) in Pten null T-ALL after 2 hour labeling in vitro. D, quantitation of Mychigh and Myclow populations in Pten null T-ALL after 24 hour JQ1 treatment in vitro using flow cytometric analysis. Data in B and D are represented as mean ± SEM.
Our cell cycle analysis suggests that deregulated Myc expression in T-ALL LICs may promote their cell cycle entry and progression, which distinguishes T-ALL LICs from AML and CML LICs. Consistent with this notion, we found that 86% of cells with high c-Myc levels (Mychigh) are also BrdU+ while only 33% of cells with low c-Myc levels (Myclow) cells are BrdU+ (Fig. 5C). Importantly, JQ1 treatment could effectively eliminate the Mychigh population (Fig. 5D), supporting the idea that high c-Myc levels in the LIC subpopulation may drive LIC entry into the cell cycle. These unique characteristics of Pten null T-ALL LICs may render them sensitive to cell cycle inhibitors.
Pten null T-ALL LICs Can Be Effectively Eliminated by Aurora Kinase Inhibition in Combination with Rapamycin
Myc upregulation of Aurora kinases, which have essential roles in many aspects of cell division (38,39), is reported to be essential for the maintenance of Myc-driven malignancies (40). Interestingly, we found that JQ1-induced c-Myc down-regulation is associated with a significant reduction of Aurora B levels (Fig. 6A), suggesting that c-Myc regulates Aurora B in Pten null TALL. This node may be critical for leukemia maintenance and further implicates potential utility of cell cycle inhibitors to eradicate LICs.
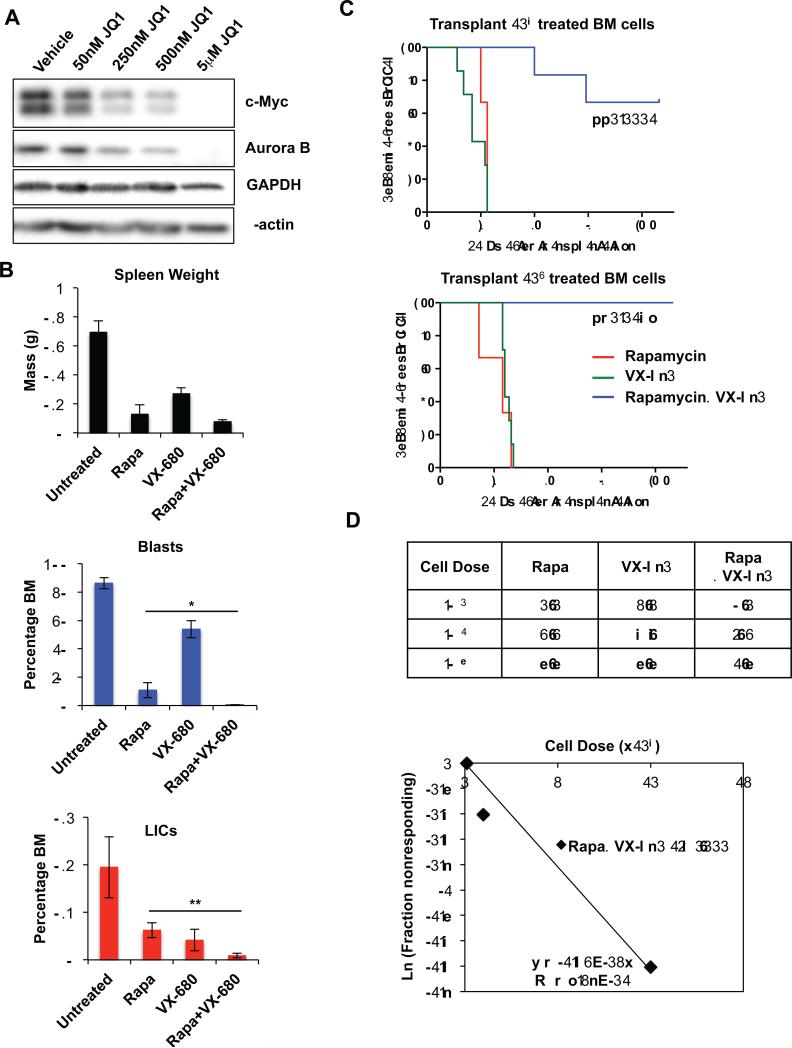
Figure 6.
Potent effects of combination therapy using rapamycin and VX-680 to eliminate blast and LIC populations in Pten null T-ALL. A, immunoblot for c-Myc and Aurora B in Pten null TALL after 24 hour treatment with JQ1 at indicated doses. B, measurement of splenic mass and quantitation of blast and LIC populations in bone marrow of Pten null T-ALL by flow cytometric analysis after 7 day treatment in vivo. C, Kaplan-Meier survival curve representing morbidity of NSG recipients transplanted with 104 and 103 cell doses of 14 day treated bone marrow (BM) cells. D, fraction of secondary recipients that developed leukemia when transplanted with limiting dilutions of rapamycin, VX-680, or rapamycin and VX-680 combination treated donor cells, and logarithmic plot and limiting dilution analysis to calculate LIC frequency after rapamycin and VX-680 combination treatment (~1/60,000). Data in B are represented as mean ± SEM (*p<0.05; **p<0.01).
In support of this hypothesis, the Aurora kinase inhibitor VX-680 has been reported to show synthetic lethality with Myc overexpression (41,42). Given the effects of JQ1 in reducing c-Myc and Aurora B levels in Pten null T-ALL, we were prompted to evaluate the effects of VX-680 in combination with rapamycin on Pten null T-ALL in vivo to further delineate the effects of anti-Myc therapy to eliminate Pten null LICs. We treated a cohort of transplanted Pten null T-ALL mice with VX-680 and rapamycin, alone and in combination, and found that combination treatment caused nearly complete elimination of leukemia burden and a robust reduction in the LIC population (Fig. 6B). Functional studies for LIC activity using transplantation of treated bone marrow into secondary NSG recipients showed again that dual treatment significantly reduced LIC activity in bone marrow compared to single agent alone (Fig. 6C and 6D). Limiting dilution analysis estimated that combination treatment reduced the frequency of the LIC to ~1/60,000, similar to but less potent than the JQ1 and rapamycin combination (Fig. 6C and 6D). These data suggest that an essential role of the deregulated Myc pathway in Pten null T-ALL LICs is regulating cell cycle progression and such a property can be targeted directly by JQ1 or indirectly with cell cycle inhibitors.
VX-680 causes Mitotic Block, Polyploidy, and Apoptosis in Pten null T-ALL
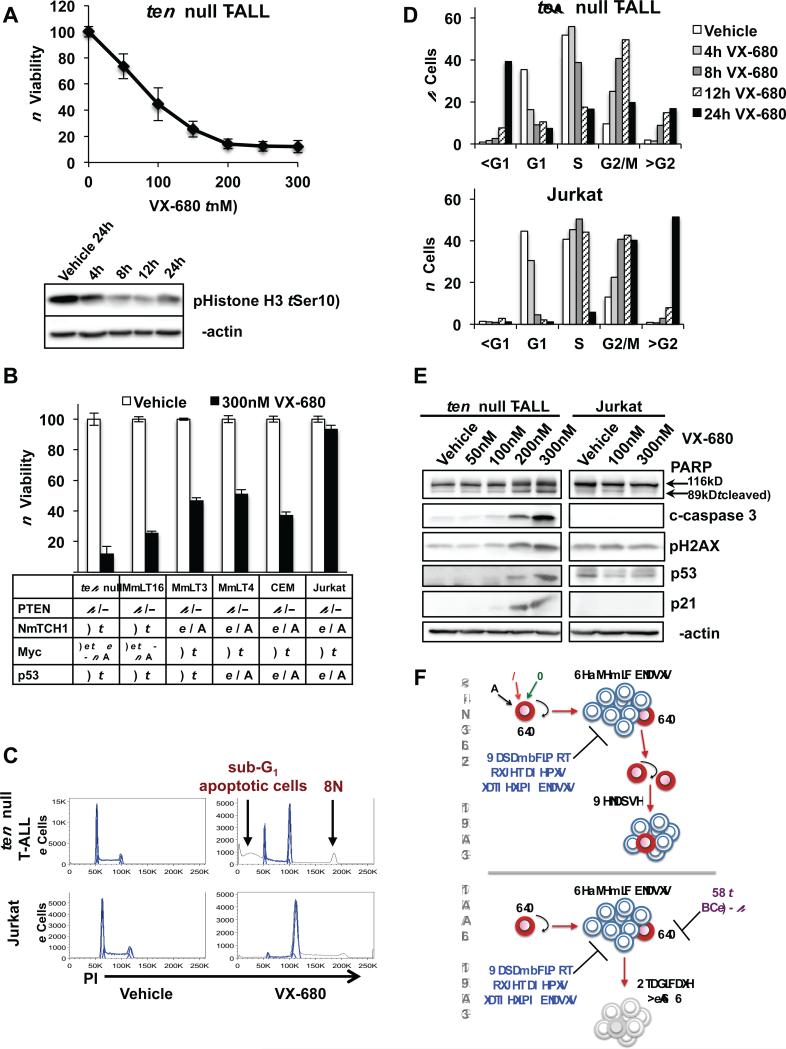
We used our in vitro model to evaluate the cellular and biochemical effects of VX-680 on Pten null T-ALL and determine potential mechanisms underlying T-ALL elimination. Pten null T-ALL cells were highly sensitive to VX-680 (IC50 ~83 nM) and treatment decreased levels of phosphorylation of Histone H3 at Ser10, a surrogate for Aurora B kinase activity, within 8 hours (Fig. 7A). We further tested the sensitivity of several human PTEN null T-ALL lines to VX-680. Consistent with the notion that Aurora B is a major target of c-Myc function, sensitivity to VX-680 mirrored that of JQ1 with MOLT-16 cells, harboring similar genetic alterations as Pten null TALL, showing greatest sensitivity, and Jurkat cells, harboring NOTCH1 (43) and p53 mutations, most resistant to treatment (Fig. 7B). Further analysis demonstrated that VX-680 treatment caused a block in mitosis and induced apoptosis in Pten null T-ALL as indicated by accumulation of cells in G2/M, formation of a polyploid population (8N), and a pronounced increase in the sub-G1 population with induction of cleaved PARP and caspase 3 (Fig. 7C-E). We also detected increased levels of phosphorylated H2AX and induction of p53 and p21 in Pten null T-ALL, suggesting the p53 pathway may be involved in the DNA damage response and subsequent apoptosis (Fig. 7E). Although VX-680 causes mitotic block and polyploidy in Jurkat cells, it does not induce apoptosis as illustrated by the lack of a sub-G1 population and absence of cleaved PARP and caspase 3 (Fig. 7C-E), possibly due to the presence of a p53 mutation. These results suggest that therapeutic approaches using VX-680 may be most effective in human T-ALL with PTEN loss, functional p53, and genetic or molecularly deregulated Myc.
Figure 7.
VX-680 induces mitotic block and causes polyploidy and apoptosis in Pten null TALL. A, viability of Pten null T-ALL measured using an MTT assay after 48 hour VX-680 treatment and immunoblot of phosphorylation of Histone H3 at Ser-10, a surrogate assay for the activity of Aurora kinase B, in Pten null T-ALL after 300 nM VX-680 treatment over 4 to 24 hour (h) time course. B, viability of T-ALL lines measured using an MTT assay after 48 hour VX-680 treatment. Table below indicates mutational status of T-ALL lines. C, representative histogram plots of cell cycle profiles of Pten null T-ALL and Jurkat cells after 24 hour treatment with 300nM VX-680. D, quantitation of cell cycle effects in Pten null T-ALL and Jurkat cells using flow cytometric analysis after 300 nM VX-680 treatment at indicated time points. E, immunoblot of PARP, cleaved caspase 3 (c-caspase 3), phosphorylation of H2AX, p53 and p21 in Pten null T ALL and Jurkat cells after 24 hour treatment with VX-680. F, model of combination drug therapy as an approach to target LICs and blasts and thereby eradicate T-ALL. Data in A and B are represented as mean ± SEM.
Discussion
Our work provides insight into the molecular and cellular mechanisms underlying relapse in Pten null T-ALL and uncovers novel biological and functional properties of LICs that can be exploited for therapeutic intervention. We demonstrate that Pten null T-ALL LICs 1) are active in the cell cycle, which is most likely driven by the Tcrα/δ-c-Myc translocation induced c-Myc overexpression; 2) are responsible for therapeutic resistance; 3) are biologically distinct and respond differently to targeted therapies in comparison to their differentiated progeny; and 4) can be eliminated by co-targeting deregulated PI3K and Myc pathways, two common alterations found in human T-ALL that are associated with the formation of LICs (Figure 7F). Our study further demonstrates that LICs are addicted to the molecular and genetic events associated with their formation and co-targeting these events may provide a potent strategy to reduce leukemia burden and overcome LIC-mediated therapeutic resistance.
Clinical data suggests that conventional chemotherapies may reduce the “bulk” of the leukemia (blast cells), but fail to eradicate the LICs, leading to relapse (15). In this study, our Pten null TALL model with functionally defined and distinguishable blast and LIC-enriched populations provides a valuable platform to develop novel targeted molecular therapies in which efficacy can be evaluated in both LICs and the bulk of the disease. Our biochemical and molecular analysis of drug effects on the blast and LIC-enriched compartments revealed that LICs and blasts show remarkably distinct responses to targeted therapies. While the bulk of Pten null T-ALL shows rapid and robust inhibition of the mTOR pathway by rapamycin, even after multiple rounds of therapy, rapamycin fails to inhibit mTOR and down-regulate nutrient receptor expression in the LIC compartment, which retains LIC activity after treatment. These data suggest that relapse of Pten null T-ALL in response to rapamycin is caused by the failure to eliminate LICs.
Intriguingly, we found that the Pten null LIC-enriched population is markedly more sensitive to down-regulation of c-Myc by JQ1 than blast cells. Although more studies are needed to determine the mechanisms underlying this differential response, we hypothesize that bromodomains or c-Myc may be differentially regulated in LIC and blast populations. AKT mediated stabilization of Myc is unlikely a mechanism to account for the reduced down-regulation of Myc by JQ1 in blasts since this population shows lower levels of phospho-AKT following JQ1 treatment in comparison to LICs (Supplementary Fig. S6A and S6B). Interestingly, Myc expression and protein levels are highest in the DN3-4 stages of T-cell development, corresponding to the stage of Pten null LIC formation, and are greater than levels in DP and SP populations, corresponding to the immunophenotype of the relatively more mature blast population (https://gexc.stanford.edu/ and Supplementary Fig. S7A and S7B) (24).
While many studies have provided evidence that LICs in AML and CML reside in a quiescent state (13,19,21), to our knowledge the cell cycle status of LICs in T-ALL has thus far never been characterized. We demonstrated that a large portion of LICs in T-ALL are actively cycling and hypothesized that deregulated Myc is responsible for promoting cell cycle entry and progression of T-ALL LICs. We found JQ1 preferentially targets cells that have high Myc levels and are actively cycling. Importantly, the active cycling nature of T-ALL LICs suggests that cell cycle inhibitors may be useful to target T-ALL LICs and offer therapeutic benefits to eliminate T-ALL. Indeed, rapamycin in combination with the Aurora kinase inhibitor VX-680 effectively reduced Pten null LICs and T-ALL in vivo. While we attribute the active cycling of Pten null LICs to deregulated Myc expression, it is possible that activation of other signaling pathways may also contribute to cycling. Notably, NrasG12D mutant hematopoietic stem cells have been recently reported to show increased proliferation and self-renewal (37).
Our study provides support that LICs are responsible for relapse and uncovers a role for anti-MYC directed therapies to target LICs, similar to a recent study (44). We also reveal a role for rapamycin as an effective agent to rapidly reduce the bulk of the leukemia, the blast population. The most effective anticancer regimens will likely involve combination approaches using therapies that kill both bulk tumor cells and disease-initiating cells (15). Our novel combination treatment strategy using agents to target the PI3K pathway and deregulated Myc is consistent with this paradigm to provide anticancer therapies that quickly reduce disease burden and eliminate the root of the cancer.
Loss of PTEN and deregulation of Myc are common in human T-ALL and associated with the formation of LICs (3-6,23,25). The remarkable efficacy of co-targeting the PI3K pathway and Myc to eliminate Pten null/Tcrα/δ-c-Myc translocation-induced LICs and blasts, and prevent leukemic transmission suggests that LICs are addicted to molecular events critical for their formation and can be effectively eliminated by targeting these alterations. The sensitivity of our human pediatric T-ALL samples with activated AKT to rapamycin supports this hypothesis and examination of primary human T-ALL samples with c-Myc translocation, similar to the MOLT-16 patient derived cell line, will further our understanding of patient response. Overall our study strongly suggests that human T-ALL needs to be stratified by the molecular genetic events associated with LIC formation and treated by co-targeting pathways of addiction.
Supplementary Material
Acknowledgments
We thank Drs. Kevin Shannon and Monique Dail for helpful comments and suggestions and are grateful to colleagues in our laboratories for helpful discussion and comments. We thank Ying Wang, Christopher Wei, Lynda Ma, and Thu Giao for assistance with experiments and assays. We thank the UCSF Hematopoietic Tissue Cell Bank and the Children's Oncology Group for providing primary human T-ALL specimens and thank Dr. Yong-Mi Kim and her laboratory for technical assistance with human samples.
Grant Support
SS was supported by a fellowship from the Leukemia and Lymphoma Society and National Cancer Institute training award T32-CA009120-36. AC was supported by the CSUN-UCLA Bridges to Stem Cell Research Program TB1-01183. This work was supported by a grant from the William Lawrence & Blanche Hughes Foundation to HW.
Footnotes
Conflict-of-interest disclosure: Drug-like derivatives of the JQ1 chemical probe have been out-licensed by the Dana-Farber Cancer Institute to Tensha Therapeutics, which was founded by J.B. and the Dana-Farber for clinical development. The remaining authors declare no competing financial interests.
Supplementary Data
Supplementary data includes Supplementary materials and methods, seven Supplementary figures, and one Supplementary Table.
References
- 1.Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov. 2007;6(2):149–65. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 2.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114(3):647–50. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson Gedman A, Chen Q, Kugel Desmoulin S, Ge Y, LaFiura K, Haska CL, et al. The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and downstream signaling in pediatric T-cell acute lymphoblastic leukemia: a report from the Children's Oncology Group. Leukemia. 2009;23(8):1417–25. doi: 10.1038/leu.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–71. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jotta PY, Ganazza MA, Silva A, Viana MB, da Silva MJ, Zambaldi LJ, et al. Negative prognostic impact of PTEN mutation in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2010;24(1):239–42. doi: 10.1038/leu.2009.209. [DOI] [PubMed] [Google Scholar]
- 8.Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengline E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol. 2013;31(34):4333–42. doi: 10.1200/JCO.2012.48.5292. [DOI] [PubMed] [Google Scholar]
- 9.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762–74. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva A, Jotta PY, Silveira AB, Ribeiro D, Brandalise SR, Yunes JA, et al. Regulation of PTEN by CK2 and Notch1 in primary T-cell acute lymphoblastic leukemia: rationale for combined use of CK2- and gamma-secretase inhibitors. Haematologica. 2010;95(4):674–8. doi: 10.3324/haematol.2009.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 12.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27(1):44–6. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 13.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 14.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Wang JC. Evaluating therapeutic efficacy against cancer stem cells: new challenges posed by a new paradigm. Cell Stem Cell. 2007;1(5):497–501. doi: 10.1016/j.stem.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 17.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355(12):1253–61. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 18.Guan Y, Gerhard B, Hogge DE. Detection, isolation, and stimulation of quiescent primitive leukemic progenitor cells from patients with acute myeloid leukemia (AML). Blood. 2003;101(8):3142–9. doi: 10.1182/blood-2002-10-3062. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–21. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Uchida N, Tanaka S, Suzuki N, Tomizawa-Murasawa M, Sone A, et al. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat Biotechnol. 2010;28(3):275–80. doi: 10.1038/nbt.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8(5):341–50. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 22.Holtz M, Forman SJ, Bhatia R. Growth factor stimulation reduces residual quiescent chronic myelogenous leukemia progenitors remaining after imatinib treatment. Cancer Res. 2007;67(3):1113–20. doi: 10.1158/0008-5472.CAN-06-2014. [DOI] [PubMed] [Google Scholar]
- 23.Guo W, Lasky JL, Chang CJ, Mosessian S, Lewis X, Xiao Y, et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453(7194):529–33. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo W, Schubbert S, Chen JY, Valamehr B, Mosessian S, Shi H, et al. Suppression of leukemia development caused by PTEN loss. Proc Natl Acad Sci U S A. 2011;108(4):1409–14. doi: 10.1073/pnas.1006937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153(7):1552–66. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–52. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeithan TW, Shima EA, Le Beau MM, Minowada J, Rowley JD, Diaz MO. Molecular cloning of the breakpoint junction of a human chromosomal 8;14 translocation involving the T-cell receptor alpha-chain gene and sequences on the 3′ side of MYC. Proc Natl Acad Sci U S A. 1986;83(17):6636–40. doi: 10.1073/pnas.83.17.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–87. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 34.Tran LM, Chang CJ, Plaisier S, Wu S, Dang J, Mischel PS, et al. Determining PTEN functional status by network component deduced transcription factor activities. PLoS One. 2012;7(2):e31053. doi: 10.1371/journal.pone.0031053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117(6):2022–32. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Bohin N, Wen T, Ng V, Magee J, Chen SC, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature. 2013;504(7478):143–7. doi: 10.1038/nature12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 39.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4(11):842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 40.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116(9):1498–505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10(3):262–7. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 42.Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A. 2010;107(31):13836–41. doi: 10.1073/pnas.1008366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112(3):733–40. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood. 2014;123(7):1040–50. doi: 10.1182/blood-2013-08-522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.