Abstract
Detection of microbes by TLRs on the plasma membrane leads to the induction of pro-inflammatory cytokines such as TNF-α, via activation of NF-κB. Alternatively, activation of endosomal TLRs leads to the induction of type I IFNs via IFN regulatory factors (IRFs). TLR4 signaling from the plasma membrane to NF-κB via the Toll/IL-1R (TIR) adaptor protein MyD88 requires the TIR sorting adaptor Mal, while endosomal TLR4 signaling to IRF3 via TRIF requires the TIR sorting adaptor TRAM. Similar to TLR4 homodimers, TLR2 heterodimers can also induce both pro-inflammatory cytokines and type I IFNs. TLR2 plasma membrane signaling to NF-κB is known to require MyD88 and Mal, while endosomal IRF activation by TLR2 requires MyD88. However whether, like TLR4, TLR2 requires a sorting adaptor for endosomal signaling was unclear. Here we show that TLR2-dependent IRF7 activation at the endosome is both Mal- and TRAM-dependent, and that TRAM is required for the TLR2-dependent movement of MyD88 to endosomes following ligand engagement. TRAM interacted with both TLR2 and MyD88 suggesting that TRAM can act as a bridging adapter between these two molecules. Furthermore, infection of macrophages lacking TRAM with herpes viruses or the bacterium Staphylococcus aureus led to impaired induction of type I IFN, indicating a role for TRAM in TLR2-dependent responses to human pathogens. Our work reveals that TRAM acts as a sorting adaptor not only for TLR4, but also for TLR2, to facilitate signaling to IRF7 at the endosome, which explains how TLR2 is capable of causing type I IFN induction.
Introduction
The mammalian innate immune system responds to invading pathogens by using pattern recognition receptors such as TLRs to detect conserved pathogen associated molecular patterns. The activation of TLRs initiates signal transduction pathways that determine the type and duration of the host anti-pathogen and inflammatory response (1-3). Upon encountering their cognate PAMP, TLR homo- or heterodimers become active and recruit downstream signaling proteins.
For example, LPS binding to the TLR4 complex causes recruitment of the adaptor proteins MyD88 adaptor-like protein (Mal) and TRIF-related adaptor molecule (TRAM). Mal and TRAM are bridging and sorting adaptors that recruit, and control the localization of, the signaling adaptors MyD88 and TIR domain-containing adaptor inducing IFN-β (TRIF) respectively to TLR4 (4-8). A TLR4/Mal/MyD88 complex is formed at the plasma membrane, due to an amino-terminal localization domain in Mal that interacts with phosphatidylinositol-4,5 bisphosphate in the plasma membrane (6). This complex mediates MyD88-dependent signaling from the plasma membrane, via IL-1R-associated kinases (IRAKs) and TNFR associated factor 6 (TRAF6), leading to activation of MAPKs and of the transcription factors AP-1 and NF-κB. In contrast to Mal, TRAM contains a bipartite amino-terminal myristoylation motif and polybasic domain that regulates the intracellular location of TRAM (7). Both domains are required for plasma membrane targeting of TRAM, while the myristoylation motif is required for TRAM to localize at endosomes (7, 9). Thus a TLR4/TRAM/TRIF complex is formed at the membrane of endosomal compartments, and this signals via TRAF3 to activate the transcription factor IFN regulatory factor 3 (IRF3) (7). For TLR4 signaling, Mal-dependent NF-κB activation upregulates inflammatory genes such as TNF-α, while TRAM-dependent IRF3 activation causes induction of IFN-β.
Apart from TLR4, several other TLRs can signal from endosomes to induce type I IFNs (IFN-α and IFN-β), in response to the detection of viral nucleic acids (10). Thus TLR3 recognizes dsRNA; TLR7 and TLR8 recognize single-stranded RNA; and TLR9 recognizes CpG motifs in DNA (11). For TLR3, type I IFN induction is achieved via TRIF and IRF3, while for TLR7, 8 and 9 the induction pathway involves MyD88-dependent IRF7 activation (10).
Whereas TLR4 responds to LPS from Gram-negative bacteria, recognition of cell surface components of Gram-positive bacteria, such as lipoproteins and lipoteichoic acids, require TLR2 (12). The fatty acid groups of triacylated lipopeptides are the ligand for TLR2/TLR1 heterodimers (13), and the fatty acid groups of diacylated lipopeptides and LTA are ligands for TLR2/TLR6 heterodimers (14, 15). Similar to TLR4 signaling, Mal acts as a bridging adaptor between the TLR2 receptor complex and MyD88, although high TLR2 ligand concentrations can overcome the requirement for Mal in the signaling pathway, while some downstream TLR2 signals are entirely Mal-independent (16, 17).
Although TLR2 is best known for its role in recognizing bacterial and fungal cell wall components, it also plays a role in the immune response to viruses. Such responses could be due to direct recognition of viral PAMPs by TLR2, or production of virally-induced endogenous TLR2 ligands. Thus glycoprotein B from human CMV activates TLR2 signaling (18, 19), while mouse CMV (20), HSV types 1 and 2 (21, 22), hepatitis C virus (23), lymphocytic choriomeningitis virus (24), measles virus (25) and vaccinia virus (VACV) (26) are also able to elicit TLR2-dependent responses. Activation of TLR2 may benefit the virus, for example measles virus may have evolved the ability to activate TLR2 as a means of upregulating the viral entry receptor CD150 (25). However in other instances TLR2 activation contributes to protection, for example mice lacking TLR2 are impaired in their ability to mount an innate or adaptive immune response to VACV (26).
It was originally thought that TLR2/TLR1 and TLR2/TLR6 heterodimers elicited pro-inflammatory, but not type I IFN responses, after ligand engagement (7, 27-30). However, later studies demonstrated that bacterial TLR2 ligands can induce type I IFN responses, while live virus-induced type I IFN has been shown to be at least partially TLR2-dependent in the case of VACV, mouse CMV and murine gammaherpesvirus-68 (MHV68) (31-33). Compared to the mechanism whereby TLR4 and the endosomal TLRs signal to type I IFN induction, much less is known about how TLR2 achieves this. Dietrich and colleagues showed that, upon stimulation with bacterial TLR2 ligands, the receptor is internalized and transported into endolysosomal compartments from where it induces IFN-β via MyD88 and IRF7 (31). Inhibition of receptor internalization or endosomal acidification could block the induction of IFN-β and IFN-inducible genes but not proinflammatory cytokines like TNF-α. This suggests that TLR2 activation, similar to TLR4, induces pro-inflammatory and type I IFN responses from distinct sub-cellular sites: the plasma membrane and endolysosomal compartments, respectively (31). However, apart from the role of MyD88 and IRFs, the signaling pathway of TLR2-dependent production of type I IFNs has yet to be elucidated, and in particular it is unclear whether other TIR adaptor proteins apart from MyD88 are required.
In this paper we show that as well as MyD88, both Mal and TRAM are required for TLR2-stimulated IFN-β induction. TLR2-induced IFN-β but not TNF was sensitive to VIPER (Viral Inhibitory Peptide of TLR4), a viral peptide inhibitor of Mal and TRAM. VIPER is derived from the VACV protein A46 which inhibits TLR4 signaling in the context of a virus infection by disrupting TLR4:Mal and TLR4:TRAM interactions (34, 35). Previously VIPER was shown to associate with Mal and TRAM but not MyD88 or TRIF (36), and it is derived from a region of A46 shown to be essential for TLR4 inhibition, and to mediate an A46:TRAM interaction (35, 37). Here, TLR2-induced IFN-β but not TNF-α required endocytosis, and was impaired in cells lacking Mal or TRAM, while TLR2-stimulated IRF7 activation was blocked by VIPER, and shown to require both Mal and TRAM. We demonstrate that stimulation of cells with a TLR2 ligand led to mobilization of MyD88 to intracellular punctate structures in a TRAM-dependent manner. Also TRAM interacted with TLR2 and MyD88 suggesting that TRAM acts as a bridging adapter between these two molecules. Furthermore, infection of macrophages lacking TRAM with herpes viruses led to impaired induction of type I IFN, indicating a role for TRAM in this TLR2-dependent pathway, while Staphylococcus aureus stimulated IFN-β was also TRAM-dependent. Thus TRAM acts as a sorting adaptor not only for TLR4, but also for TLR2, to facilitate signaling to IRF7 at the endosome, which explains how TLR2 is capable of causing type I IFN induction in response to both viral and bacterial pathogens.
Materials and Methods
Cell culture
HEK293T cells were purchased from European Collection of Animal Cell Cultures (Salisbury, UK). HEK293 cells stably transfected with TLR4, MD2 and CD14 (HEK293-TLR4) were purchased from InvivoGen (San Diego, CA). HEK293 cells stably transfected with TLR2 (HEK293-TLR2) were a gift from Dr. K. Fitzgerald (University of Massachusetts Medical School, Worcester, MA). Immortalized murine wild type (WT), MyD88−/−, Mal−/−, TRIF−/− and TRAM−/− bone marrow-derived macrophages (iBMDMs) were generated from corresponding knockout mice using J2 recombinant retrovirus carrying v-myc and v-raf/mil oncogenes as previously described in (17, 38) and were a kind gift from K. Fitzgerald and D. Golenbock (University of Massachusetts Medical School, Worchester, MA). Cells were maintained in DMEM containing 10% (v/v) FCS, 10 μg/ml Ciproflaxin and 2 mM L-glutamine. Selection agents were used as follows: HEK293-TLR4 cells, 10 μg/ml Blasticidin (Sigma) and 50 μg/ml of HygroGold (Invivogen); HEK293-TLR2 cells, 1 mg/ml G-418 (Sigma).
Receptor agonists and reagents
Ultrapure LPS from Escherichia coli (99.9% pure in respect to contaminating protein, DNA, and TLR2 agonists) was purchased from Alexis Biochemicals (Plymouth Meeting, PA). N-palmitoyl-S-dipalmitoylglyceryl Cys-Ser-(lys)4 (Pam3CSK4) and macrophage-activating lipopeptide-2 (Malp2) were purchased from Invivogen (San Diego, CA). Bafilomycin A was purchased from Sigma. Synthetic dsDNA 60-mer derived from nucleotides 144107-144166 the HSV-1 genome (HSV 60 mer), was obtained from DNA Technology (Aarhus, Denmark). Mouse anti-Flag M2 and anti-β-actin Abs were from Sigma and rabbit anti-EEA1 was from Abcam (Cambridge, UK).
Peptide synthesis and reconstitution
Peptides were synthesized by GenScript (Piscataway, NJ) and were >95% pure as confirmed by HPLC. Lyophilized peptides were reconstituted aseptically with molecular biology-grade water to a concentration of 10 mM and stored at −80°C. Working stocks of 0.2 or 1 mM were stored at −20°C or kept at 4°C for a maximum of 2 weeks.
Plasmids
Sources of expression plasmids were as follows: pCMV-myc empty vector (Clontech, Mountain View, CA), phRL-TK vector (Promega, Madison, WI), pFR-luciferase reporter gene (Stratagene/Agilent Technologies, Cork, Ireland), Gal4-IRF3, Gal4-IRF7, Flag-MyD88 and Flag-Mal (K.A. Fitzgerald, The University of Massachusetts Medical School, Worcester, MA), Flag-TRIF (S. Sato, Research Institute for Infectious Diseases, Osaka University, Japan), Flag-TLR2, CFP-MyD88, GFP-TRAM, YFP-TLR2, YFP-TLR6, GST and the GST fusion of TRAM (L.A. O’Neill, Trinity College Dublin, Dublin, Ireland) and the NF-κB reporter gene (described in (39)). Flag-TRAM G2A, which contains a point mutation in the myristoylation motif, was generated from WT Flag-TRAM plasmid using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene).
Quantitative RT-PCR
For mRNA analysis iBMDMs were seeded at 2 × 105 cell/ml in 24-well plates 24 h prior to treatment. RNA was isolated using High Pure RNA isolation kit from Roche Applied Science (Burgess Hill, U.K.) according to the manufacturer’s instructions. RT-PCR was performed using Qiagen’s One-Step RT-PCR Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Quantitative real-time PCR was done using GoTaq qPCR Master Mix (Promega, Madison, WI) and the 7500 Fast Real-time PCR System (Applied Biosystems) with the following primers: mTNFα forward, 5′-TCCCCAAAGGGATGAGAAGTT-3′, and reverse, 5′-GTTTGCTACGACGTGGGCTAC-3′; mIFNβ forward, 5′-ATGGTGGTCCGAGCAGAGAT-3′, and reverse, 5′-CCACCACTCATTCTGAGGCA-3′. To measure knockdown of Mal or TRAM mRNA by treatment with siRNA (described below), cDNA from HEK293-TLR2 cells was prepared as above and analyzed by qPCR using the following primers: hTRAM forward, 5′-TTCCTGCCCTCTTTCTCTCTC-3′, and reverse 5′-AACATCTCTTCCACGCTCTGA-3; hTIRAP/Mal forward, 5′-CCAGCCTTTCACAGGAGAAG-3′, and reverse, 5′-ATATTCGGGATCTGGGGAAG-3′. Relative mRNA expression was calculated using the comparative CT method, normalizing the gene of interest to the housekeeping gene β-actin, and comparing it to an untreated sample as calibrator.
Cytokine analysis
For cytokine production, iBMDMs were seeded at 2 × 105 cell/ml in 96-well plates 24 h prior to treatment. The supernatants were collected and assessed for TNF-α by ELISA (R&D Systems, Minneapolis, MN). IFN-β protein in cell culture supernatants was measured using a custom ELISA originally described elsewhere (40), with a few modifications. In brief, high-binding 96-well polystyrene plates were coated overnight with a 1:1000 dilution of rat anti-mouse IFN-β mAb (Santa Cruz, Heidelberg, Germany) in carbonate buffer (10 mM NaHCO3, 3.4 mM Na2CO3, pH 9.4-9.6) at 4°C. Plates were washed three times and then blocked with 10% FCS/PBS for 2 h at 37°C. The blocking solution was removed and 40μl of recombinant IFN-β standard (PBL Biomedical Laboratories, Piscataway, NJ) in triplicates and 1:2 serial dilutions in 10% FCS/PBS starting at 20U/ml was applied to the plate. 40μl sample supernatant (undiluted or diluted 1:2 with 10% FCS/PBS) were added to each well and incubated at 4°C over night. The following day the plates were washed and incubated over night with 50μl/well of rabbit anti-mouse IFN-β pAb (PBL Biomedical Laboratories, Piscataway, NJ) diluted 1:2000 in 10% FCS/PBS. After washing, the plates were incubated for 2h with 50μl/well of anti-rabbit horseradish peroxidase (HRP; Sigma-Aldrich) diluted 1:2000 in 10% FCS/PBS. The plates were washed again and developed with TMB substrate as usual. Experiments were performed three times in triplicate, and data are expressed as mean ± S.D. from one representative experiment.
Reporter gene assays
HEK293-TLR4 cells (4 × 104 cells per well) or HEK293-TLR2 cells (2 × 104 cells per well) were seeded into 96-well plates and transfected 24 h later with expression vectors and luciferase reporter genes using GeneJuice (Novagen/Merck, Nottingham, UK). For the NF-κB assays, 60 ng of κB-luciferase reporter gene was used. For the IRF3 and IRF7 assays, IRF3-Gal4 and IRF7-Gal4 fusion vectors (1-3 ng) were used in combination with 60 ng pFR luciferase reporter as described previously (34). In all cases, 20 ng/well of phRL-TK reporter gene was cotransfected to normalize data for transfection efficiency. The total amount of DNA per transfection was kept constant at 230 ng by addition of pCMV-Myc. After 24 h, cells were stimulated with the indicated TLR ligands. After a further 6 h, cells were lysed in Passive Lysis Buffer (Promega, Madison, WI) and whole cell lysates were analyzed for luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity, and data are expressed as the mean fold induction, relative to control levels, for a representative experiment from a minimum of three separate experiments, each performed in triplicate.
Transfection of HEK293-TLR cells with siRNA
HEK293-TLR4 cells (1 × 105 cells/ml) or HEK293-TLR2 (0.5 × 105 cells/ml) cells were seeded into 96-well plates and transfected 24 h later with 50 nM control siRNA or siRNA targeting TRAM or Mal (ON-TARGETplus Dharmacon siRNA from Thermo Scientific) using Lipofectamine (Invitrogen). A second transfection was then done 24 h later with relevant reporter assay constructs as described above. NF-κB-luciferase activity or IRF7-Gal4 transactivation was measured 24 h later after the relevant stimulations. To measure knockdown of Mal or TRAM mRNA after siRNA treatment, HEK293-TLR2 cells (0.5 × 105 cells/ml) were seeded into 24-well plates and transfected with 50 nM siRNA as above. After 48 h transfection, cDNA was prepared from these cells and Mal or TRAM mRNA measured by qPCR. To measure knockdown of Mal or TRAM protein after siRNA treatment, HEK293-TLR2 cells (0.5 × 105 cells/ml) were seeded into 6-well plates and transfected with 50 nM siRNA as above. A second transfection was then done 24 h later with 1 μg of either Flag-Mal or Flag-TRAM. Cell lysates were prepared 24 h later and analyzed by SDS-PAGE and immunoblotting.
Confocal microscopy
HEK293 cells were transfected with plasmids expressing YFP and CFP fusion proteins using GeneJuice (Novagen/Merck, Nottingham, UK)) according to the manufacturer’s protocol. iBMDM cells were transfected with plasmids using Lipofectamine (Invitrogen) according to the manufacturer’s protocol. Media was replaced 24 h after transfection and cells were left to recover for a further 24 h. Cells were treated with TLR agonists as required. For intracellular staining, the cells were fixed with 2% paraformaldehyde in PBS, incubated for 15 min on ice, permeabilized with PEM buffer (80 mM K-Pipes [pH 6.8], 5 mM EGTA, 1 mM MgCl2, 0.05% saponin) for 15 min on ice, quenched of free aldehyde groups in 50 mM NH4Cl with 0.05% saponin for 5 min, and blocked in PBS with 10% FCS and 0.05% saponin for 20 min. The cells were incubated with 5 μg/ml primary Ab in PBS with 0.05% saponin for 60 min at room temperature. Alexa Fluor-labeled secondary Abs (Invitrogen) were added for 30 min at room temperature and cells washed three times in PBS with 0.05% saponin. Images were captured using a confocal laser scanning microscopy (Olympus FluoView TM FV1000), at 60× original magnification.
GST pulldown assays
Empty pGEX.4T2, or pGEX.4T2 plasmid containing TRAM were transformed into Escherichia coli Rosetta-Gami B Host Strains (Novagen/Merck, Nottingham, UK) and grown in LB Broth. Protein expression was induced with 0.5 mM IPTG at 30°C. Bacterial cells were harvested by centrifugation after 6 h induction and lysed using BugBuster (Merck/Millipore, Nottingham, UK). Insoluble fractions were removed by centrifugation. The remaining soluble fractions were cleared by glutathione sepharose 4B affinity chromatography (Amersham Biosciences) and levels of protein expression confirmed by SDS-PAGE and Coomassie staining of the gel. HEK293T cells were seeded into 15-cm dishes (3 × 106 cells) 24 h before transfection with GeneJuice. Cells were transfected with the relevant signaling molecule or pCMV-myc plasmid. Cells were harvested after 48 h in 850 μl lysis buffer (50 mM Hepes, pH 7.5, 250 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40 containing 0.01% aprotinin, 1 mM sodium orthovanadate, and 1 mM PMSF) for 30 mins on ice. Whole cell lysates were clarified by centrifugation. 50 μl cleared lysate was retained for analysis of protein expression (i.e. input lysate), the remainder was divided in two and was added to either purified GST or purified GST-fusion protein coupled to glutathione-sepharose and incubated for 2 h at 4°C. The immune complexes were precipitated and washed four times in lysis buffer. Pulldowns were analyzed by SDS-PAGE and immunoblotting.
IFN-α/β bioassay of virally infected BMDMs
The viruses used were HSV-1 (F+ strain), HSV-2 (333 strain), MHV68 and Sendai virus (SeV, Cantell strain). HSV-2 and HSV-1 were amplified in Vero cells whereas MHV68 was amplified in BHK-21 cells. Immortalized BMDMs were seeded in 48-well plates at a density of 4 × 105 cells per well and were infected with viruses, or transfected with lipofectamine alone or with dsDNA as indicated. IFN-α/β bioactivity was measured by a L929 cell-based bioassay. L929 cells (2 × 104 cells/well in 100 μl) in MEM with 5% FCS were incubated overnight at 37°C in successive 2-fold dilutions of samples or murine IFN-α/β as standard. Subsequently, vesicular stomatitis virus (VSV/V10) was added to the wells and the cells were incubated for 2-3 days. The dilution mediating 50% protection was defined as 1 U/ml of IFN-α/β (41).
Infection of BMDMs with S.aureus
S.aureus strain SH1000 has been previously described (42, 43). Bacteria were cultivated from frozen stocks for 24 h at 37°C on agar plates. Bacterial suspensions were then prepared in PBS, and the concentrations estimated by measuring the absorbance of the suspension at 600 nm. iBMDM cells were infected with live S.aureus at multiplicities of infection (MOIs) of 10 and 100 for the indicated times as previously described (43). Supernatants were then collected and assayed for IFN-β by ELISA.
Statistical analysis
Statistical analysis was carried out using paired Student’s t test.
Results
VIPER inhibits TLR2-dependent IFN-β production
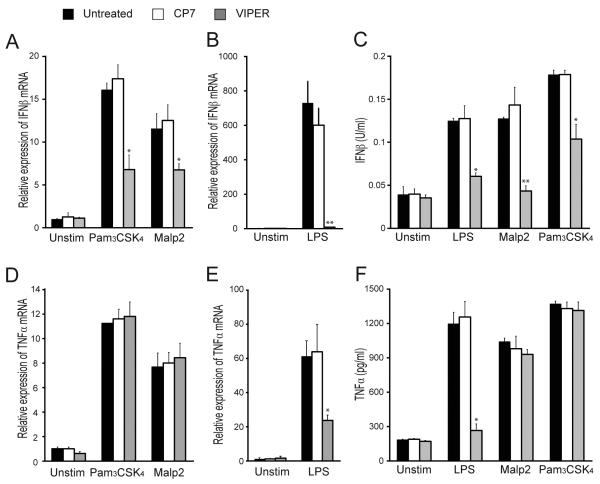
We previously showed that the peptide VIPER derived from the VACV protein A46 could block TLR4-dependent gene induction, by antagonizing TRAM and Mal, but not MyD88 nor TRIF (35-37). Consistent with this, VIPER did not inhibit TNF-α production mediated by TLRs 2, 3, and 9 (36). Since stimulation of TLR2/1 and TLR2/6 heterodimers with Pam3CSK4 or Malp2 respectively leads to production of type I IFNs via a signaling pathway that has yet to be fully elucidated, we tested the effect of VIPER on this pathway. Stimulation of iBMDM cells with either Pam3CSK4 or Malp2 led to induction of IFN-β mRNA, which interestingly was significantly inhibited by pretreatment of cells with VIPER, but not with CP7 control peptide (Fig. 1A). As expected from previously published work (36), VIPER, but not CP7, could also impair induction of LPS-induced IFN-β mRNA (Fig. 1B). Although LPS stimulation induces a much higher level of IFN-β mRNA in WT iBMDMs than either Pam3CSK4 or Malp2, a similar level of IFN-β protein was produced by these cells in response to all three ligands, and VIPER significantly inhibited production of IFN-β for each of these ligands (Fig. 1C). In contrast, only LPS-induced TNF-α mRNA and protein (Fig. 1E and 1F), and not Pam3CSK4- or Malp2-induced TNF-α mRNA and protein (Fig. 1D and 1F), were inhibited by VIPER treatment of cells. These data demonstrate that VIPER can impair TLR2-dependent production of IFN-β but not TNF-α, which was suggestive of a role for Mal and/or TRAM in TLR2-induced type I IFN.
Figure 1. VIPER inhibits TLR2-dependent IFN-β induction.
(A, B, D, E) WT iBMDMs were treated with 5 μM VIPER or CP7 peptide 1 h prior to being stimulated with 20 nM Malp2, 20 ng/ml Pam3CSK4 or 100 ng/ml LPS for 3 h. Induction of IFN-β (A, B) and TNF-α (D, E) mRNA expression was assayed by quantitative RT-PCR, normalized to β-actin and presented relative to untreated, unstimulated cells. (C, F) Cells were treated as described above except TLR stimulations were for 24 h. Production of IFN-β (C) and TNF-α (F) protein was assayed by ELISA. For (A-F) the data are mean ± SD of triplicate samples and are representative of at least three independent experiments. *p<0.05, **p<0.005 or ***p<0.0005 compared to samples treated with control peptide (CP7).
Mal and TRAM are required for TLR2-dependent IFN-β production
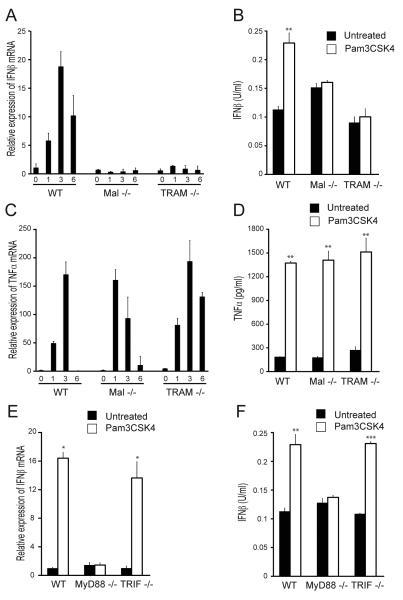
In order to determine whether TRAM and/or Mal are required for TLR2-mediated IFN-β production, iBMDMs deficient in either Mal or TRAM were examined for their ability to produce IFN-β mRNA and protein in response to Pam3CSK4. Time course analysis of mRNA induction showed that Pam3CSK4 induced IFN-β transcription in WT iBMDMs with a rapid kinetic, peaking at 3 h (Fig. 2A), as has been previously shown (31). However, Mal−/− or TRAM−/− iBMDMs were incapable of IFN-β mRNA induction in response to Pam3CSK4 (Fig. 2A), indicating that both Mal and TRAM are required for TLR2-dependent IFN-β induction. In agreement with these data, 24 h stimulation with Pam3CSK4 led to IFN-β protein production in WT, but not Mal−/− or TRAM−/−, iBMDMs (Fig. 2B). Both Mal−/− and TRAM−/− iBMDMs were responsive to Pam3CSK4, as evidenced by the induction of TNF-α mRNA and protein in these cells, which was comparable to WT iBMDMs (Fig. 2C, D). It has previously been shown that MyD88 is required for the production of TLR2-dependent type I IFN (31, 32, 44), and results obtained here agree with those findings since compared to WT cells stimulation of MyD88−/− iBMDMs with Pam3CSK4 did not lead to induction of either IFNβ mRNA (Fig. 2E) or protein (Fig. 2F). It has previously been shown that TRIF is not involved in this pathway (32), and this was also confirmed here since the Pam3CSK4 -IFNβ response was not impaired in TRIF−/− iBMDMs (Fig. 2E, F). These data confirm that TLR2-dependent induction of IFN-β requires MyD88 and not TRIF, and reveal a role for Mal and TRAM in this pathway.
Figure 2. Mal and TRAM are required for TLR2-dependent IFNβ induction.
(A, C) WT, Mal−/− and TRAM−/− iBMDMs were stimulated with 20 ng/ml Pam3CSK4 for 1, 3 and 6 h. Induction of IFN-β (A) and TNF-α mRNA (C) expression was assayed by quantitative RT-PCR, normalized to β-actin and presented relative to untreated, unstimulated cells. (B, D, F) WT, Mal−/−, TRAM−/−, MyD88−/− and TRIF−/− iBMDMs were stimulated with 20 ng/ml Pam3CSK4 for 24 h. Production of IFN-β (B, F) and TNF-α (D) protein was measured by ELISA. (E) WT, MyD88−/− and TRIF−/− iBMDMs were stimulated with 20 ng/ml Pam3CSK4 for 3 h. Induction of IFN-β mRNA expression was assayed by quantitative RT-PCR, normalized to β-actin and presented relative to untreated, unstimulated cells. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. *p<0.05, **p<0.005 or ***p<0.0005 compared to untreated samples.
TLR2 activates IRF7 via Mal and TRAM
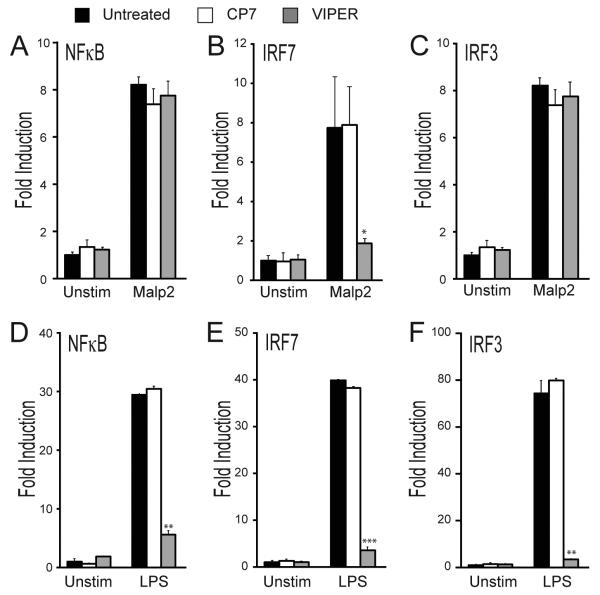
IFN-β mRNA induction by TLRs is regulated by IRF3, IRF7 and NF-κB (45). To assess the involvement of these transcription factors in the TLR2-IFN-β signal transduction pathway, HEK293 cells stably transfected with TLR2 were utilized (HEK293 cells naturally express the TLR2 coreceptors TLR1 and TLR6 (46)). The ability of Malp2 to activate IRF3, IRF7 and NF-κB, and the effect of VIPER on transcription factor activation, was measured by reporter gene assay (34, 47). Fig. 3A-C shows that Malp2 treatment stimulated activation of all three transcription factors, however VIPER only inhibited IRF7 activation (Fig 3B). The lack of effect of VIPER on TLR2-stimulated NF-κB activation was consistent with the inability of VIPER to inhibit TNF mRNA induction by TLR2 (Fig. 1D), which is strongly NF-κB-dependent. Further, the effect of VIPER on TLR2-stimulated IRF7 activation and not on IRF3 implicated IRF7 in the VIPER-sensitive TLR2-mediated IFN-β mRNA induction (Fig. 1A). In agreement with a previous study showing that VIPER inhibits all signals downstream of TLR4 (36), pretreatment of HEK293-TLR4 cells with VIPER potently blocked LPS-dependent activation of NF-κB, IRF7 and IRF3 (Fig. 3, D-F).
Figure 3. VIPER inhibits TLR2-mediated IRF7 transactivation.
HEK293-TLR2 cells (A-C) or HEK293-TLR4 cells (D-F) were transfected for 24 h with the NF-κB luciferase reporter gene (A, D), or the pFR luciferase reporter gene along with plasmid expressing either IRF7-Gal4 (B, E) or IRF3-Gal4 (C, F). Cells were pretreated with 5μM VIPER or CP7 peptide 1 h prior to being stimulated with 20 nM Malp2 (A-C) or 10 ng/ml LPS (D-F) for 6 h and luciferase reporter gene activity was measured. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. *p<0.05, **p<0.005 or ***p<0.0005 compared to stimulated samples treated with control peptide (CP7).
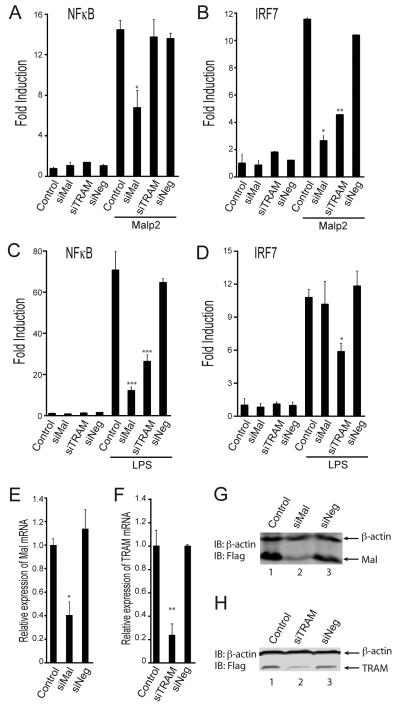
Since TLR2-stimulated IRF7 was sensitive to VIPER we next determined whether TRAM and/or Mal were involved in this pathway using siRNA. We used a combination of four anti-Mal or anti-TRAM siRNAs, provided as single reagents, and a combination of four non-targeting siRNAs as a control, and assessed the role of Mal and TRAM in IRF7 activation, compared to NF-κB activation. This demonstrated a role for Mal in both NF-κB and IRF7 activation by TLR2 (Fig. 4A, B), while TRAM was required for TLR2-stimulated IRF7 but not NF-κB activation. Similar results were obtained for TLR4-stimulated transcription factor activation, whereby both Mal and TRAM were required for NF-κB activation (Fig. 4C) while TRAM and not Mal was required for IRF7 activation (Fig. 4D). We confirmed the efficacy of the siRNAs by showing reduced mRNA induction of the targets (Fig. 4E, F) and also reduced protein expression of overexpressed Mal (Fig. 4G) and TRAM (Fig. 4H), since commercially available anti-Mal and anti-TRAM antibodies were not successful in our hands for detecting endogenous Mal or TRAM in HEK 293 cells. These data show that both Mal and TRAM are required for TLR2 signaling to IRF7.
Figure 4. TLR2 activates IRF7 via Mal and TRAM.
(A-D) HEK293-TLR2 cells (A, B) or HEK293-TLR4 cells (C, D) were transfected for 24 h with siRNA targeting Mal (siMal), TRAM (siTRAM) or control siRNA (siNeg). 24 h later cells were transfected with the NF-κB luciferase reporter gene (A, C), or the pFR luciferase reporter gene along with plasmid expressing IRF7-Gal4 (B, D). Cells were stimulated with either 20 nM Malp2 (A, B) or 10 ng/ml LPS (C, D) for 6 h and luciferase reporter gene activity was measured. (E, F) HEK-293 TLR2 cells were transfected for 48 h with siNeg and siMal (E) or siTRAM) (F) for 48 h. Mal and TRAM mRNA expression was assayed by quantitative RT-PCR, normalized to β-actin and presented relative to untreated (Control) cells. (G, H) HEK-293 TLR2 cells were transfected with siRNAs and 24 h later transfected with 1 μg plasmid expressing Flag-Mal (G) or Flag-TRAM (H). Cell lysates were prepared 24 h later and analyzed by SDS-PAGE and immunoblotting. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. *p<0.05 or **p<0.005 compared to stimulated samples treated with control siRNA (siNeg).
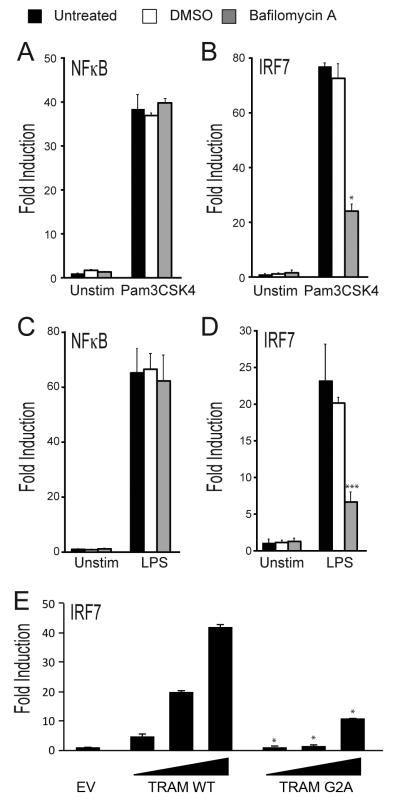
TLR2 mediated IRF7 activation requires endocytosis
TLRs appear to signal from the endosome in order to stimulate IRF activation. TLR3, TLR7, TLR8 and TLR9 are naturally located at endosomal compartments, whereas for TLR4 stimulation of IRF activity, it is necessary for the receptor to move to endosomal compartments after ligand engagement (7). For TLR2, Dietrich et al showed that upon activation, TLR2 is internalized and transported into endolysosomal compartments from where it induces IFN-β, since Malp2 or Pam3Csk4-stimulated IFN-β (but not TNF-α) induction was blocked by BafilomycinA (BafA), an inhibitor of the endosomal proton pump (31). Here BafA inhibited Pam3Csk4-stimulated IRF7 but not NF-κB activation (Fig. 5A, B), and as expected LPS-stimulated IRF7 and not NF-κB (Fig. 5C, D). Hence, similar to TLR4, the TLR2 signaling pathway to IRF7 requires both TRAM and endocytosis. Consistent with this, when the myristoylation motif of TRAM, which is required for TRAM to localize at endosomes (7, 9), was mutated at a single amino acid residue, the ability of TRAM to activate IRF7 was impaired: Fig. 5E shows that expression of TRAM but not TRAM G2A led to activation of IRF7.
Figure 5. TLR2 mediated IRF7 activation requires endocytosis.
HEK293-TLR2 cells (A, B) or HEK293-TLR4 cells (C, D) were transfected for 24 h with the NF-κB luciferase reporter gene (A, C), or the pFR luciferase reporter gene along with plasmid expressing IRF7-Gal4 (B, D). Cells were pretreated with 100 nM Bafilomycin A or DMSO (vehicle) 1 h before stimulation with either 20 ng/ml Pam3CSK4 (A, B) or 10 ng/ml LPS (C, D). Luciferase reporter gene activity was measured 6 h later. (E) HEK293-TLR2 cells were transfected with empty vector (EV) or 10, 60 or 120 ng (wedge) of plasmid encoding TRAM WT or TRAM G2A, and the pFR luciferase reporter gene along with plasmid expressing IRF7-Gal4. Luciferase reporter gene activity was measured 30 h later. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. *p<0.05 or ***p<0.0005 compared to stimulated samples treated with DMSO (B, D) or compared to TRAM WT (E).
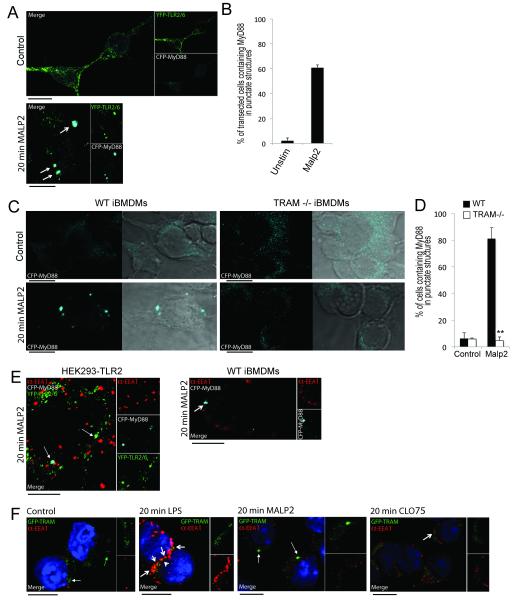
TLR2-stimulated intracellular mobilization of MyD88 is TRAM-dependent
The data so far demonstrated that TLR2 signaling to IFN induction requires MyD88, TRAM, endocytosis and IRF7 activation, and that TLR2 stimulation causes TRAM to relocate to endosomes. This suggested that, similar to the case for TLR4, TRAM could be required for TLR2 as a sorting adaptor to mobilize a signaling adaptor to an endosome (which is TRIF in the case of TLR4 (7)). If TRAM is a sorting adaptor for the TLR2-IRF7 pathway, TLR2 ligand stimulation should cause relocalization of MyD88 within the cell in a TRAM-dependent manner. In order to test this prediction, HEK293 cells were transfected with TLR2-YFP, TLR6-YFP and MyD88-CFP and then treated with Malp2. In untreated cells TLR2/6 was observed at the periphery of the cells whereas MyD88 was spread diffusely throughout the cell (Fig 6A, upper panel). After 20 min Malp2 treatment, TLR2/6 was observed in punctate structures throughout the cell and MyD88 and TLR2/6 were observed to co-localize in large intracellular structures (Fig 6A, lower panel, see arrows). After 20 min Malp2 treatment, an average of 61 % of transfected cells contained MyD88 in large intracellular structures compared with 2 % of unstimulated cells (Fig 6B), strongly suggestive of TLR2-stimulated re-localization of MyD88 to intracellular compartments.
Figure 6. Endosomal localization of MyD88 after TLR2 stimulation is TRAM-dependent.
(A) Confocal image of YFP-TLR2 and YFP-TLR6 (green) in HEK293T cells coexpressing CFP-MyD88 (cyan). Cells were left untreated (top panel) or treated with 50 nM Malp2 for 20 min (bottom panel). The arrows indicate areas of overlap in the overlay panels. Data are representative of two independent experiments. (B) Transfected cells from (A) were observed by confocal microscopy and scored for CFP-MyD88-containing punctate structures. At least 100 cells were counted for each sample. Values shown are mean ± spread from the two independent experiments. (C) Confocal images of iBMDMs expressing CFP-MyD88. WT (left panels) or TRAM−/− (right panels) iBMDMs were left untreated (top panels) or treated with 50 nM Malp2 for 20 min (bottom panels). Data are representative of three independent experiments. (D) Transfected cells from (C) were observed by confocal microscopy and scored for CFP-MyD88-containing punctate structures. At least 100 cells were counted for each sample. Values shown are mean ± SD from the three independent experiments **p<005 compared to WT cells. (E) Confocal images of (left panel) YFP-TLR2 and YFP-TLR6 (green) in HEK293T cells co-expressing CFP-MyD88 (cyan) and stained for EEA1 (red), or (right panel) WT BMDMs expressing CFP-MyD88 (cyan) and stained for EEA1 (red). Cells were treated with 50 nM Malp2 for 20 min. Arrows indicate areas of overlap in the overlay panels between MyD88 and TLR2/6 (left panel) or MyD88 accumulation (right panel). Data are representative of three independent experiments. (F) Confocal images of GFP-TRAM (green) in WT iBMDMs co-stained for EEA1 (red). WT iBMDMs were left untreated (first panel) or treated with LPS (100ng/ml, second panel), Malp2 (50nM, third panel) or CLO75 (2.5 μg/ml, fourth panel) for 20 min. Closed arrows indicate regions of TRAM localized to EEA1 negative compartments; open arrows indicate overlapping TRAM and EEA1 regions which appear as yellow in the overlay panels. Data are representative of two independent experiments. All scale bars represent 10 μm.
In order to determine if the observed Malp2-induced mobilization of MyD88 was dependent on the presence of TRAM, we transfected WT iBMDM or TRAM −/− iBMDM with MyD88-CFP. Prior to TLR2 stimulation, MyD88 was spread diffusely throughout the cytoplasm of both WT and TRAM −/− iBMDMs (Fig 6C, top panels). After 20 min Malp2 treatment MyD88 could be observed in endosome-like structures, reminiscent of those observed in the HEK293 cells. Compellingly, this mobilization of MyD88 could only be observed in the WT and not in the TRAM −/− iBMDMs (Fig 6C, lower panels). Further, after 20 min Malp2 treatment, on average 81 % of WT iBMDMs contained MyD88 in punctate structures compared with 5 % of TRAM −/− iBMDMs (Fig 6D). Thus intracellular movement of MyD88 after TLR2 stimulation is TRAM-dependent.
Under basal conditions TRAM is known to localize to the plasma membrane and to endosomal compartments. In response to LPS, TRAM is initially mobilized to EEA1-positive early endosomes and can also be found in Rab11-positive sorting/recycling endosomes (48, 49). Thus we next investigated if the compartments to which MyD88 localized after Malp2 stimulation were early endosomes. As seen in Fig 6E, left panel, the MyD88-positive, TLR2/6-positive compartments (white arrows) did not correlate with the EEA1-positive early endosomes. This observation was paralleled in WT BMDMs (Fig 6E, right panel, see arrow). In order to ensure the cells were responding normally, TRAM-GFP was transfected into WT iBMDMs and cells were stained for EEA1. As expected, TRAM localized to the membrane (Fig 6F, first panel, green arrow) and to endosomal structures (Fig 6F, first panel, white arrow) in untreated cells. After 20 min LPS treatment, TRAM was found to co-localize with EEA1-positive early endosomes (Fig 6F, second panel, see arrows). Interestingly, after 20 min Malp2 treatment TRAM localization was distinctly different from that observed after LPS treatment, as TRAM was observed in large EEA1-negative endosome-like structures reminiscent of the Malp2-inducible MyD88-TLR2/6-positive compartments (Fig 6F, third panel, see arrows). As a control we showed that ligand stimulation of TLR7 with CLO75, while increasing the number of EEA1-positive endosomes, did not induce observable TRAM mobilization, with the vast majority of TRAM localized in a similar pattern to the untreated cells (Fig 6F, fourth panel). Thus TLR2 causes relocalization of both MyD88 and TRAM to large intra-cellular compartments that are distinct from the EEA1-positive endosomes that TLR4 stimulates TRAM to locate to.
Together these data indicated that MyD88 and TRAM likely localize together at the same TLR2/6-positive compartment in response to Malp2, a structure that is distinct from the early endosomes involved downstream of LPS signaling, and that the TLR2 stimulation of MyD88 movement to these compartments requires the sorting adaptor TRAM.
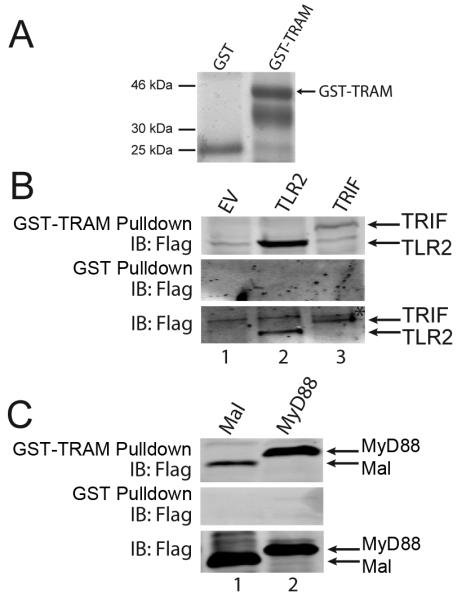
TRAM interacts with TLR2 and MyD88
To gain further evidence that TRAM acts as a sorting adaptor for TLR2, and controls MyD88 location after TLR2 stimulation, we tested whether TRAM interacted with TLR2 and MyD88, as is the case for TLR4 and TRIF. For this, a GST pulldown assay was used, whereby the ability of the TIR proteins expressed in cells to interact with a GST fusion of TRAM was examined. Cell lysates from TLR2- or TIR adaptor-expressing cells were incubated with equal amounts of either GST or GST-TRAM (Fig. 7A). Immunoblot of GST pulldowns demonstrated that TLR2 interacted with GST-TRAM (Fig. 7B, top panel, lane 2), but not with GST alone (Fig. 7B, middle panel). The known TRAM-interaction partner in the TLR4 receptor complex, TRIF, also associated with GST-TRAM in this assay (Fig. 7B, top panel, lane 3). Consistent with the notion that TRAM would provide a link between TLR2 and MyD88, GST-TRAM also interacted with MyD88 (Fig. 7C, top panel, lane 3). In agreement with published data (47), GST-TRAM also interacted with Mal in this assay (Fig. 7C, top panel, lane 2). Thus TRAM can interact with all the known components of the TLR2 complex, namely Mal, MyD88 and TLR2 itself.
Figure 7. TRAM interacts with TLR2 and MyD88.
(A) GST and GST-TRAM were analyzed by SDS-PAGE and Coomassie staining to demonstrate equal inputs for GST pulldown assays in (B) and (C). (B, C) HEK293T cells were transfected with 8 μg empty vector (EV), Flag-TLR2 or Flag-TRIF (B) or with 4 μg Flag-Mal or Flag-MyD88 (C). After 48 h, lysates were incubated with GST-TRAM or GST alone as indicated, and together with input lysates, were analyzed by SDS-PAGE and immunoblotting. Data are representative of at least three independent experiments * = nonspecific band.
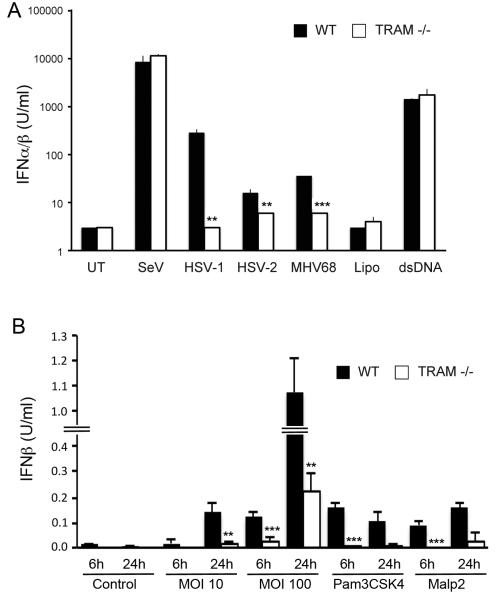
Pathogen-induced type I IFN is TRAM-dependent
Since TRAM-dependent TLR2-stimulated type I IFN would likely be relevant for anti-viral innate immune sensing, we tested whether herpes virus-induced type I IFN was TRAM-dependent in mouse macrophages. HSV, an alphaherpesvirus, is detected by TLR2 and TLR9 through recognition of a still unidentified viral surface component and viral genomic DNA, respectively (22, 50-52), while MHV68 induces type I IFN via TLR2 in MEFs and in vivo (33). Here, infection of WT iBMDMs with HSV-1, HSV-2 or MHV68 led to induction of type I IFN as measured by bioassay (Fig. 8A). This herpes virus-stimulated type I IFN induction was strongly impaired in TRAM−/− cells (Fig. 8A). In contrast, type I IFN produced by Sendai virus infection, or dsDNA transfection, of BMDMs was identical in WT or TRAM−/− BMDMs (Fig. 8A). These data suggest a role for TRAM in the TLR2-dependent type-I IFN response to HSV and MHV68. We also tested the ability of S.aureus, a gram positive bacterium for which TLR2 is a major PRR in macrophages and in vivo (53-58), to stimulate IFN-β production BMDMs. Fig. 8B shows that infection of WT BMDMs with S.aureus strain SH1000 at an MOI of 10 produced a comparable amount of IFN-β production as the pure TLR2 ligands Pam3CSK4 and Malp2, and even higher IFN-β production than the pure TLR2 ligands at an MOI of 100. Similar to the case for Pam3CSK4 and Malp2, live S.aureus-induced IFN-β was significantly impaired in TRAM−/− BMDMs compared to WT cells, for all MOIs and time points tested (Fig. 8B), indicating that in addition to pure TLR2 ligands, S.aureus-induced IFN-β in macrophages is TRAM-dependent.
Figure 8. TRAM-dependent pathogen-induced type I IFN.
(A) WT or TRAM-deficient iBMDMs were left untreated (UT) or infected with herpes viruses HSV-1 (MOI of 3), HSV-2 (MOI of 3), MHV68 (MOI of 50), or with SeV (MOI of 0.001), or transfected with lipofectamine (Lipo) alone or with 1 μg of dsDNA, all for 10 h. Supernatants were harvested and the amount of type I IFN produced was determined by bioassay. (B) WT or TRAM-deficient iBMDMs were left untreated (Control), stimulated with Pam3CSK4 or Malp2, or infected with S. aureus strain SH1000 at MOIs of 10 and 100 for the indicated times. Supernatants were harvested and IFN-β production measured by ELISA. The data are mean ± SD of triplicate samples and are representative of two independent experiments. **p<0.005 or ***p<0.0005 compared to samples from WT iBMDMs.
Discussion
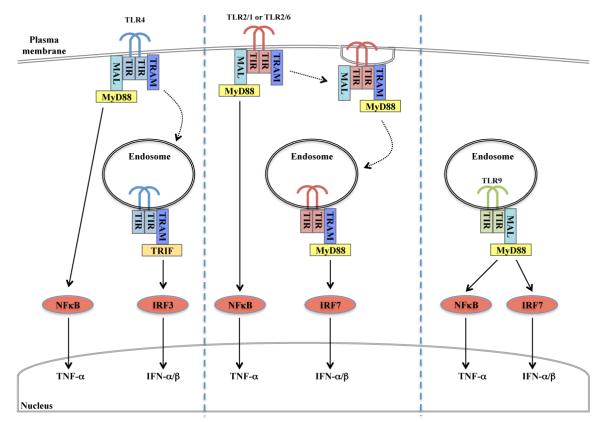
In recent years there has been a growing appreciation that upon encountering either viral or bacterial ligands, TLR2 heterodimers not only signal induction of pro-inflammatory cytokines, but also of type I IFN. This implied that similar to TLR4, and to the well-characterized endosomal TLRs such as TLR3 and TLR7, TLR2 signaling would activate IRFs. Indeed, Dietrich et al showed that induction of IFN-β by bacterial ligands of TLR2 was IRF7-dependent in BMDMs (31). They also showed that similar to other TLRs, TLR2 signaling to IRFs occurred at endosomes, and not at the plasma membrane. However, apart from the requirement of MyD88 for TLR2-stimulated IRF7 activation, it was unclear how TLR2 signaling at the endosome would be enabled, and in particular whether other TIR adaptor proteins apart from MyD88 are required. For TLR4, TRAM is known to be the key sorting adaptor that facilitates endosomal-dependent signaling, in that case via TRIF.
Here we show that TLR2-dependent IRF activation at the endosome requires both Mal and TRAM, and that TRAM is required for the TLR2-stimulated movement of MyD88 to endosomes following ligand engagement. This pathway operates for both TLR2/1 and TLR2/6 receptor complexes since we always obtained almost identical results for Pam3CSK4 and Malp2. Furthermore we demonstrate that TRAM interacts with both TLR2 and MyD88, and that both herpes virus-and S. aureus-stimulated type I IFN induction in BMDMs is TRAM-dependent. Thus our work reveals a novel and broader role for TRAM than was previously appreciated, in that TRAM acts as a sorting adaptor not only for TLR4, but also for TLR2, to facilitate signaling to IRF7 at the endosome. Our data do not exclude the possibility that other IRFs apart from IRF7 are also involved in TRAM-dependent induction of type I IFNs by TLR2. Indeed, Dietrich et al showed that in BMDMs lacking either IRF7 or IRF1 (but not IRF3), TLR2-dependent IFN-β production was impaired (31), while Liljeroos et al showed that in RAW264.7 macrophages both IRF1 and IRF2 had a role in TLR2-dependent IFN-α production (44). Whether other IRFs such as IRF5 might also be involved in TLR2 response pathways remains to be determined, as does the exact mechanism whereby TRAM would regulate a specific IRF downstream of TLR2.
Interestingly, a role for TRAM in TLR2 signaling was previously hinted at through the use of dominant-negative TRAM (59). Sacre et al used an adenoviral construct expressing TRAM mutated in the key signaling BB loop domain (TRAM-C117H) and observed that this mutant inhibited LPS- and LTA- (i.e. TLR2-) induced NF-κB activation and cytokine production in human synovial fibroblasts, human umbilical endothelial cells and MEFs, but not in human macrophages. Thus they concluded that TRAM is an adaptor protein for both TLR4 and TLR2/6 signaling in specific cell types (59). Here we used three different approaches to examine the role of TRAM in TLR2 signaling, namely the VIPER peptide inhibitor of TRAM, TRAM siRNA and TRAM−/− iBMDMs. These approaches revealed that TRAM was required for IRF7 activation and IFNβ induction, but not for TLR2-dependent NF-κB activation or pro-inflammatory cytokine production. Other studies using TRAM-deficient murine peritoneal macrophages (60) or TRAM dominant negative in HEKTLR2 cells (61) concur with our conclusion of no role for TRAM in pro-inflammatory signaling initiated by TLR2. However, the exact role of TRAM in TLR2 responses may vary in different cell types.
The role of TRAM in TLR2-stimulated IRF activation revealed here is reminiscent of a similar requirement of TLR4 for TRAM. TLR4 initially signals at the plasma membrane (via Mal and MyD88) and is then internalized into early endosomes, where signaling switches to utilizing TRAM and TRIF (Fig. 9) (7, 62). Thereafter, TLR4 moves to late endosomes where TRIF-dependent signaling is suppressed by TRAM adaptor with GOLD domain, a splice variant of TRAM (48). Endosomal location and IRF signaling for TLR4 are both dependent on TRAM, and compromised when the myristoylation motif of TRAM is mutated. Here we showed TLR2-stimulated re-localization of MyD88 to intracellular compartments was TRAM-dependent, in the same way that TLR4-stimulated relocalization of TRIF to endosomes is TRAM-dependent. Further, TRAM interacted with both TLR2 and MyD88, consistent with the notion that it acts as a bridging adaptor to link TLR2 and MyD88, as well as a sorting adaptor to facilitate TLR2 and MyD88 locating at the endosome.
Figure 9. Role of TRAM in endosomal TLR2-stimulated IRF activation.
Model for TRAM-dependent endosomal signaling to IRF7 for TLR2 (center), compared to endosomal TLR4 (left) and TLR9 (right) signaling. See text for details.
That TRAM and MyD88 interact was also recently demonstrated by Ohnishi et al, who also showed that TRAM was required for IL-18 signaling and thus suggested a role for TRAM as a bridging adapter between MyD88 and the IL-18R (63). How exactly TRAM engages with MyD88 remains to be determined, although those authors showed that the binding site for TRAM on MyD88 overlaps with the binding site used by MyD88 to engage Mal (63). Therefore during TLR2 signaling, it is conceivable that MyD88 may be directly transferred from Mal to TRAM as the TLR2 complex moves from the plasma membrane (Fig. 9), or alternatively the TLR2 signaling complex may engage MyD88 via TRAM from the outset of initiation of signaling. Discriminating between these possibilities will require further studies.
TLR2 has previously been shown to be internalized after ligand engagement (64). In that study the authors concluded that TLR2 internalization was not actually required for signaling; however they only examined NF-κB activation, which we showed here does not require TRAM or endocytosis. Apart from TRAM, we also showed that Mal, which is required for some but not all TLR2-dependent signaling events (16, 17), is essential for TLR2-dependent IRF7 activation and IFNβ induction. It has been hypothesized that the reason TLRs signal to IRFs from the endosome and not the plasma membrane is because TRAF3 can only engage with TLR signaling at the former location (7). Whether TRAF3 is required for the endosomal TLR2-TRAM-IRF pathway remains to be confirmed. Which downstream kinases activate IRFs after TLR2-TRAM signaling at the endosome also remains to be determined.
The data lead to the following model for TRAM involvement in TLR2 signaling to IRFs for type I IFN induction (Fig 9): Upon ligand engagement at the plasma membrane, TLR2 recruits MyD88 via Mal, giving rise to NF-κB activation. This part of the signaling process for TLR2 is TRAM-independent (Fig. 4A). Thereafter, a TLR2-TRAM-MyD88 complex would be endocytically internalized. This leads to formation of a TLR2-signaling competent endosome to affect IRF7 activation. Within this part of the signaling process, Mal might directly transfer MyD88 to TRAM to license the complex for endosomal localization and subsequent signaling. Alternatively, Mal itself may also be retained in the endosomal TLR2 signaling complex. Since our data show that both Mal and TRAM were required for IRF7 activation, further work will be required to distinguish these two possibilities. Interestingly, it was shown very recently that for natural ligands, TLR9 also needs a sorting adaptor for endosomal signaling, apart from the signaling adaptor MyD88, which in that case was shown to be Mal (Fig 9) (65). Thus it will be of interest to re-examine whether other endosomal TLRs (TLR3, TLR7, TLR8) have any requirement for TRAM and/or Mal for signaling, which might be the case in certain cell types or for particular ligands.
Acknowledgements
We thank Drs K. Fitzgerald, D. Golenbock, S. Sato, L. O’Neill and J. McCarthy for the kind gift of expression plasmids and cell lines.
Grant support: This work was supported by Science Foundation Ireland grants 11/PI/1056 and 07/SRC/B1144 (to A.G.B.), grants from The National Children’s Research Centre (Paediatric Research Immunology Programme) and The BrightFocus Foundation (to S.L.D), the Lundbeck Foundation (S.R.P.), and a Welcome Trust RCDF (WT086515MA) to RMcL.
Abbreviations used
- BMDM
bone marrow-derived macrophage
- IRAK
IL-1R associated kinase
- IRF
IFN regulatory factor
- Mal
MyD88-adaptor like
- MHV68
murine gammaherpesvirus 68
- TIR
Toll/IL-1R
- TRAF
TNFR-associated factor
- TRIF
TIR domain-containing adaptor inducing IFN-β
- TRAM
TRIF-related adaptor molecule
- TRIF
TIR domain containing adaptor inducing IFN-β
- VACV
Vaccinia Virus
- VIPER
Viral inhibitory peptide of TLR4
- WT
wild type
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14:103, 110. doi: 10.1016/s0952-7915(01)00304-1. [DOI] [PubMed] [Google Scholar]
- 4.Gay NJ, Gangloff M, O’Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32:104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 7.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill LA, Bowie AG. The family of five: TIR-domaincontaining adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 9.Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O’Neill LA, Fitzgerald KA, Golenbock DT. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci U S A. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends in microbiology. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 12.Kirschning CJ, Schumann RR. TLR2: cellular sensor for microbial and endogenous molecular patterns. Curr Top Microbiol Immunol. 2002;270:121–144. doi: 10.1007/978-3-642-59430-4_8. [DOI] [PubMed] [Google Scholar]
- 13.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Morath S, Stadelmaier A, Geyer A, Schmidt RR, Hartung T. Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J Exp Med. 2002;195:1635–1640. doi: 10.1084/jem.20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deininger S, Stadelmaier A, von Aulock S, Morath S, Schmidt RR, Hartung T. Definition of structural prerequisites for lipoteichoic acidinducible cytokine induction by synthetic derivatives. J Immunol. 2003;170:4134–4138. doi: 10.4049/jimmunol.170.8.4134. [DOI] [PubMed] [Google Scholar]
- 16.Santos-Sierra S, Deshmukh SD, Kalnitski J, Kuenzi P, Wymann MP, Golenbock DT, Henneke P. Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 2009;28:2018–2027. doi: 10.1038/emboj.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny EF, Talbot S, Gong M, Golenbock DT, Bryant CE, O’Neill LA. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J Immunol. 2009;183:3642–3651. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- 18.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 19.Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, Finberg RW. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J Virol. 2003;77:4588–4596. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szomolanyi-Tsuda E, Liang X, Welsh RM, Kurt-Jones EA, Finberg RW. Role for TLR2 in NK cell-mediated control of murine cytomegalovirus in vivo. J Virol. 2006;80:4286–4291. doi: 10.1128/JVI.80.9.4286-4291.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato A, Linehan MM, Iwasaki A. Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells. Proc Natl Acad Sci U S A. 2006;103:17343–17348. doi: 10.1073/pnas.0605102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci U S A. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang S, Dolganiuc A, Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. Journal of leukocyte biology. 2007;82:479–487. doi: 10.1189/jlb.0207128. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Kurt-Jones EA, Mandell L, Cerny A, Chan M, Golenbock DT, Finberg RW. MyD88 is critical for the development of innate and adaptive immunity during acute lymphocytic choriomeningitis virus infection. Eur J Immunol. 2005;35:822–830. doi: 10.1002/eji.200425730. [DOI] [PubMed] [Google Scholar]
- 25.Bieback K, Lien E, Klagge IM, Avota E, Schneider-Schaulies J, Duprex WP, Wagner H, Kirschning CJ, Ter Meulen V, Schneider-Schaulies S. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J Virol. 2002;76:8729–8736. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Martinez J, Huang X, Yang Y. Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood. 2007;109:619–625. doi: 10.1182/blood-2006-06-027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 28.Hoshino K, Kaisho T, Iwabe T, Takeuchi O, Akira S. Differential involvement of IFN-beta in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 2002;14:1225–1231. doi: 10.1093/intimm/dxf089. [DOI] [PubMed] [Google Scholar]
- 29.Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 30.Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ, Vogel SN. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 31.Dietrich N, Lienenklaus S, Weiss S, Gekara NO. Murine tolllike receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS One. 2010;5:e10250. doi: 10.1371/journal.pone.0010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaud F, Coulombe F, Gaudreault E, Kriz J, Gosselin J. Involvement of TLR2 in recognition of acute gammaherpesvirus-68 infection. PLoS One. 2010;5:e13742. doi: 10.1371/journal.pone.0013742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med. 2005;201:1007–1018. doi: 10.1084/jem.20041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stack J, Bowie AG. Poxviral protein A46 antagonizes Toll-like receptor 4 signaling by targeting BB loop motifs in Toll-IL-1 receptor adaptor proteins to disrupt receptor:adaptor interactions. The Journal of biological chemistry. 2012;287:22672–22682. doi: 10.1074/jbc.M112.349225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lysakova-Devine T, Keogh B, Harrington B, Nagpal K, Halle A, Golenbock DT, Monie T, Bowie AG. Viral inhibitory peptide of TLR4, a peptide derived from vaccinia protein A46, specifically inhibits TLR4 by directly targeting MyD88 adaptor-like and TRIF-related adaptor molecule. J Immunol. 2010;185:4261–4271. doi: 10.4049/jimmunol.1002013. [DOI] [PubMed] [Google Scholar]
- 37.Fedosyuk S, Grishkovskaya I, de Almeida Ribeiro E, Jr., Skern T. Characterization and structure of the vaccinia virus NF-kappaB antagonist A46. The Journal of biological chemistry. 2014;289:3749–3762. doi: 10.1074/jbc.M113.512756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verstak B, Nagpal K, Bottomley SP, Golenbock DT, Hertzog PJ, Mansell A. MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-kappaB proinflammatory responses. The Journal of biological chemistry. 2009;284:24192–24203. doi: 10.1074/jbc.M109.023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O’Neill LA. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorensen LN, Reinert LS, Malmgaard L, Bartholdy C, Thomsen AR, Paludan SR. TLR2 and TLR9 synergistically control herpes simplex virus infection in the brain. J Immunol. 2008;181:8604–8612. doi: 10.4049/jimmunol.181.12.8604. [DOI] [PubMed] [Google Scholar]
- 42.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. Journal of bacteriology. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maher BM, Mulcahy ME, Murphy AG, Wilk M, O’Keeffe KM, Geoghegan JA, Lavelle EC, McLoughlin RM. Nlrp-3-driven interleukin 17 production by gammadeltaT cells controls infection outcomes during Staphylococcus aureus surgical site infection. Infection and immunity. 2013;81:4478–4489. doi: 10.1128/IAI.01026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liljeroos M, Vuolteenaho R, Rounioja S, Henriques-Normark B, Hallman M, Ojaniemi M. Bacterial ligand of TLR2 signals Stat activation via induction of IRF1/2 and interferon-alpha production. Cell Signal. 2008;20:1873–1881. doi: 10.1016/j.cellsig.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 46.Hise AG, Daehnel K, Gillette-Ferguson I, Cho E, McGarry HF, Taylor MJ, Golenbock DT, Fitzgerald KA, Kazura JW, Pearlman E. Innate immune responses to endosymbiotic Wolbachia bacteria in Brugia malayi and Onchocerca volvulus are dependent on TLR2, TLR6, MyD88, and Mal, but not TLR4, TRIF, or TRAM. J Immunol. 2007;178:1068–1076. doi: 10.4049/jimmunol.178.2.1068. [DOI] [PubMed] [Google Scholar]
- 47.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, Gong M, Golenbock D, Espevik T, O’Neill LA. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- 49.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E, Mollnes TE, Bakke O, Espevik T. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J Immunol. 2005;175:4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 51.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 52.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe I, Ichiki M, Shiratsuchi A, Nakanishi Y. TLR2-mediated survival of Staphylococcus aureus in macrophages: a novel bacterial strategy against host innate immunity. J Immunol. 2007;178:4917–4925. doi: 10.4049/jimmunol.178.8.4917. [DOI] [PubMed] [Google Scholar]
- 54.Schmaler M, Jann NJ, Ferracin F, Landolt LZ, Biswas L, Gotz F, Landmann R. Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J Immunol. 2009;182:7110–7118. doi: 10.4049/jimmunol.0804292. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 56.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 58.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 59.Sacre SM, Lundberg AM, Andreakos E, Taylor C, Feldmann M, Foxwell BM. Selective use of TRAM in lipopolysaccharide (LPS) and lipoteichoic acid (LTA) induced NF-kappaB activation and cytokine production in primary human cells: TRAM is an adaptor for LPS and LTA signaling. J Immunol. 2007;178:2148–2154. doi: 10.4049/jimmunol.178.4.2148. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 61.Ding A, Yu H, Yang J, Shi S, Ehrt S. Induction of macrophage-derived SLPI by Mycobacterium tuberculosis depends on TLR2 but not MyD88. Immunology. 2005;116:381–389. doi: 10.1111/j.1365-2567.2005.02238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 63.Ohnishi H, Tochio H, Kato Z, Kawamoto N, Kimura T, Kubota K, Yamamoto T, Funasaka T, Nakano H, Wong RW, Shirakawa M, Kondo N. TRAM is involved in IL-18 signaling and functions as a sorting adaptor for MyD88. PLoS One. 2012;7:e38423. doi: 10.1371/journal.pone.0038423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nilsen NJ, Deininger S, Nonstad U, Skjeldal F, Husebye H, Rodionov D, von Aulock S, Hartung T, Lien E, Bakke O, Espevik T. Cellular trafficking of lipoteichoic acid and Toll-like receptor 2 in relation to signaling: role of CD14 and CD36. Journal of leukocyte biology. 2008;84:280–291. doi: 10.1189/jlb.0907656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonham KS, Orzalli MH, Hayashi K, Wolf AI, Glanemann C, Weninger W, Iwasaki A, Knipe DM, Kagan JC. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–716. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]