Abstract
The Groucho transcriptional corepressor TLE1 protein has recently been shown to be a putative lung specific oncogene, but its underlying oncogenic activity in lung cancer has not been fully elucidated. In this report, we investigated whether TLE1 regulates lung cancer aggressiveness using the human lung adenocarcinoma cell line A549 as a model system. Through a combination of genetic approaches, we found that TLE1 potentiates Epithelial-to-Mesenchymal Transition (EMT) in A549 cells in part through suppression of the tumor suppressor gene E-cadherin. Exogenous expression of TLE1 in A549 cells resulted in heightened EMT phenotypes (enhanced fibroblastoid morphology and increased cell migratory potential) and in molecular alterations characteristic of EMT (downregulation of the epithelial marker E-cadherin and upregulation of the mesenchymal marker Vimentin). Conversely, downregulation of endogenous TLE1 expression in these cells resulted in reversal of basal EMT characterized by a cuboidal-like epithelial cell phenotype, reduced cell motility, and upregulated E-cadherin expression. Mechanistic studies showed that TLE1 suppresses E-cadherin expression at the transcriptional level in part by recruiting Histone Deacetylase (HDAC) activity to the E-cadherin promoter. Consistently, the HDAC inhibitor TSA partially reversed the TLE1-induced E-cadherin downregulation and cell migration, suggesting a role for HDACs in TLE1-mediated transcriptional repression of E-cadherin and EMT function. These findings uncover a novel role of TLE1 in regulating EMT in A549 cells through its repressive effect on E-cadherin and provide a mechanism for TLE1 oncogenic activity in lung cancer.
Introduction
TLE1 is a member of the Groucho (Gro)/TLE family of transcriptional co-repressors that regulate the transcriptional activity of a wide range of genes (1). As a co-repressor, the TLE1 protein does not bind to DNA directly but is recruited to target gene (s) by direct interaction with DNA-binding transcription factors to form large multi-protein complexes (2). Alternatively, it has also been shown that TLE1 may interact with chromatin through its interactions with the amino-terminal tail of histone H3 (3). The exact mechanism underlying the transcriptional repression function of TLE1 is yet to be fully elucidated. Numerous data indicate that the TLE1 gene silencing function appears to depend in part on recruitment of the histone deacetylase (HDAC) protein to target genes and the subsequent removal of acetyl groups from nearby DNA bound histones. Indeed, the interaction of TLE1 with HDAC1 in particular has been demonstrated using genetic and biochemical approaches (4,5). Importantly, inhibition of HDAC activity via chemical inhibitors partially blocks TLE1 repressive activity, suggesting the importance of HDAC activity on TLE1 repressive function (6). Based on the inability of HDAC inhibitors to completely block the TLE1 repressive function, it is likely that TLE1 may depend on other chromatin remodelling proteins to effect transcriptional repression (6).
As transcriptional corepressors, the Groucho/TLE proteins regulate diverse cellular functions including neurogenesis and a number of developmental processes (7,8). Recently, a series of independent findings indicate a survival promoting role of groucho proteins, TLE1 in particular, in several cellular models. Exogenous TLE1 expression stimulated anchorage-independent growth in chicken embryo fibroblast (9). As a negative regulator of apoptosis, TLE1 inhibited low potassium-induced apoptosis in neurons (10) and protected synovial sarcoma cells from doxorubicin-mediated apoptosis (11). In line with its anti-apoptotic function, we have found that TLE1 specifically inhibited the caspase-independent cell death induced by Bit1 (Bcl2-inhibitor of transcription 1) in several types of malignant cells (12). Recently, we showed that TLE1 effectively abrogates Bit1-induced anoikis in lung cancer cells (13). Based on the latest report indicating that TLE1 is a putative lung specific oncogene (14), it is interesting to speculate whether the anti-apoptotic function of TLE1 represents as an oncogenic stimulus. In line with its function as a regulator of a survival-promoting gene transcription program, TLE1 was shown to positively regulates Bcl2 expression (12) and ErbB1 and ErbB2 signaling (14), two survival pathways that impact cancer progression.
As a putative lung specific oncogene, TLE1 was found to be overexpressed in a subset of aggressive and advanced human lung tumors (14). In light of this finding, we examined the possibility that TLE1 may regulate lung cancer aggressiveness. Here, we have uncovered a novel function of TLE1 in regulating EMT in the human lung adenocarcinoma cell line A549. Importantly, our findings indicate that TLE1 mediates transcriptional silencing of the invasive suppressor E –cadherin gene in part through recruitment of HDAC activity, and further suggest TLE1 as a novel in vivo repressor of E-cadherin in lung cancer.
Materials and Methods
Cell culture and transfection assays
The human lung adenocarcinoma cell line A549 from American Type Culture Collection (ATCC) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with glutamine containing 10% fetal bovine serum, penicillin, and streptomycin. Stable A549 control GFP and GFP-TLE1 pool of cells were generated by transduction with the lentiviral GFP-TLE1 or the empty control GFP construct (Open Biosystems, Huntsville, AL) according to the manufacturer’s protocol. 48 hours post-transduction, cells were cultured in the 20µg/ml Blasticidin S (Invitrogen) to select for positive GFP-TLE1 expressing clones. Several Blasticidin S resistant control-GFP and GFP-TLE1 clones were harvested by ring-cloning, and the level of GFP-TLE1 expression was confirmed by immunoblotting against a specific GFP antibody. Two control GFP clones and three positive GFP-TLE1 clones were pooled together to generate the control GFP and GFP-TLE1 pools, respectively.
Chemical reagents, antibodies, and plasmids
The mouse monoclonal anti-myc, anti-GFP, and anti-B-actin antibodies were purchased from Sigma (St. Louis, MO). The mouse polyclonal anti-TLE1 and the anti-HDAC1 antibodies were obtained from Abcam (Cambridge, MA). The histone deacetylase inhibitor Trichostatin A (TSA) was purchased from Calbiochem (La Jolla, CA).
SiRNA transfection
The TLE1 specific siRNAs and the control, non-targeting siRNAs pools were purchased from Invitrogen (13, Carlsbad, CA). For transient transfection experiments, A549 cells (2 × 105) were transfected with 25 µM of each siRNA pool by using the Lipofectamine RNAiMAX transfection reagent (Invitrogen). 48 hrs post-transfection, cells were harvested and subjected to immunoblotting, migration, real-time PCR, or reporter assays as described below.
Analysis of cell proliferation and migration
To determine anchorage-dependent growth, cells were plated in a volume of 150 µl at a density of 2,000 cells per well in 96-well plates. At each indicated time, the number of metabolically active cells was measured with the use of MTT assay as described previously (13). Briefly, MTT reagent (Sigma) was dissolved in sterile PBS at 5 mg/ml. Ten microliters of this solution was added to each well in a 96-well plate (1:10 dilution). The plate was then incubated at 37°C, 5% CO2 for 3h. Afterwards, the medium was gently aspirated away and the MTT precipitate was dissolved in 100 ul of a 50% MeOH-50% DMSO solution. The precipitate was allowed to dissolve at room temperature for ten minutes with gentle shaking. The resulting 550 nm absorbance was read on a microplate reader (BioTek Instruments).
The migration potential of cells was determined by a wound closure and the Boyden chamber cell migration assays. In wound closure experiments, cell monolayers were scarred with a sterile micropipette tip and incubated for another 16 h. For each sample, three defined areas were monitored during the time period and photographs were taken at time 0h and 16 h (magnification, X100). In Boyden chamber migration assays, we seeded 2.5×104 cells/well in the presence or absence of 300nM TSA into the top chambers of a 24-well, 8-µm pore-size micropore polycarbonate membrane filter (BD Biosciences), and the lower chambers were filled with DMEM containing 10% FBS as a chemoattractant and incubated for 20h at 37°C. Cells remaining on the upper surface were carefully removed with a cotton swab, and the membranes were fixe 10% phospho-buffered formalin, permeabilized with 100% ice-cold methanol, and stained with 0.1% crystal violet in 20% methanol. Data are represented as the average number of migrated cells per field (20 random 20× magnification fields) per membrane filter.
Protein preparation and western blotting assays
Protein preparation and western blotting were performed as described previously (13). Briefly, cells were harvested by adding ice-cold NP-40 lysis buffer (1% NP-40; 20mM Tris-HCL [pH7.4]; 150 mM NaCl; 10% glycerol, 2 mM sodium vanadate; 1 mM henylmethylsulfonyl fluoride; 10 µg/ml leupeptine; and 5 µg/ml aprotinin) and incubating at 4°C for 20 min. For immunoblot analysis, equal amounts of proteins were resolved on 4–20% gradient Tris-glycine gels (Invitrogen) and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with primary antibodies overnight at 4°C followed by secondary antibodies conjugated with horseradish peroxidase. Membranes were developed using the ECL detection system.
Coimmunoprecitation assay
Coimmunoprecipitation assay was performed as described previously (13). Briefly, control GFP and GFP-TLE1 cells were harvested by washing once with PBS and resuspended in ice-cold Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 2 mm sodium vanadate, 1 mm phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, and 5 µg/ml aprotinin) followed by a 20-min incubation at 4 °C. Cell debris was removed by centrifugation. GFP-tagged TLE1 was immunoprecipitated with anti-GFP-agarose conjugate (Abcam) and thoroughly washed with lysis buffer. Bound proteins were resolved by SDS-PAGE, and Western blotting was performed using anti-HDAC1 antibody.
Histone deacetylase (HDAC) activity assay
HDAC activity was measured using the HDAC Fluorometric Activity assay (Sigma). Briefly, control GFP and GFP-TLE1 cells treated or untreated with 300 nM TSA were immunoprecipitated with the anti-GFP-agarose conjugate (Abcam) as described above. The resulting immunoprecitated fractions were used to assay for associated HDAC activity. The immunoprecipitate preparation was diluted with the assay buffer (50 ul) followed by addition of fluorogenic HDAC substrate (50 ul), and the reaction mixtures were incubated for 30 minutes at 30Cº. The developer solution was then added to each sample which was incubated for an additional 10 minutes. The fluorescence of the reaction mixture was determined at excitation 360nm/emission 460nm using the SPECTRAmax Gemini plate reader (Molecular Devices).
Total RNA extraction and Quantitative Real Time PCR
As described previously (13), total RNA was extracted from 1.0×106 cultured cells using the RNeasy kit (Qiagen) and quantified by spectrophotometry (NanoDrop 8000, Thermo Scientific). Total RNA was reversed transcribed using the Superscript First-Strand Synthesis Kit for RT-PCR (Invitrogen) as prescribed by the supplier. cDNA was quantified by real-time PCR on the ABI Prism 7900 Sequence Detection System (Applied Biosystems) with the following human E-cadherin (forward primer: AGGCTAGAGGGTCACCGCGTC and reverse primer: GCTTTGCAGTTCCGACGCCAC) using the Sybr Green PCR Core reagents (Applied Biosystems, Foster City, CA). Amplification of the same cDNAs with human GAPDH primers (forward primer: CCCACTCCTCCACCTTTGAC and reverse primer: TTGCTGTAGCCAAATTCGTTGT) was used for internal normalization.
Promoter luciferase analysis
Control GFP and GFP-TLE1 cells treated or untreated with 300nM TSA (Sigma) were cotransfected with the E-cadherin luciferase promoter-reporter construct (SwitchGear Genomics) and the GAPDH luciferase promoter-reporter vector (SwitchGear Genomics) as an internal control for luciferase activity. Following 24h of incubation, the cells were subjected to a luciferase assay (SwitchGear Genomics’ LightSwitch Luciferase Assay System) following the manufacturer’s protocol. Luciferase activity was normalized to GAPDH luciferase activity and the relative luciferase activity was presented. To determine the effect of TLE1 knockdown on E-cadherin promoter activity, A549 cells were initially transfected with control or TLE1 specific siRNA pools, and 24 h later, cells were cotransfected with the E-cadherin luciferase promoter-reporter construct (SwitchGear Genomics) and the GAPDH luciferase promoter-reporter vector (SwitchGear Genomics). Following 24h of incubation, the cells were subjected to a luciferase assay (SwitchGear Genomics’ LightSwitch Luciferase Assay System) as described above. Each of the above experiment was performed in triplicate.
ChIP assay
The control GFP and GFP-TLE1 A549 cells grown to 70% to 80 % confluence were crosslinked with 1% formaldehyde and processed using the EpiSeeker ChIP Kit – One Step (Abcam). The resulting chromatin fragments were immunoprecipitated with the anti-GFP antibody-ChIP Grade (Abcam), anti-Acetyl-Histone H3-ChIP (anti-Ac-H3) grade (EMD MILLIPORE), or non specific IgG (Santa Cruz Biotechnology). Subsequent downstream steps were conducted following the protocol from the EpiSeeker ChIP Kit (Abcam). The PCRs were conducted using the E-cadherin primers (5'-CCCACCACGTACAAGGGTC-3` (sense), 5'-ATGCCATCGTTGTTCACTGGA-3` (antisense)) with the following program: 45 cycles at 95°C for 30 s, 56°C for 30s, and 72°C for 30s as described previously (20). The amplified DNA fragment of the E-cadherin promoter was separated on 1.5% agarose gel and visualized with ethidium bromide. The sensitivity of PCR amplification was evaluated using serial dilutions of the total DNA collected after sonication (input fraction). Each ChIP experiment was repeated at least three times.
Statistical Analysis
Data are presented as means (±S.E.). For western blots and ChIP assays, experiments were performed at least three times with duplicates. Statistical differences between groups were established at a P value < 0.05 using the two-tailed Student’s t test. All calculations were done using the NCSS statistical software (NCSS, Kasville, UT).
Results and Discussion
TLE1 expression induces EMT in A549 cells
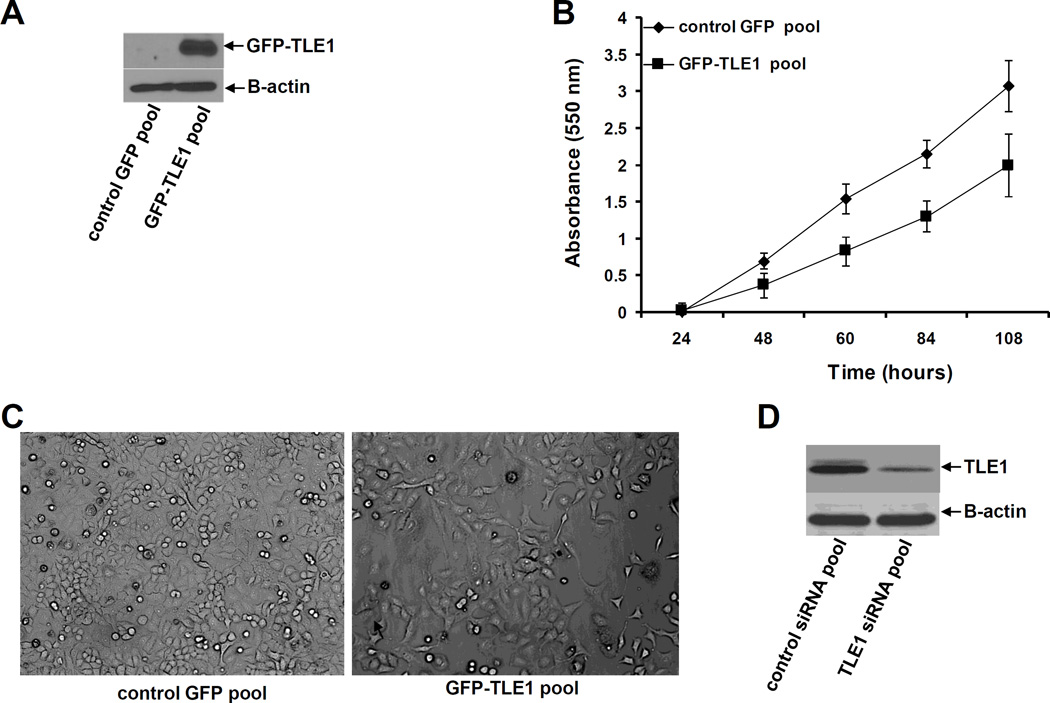
To address the hypothesis that TLE1 may play a role in aggressiveness of lung cancer cells, we generated stable TLE1 expressing clones by infecting the human adenocarcinoma A549 cell line with lentiviral expression vectors containing either the GFP-tagged TLE1 or control GFP alone construct. Several GFP-TLE1 and control GFP clones were pooled together to generate the GFP-TLE1 pool and control GFP pool, respectively. Exogenous TLE1 expression in the GFP-TLE1 pool was confirmed by immunoblotting using the anti-GFP antibody (Fig. 1A). Characterization of the control GFP and GFP-TLE1 pool showed that the exogenous TLE1 expressing cells exhibited significantly reduced anchorage-dependent growth kinetics as compare to control cells (Fig. 1B). Furthermore, we observed a striking difference in their morphology. The stable GFP-TLE1 pool showed a flatter, stretched fibroblast-like appearance and was more dispersed than the control GFP pool (Fig. 1C). To confirm the specificity of ectopic TLE1 effect on these morphological changes, we also knockdown endogenous TLE1 in A549 cells which exhibit moderate levels of TLE1 with the use of a previously validated specific TLE1 siRNA SMART pool (13). The TLE1 siRNA duplexes showed a significant 60–80% downregulation of endogenous TLE1 expression as compared to that of control siRNA cells (Fig. 1D). Importantly, targeted reduction of TLE1 resulted in a more prominent epithelial cell morphology (Fig. 1E). The TLE1-siRNA treated cells exhibited a cobblestone-like and a more compact appearance while the control-siRNA cells maintained their spindle-shaped morphology. These morphological changes associated with TLE1 alteration suggest that TLE1 promotes EMT. To further address this possibility, we examined the effect of TLE1 on molecular markers of EMT (Fig. 1F). In line with the inductive effect of TLE1 on EMT, the stable GFP-TLE1 pool exhibited a significant reduction in the basal expression of the epithelial marker E-cadherin with concomitant high expression of the mesenchymal marker Vimentin as compared to the control GFP pool. Importantly, acute ablation of TLE1 expression dramatically resulted in the induction of E-cadherin expression with no effect on the expression of Vimentin, a late stage EMT marker (Fig. 1F).
Fig. 1.
TLE1 induces EMT and suppresses E-cadherin expression in A549 cells. A. Stable control GFP and GFP-TLE1 pool of cells derived from the A549 cell line were subjected to total cell lysate isolation, SDS-PAGE, and immunoblotting against specific antibodies to GFP (to confirm the expression of exogenous GFP-TLE1) and B-actin. B. The growth of control GFP and GFP-TLE1 cells was quantified by examining the number of metabolically active cells at various indicated time points with the use of MTT assay as described previously (13). C. The morphology of control GFP and GFP-TLE1 cells was examined by phase contrast microscopy (100× magnification) under normal culture conditions. D and E. A549 cells were transfected with control or TLE1 specific siRNAs pool, and 48 h post-transfection, cells were harvested and subjected to immunoblotting (D) with antibodies against TLE1 and viewed under a phase contrast microscope (E). F. Control GFP and GFP-TLE1 cells (left panel) and A549 cells treated with control or TLE1 siRNAs (right panel) were subjected to immunoblotting against specific antibodies to E-cadherin, Vimentin, and B-actin.
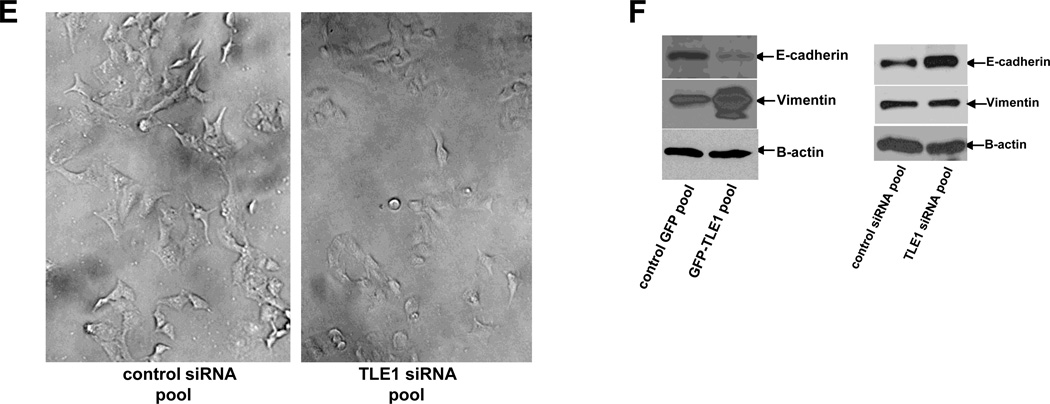
An important phenotype conferred by EMT is enhanced motility. To further examine the effect of TLE1 on EMT, the stable GFP-TLE1 and control GFP A549 cells were subjected to a wound closure assay. As shown in Fig. 2A, exogenous TLE1 accelerated the closure of the wound which remained relatively open in control cells. This result was confirmed using the Boyden Chamber migration assay. The ability of A549 cells to migrate to the bottom chamber was significantly enhanced by ectopic TLE1 (Fig. 2B). Consistently, knockdown of endogenous TLE1 in these cells reduced their basal migration potential (Fig. 2C).
Fig. 2.
TLE1 enhances motility of A549 cells. A. Stable control GFP and GFP-TLE1 pool of cells were treated to a wound healing experiment wherein cells were assessed to close a wound 16 h later by phase contrast microscopy. B and C. Control GFP and GFP-TLE1 cells (B) and A549 cells treated with control or TLE1 siRNAs (C) were subjected to a migration Boyden chamber assay wherein the number of cells that migrated to the bottom of the insert membrane was quantified. In B and C, results are representative of three independent experiments, *p<0.05 as compared with the control cells (Student’s t test).
TLE1 represses E cadherin expression at the transcriptional level
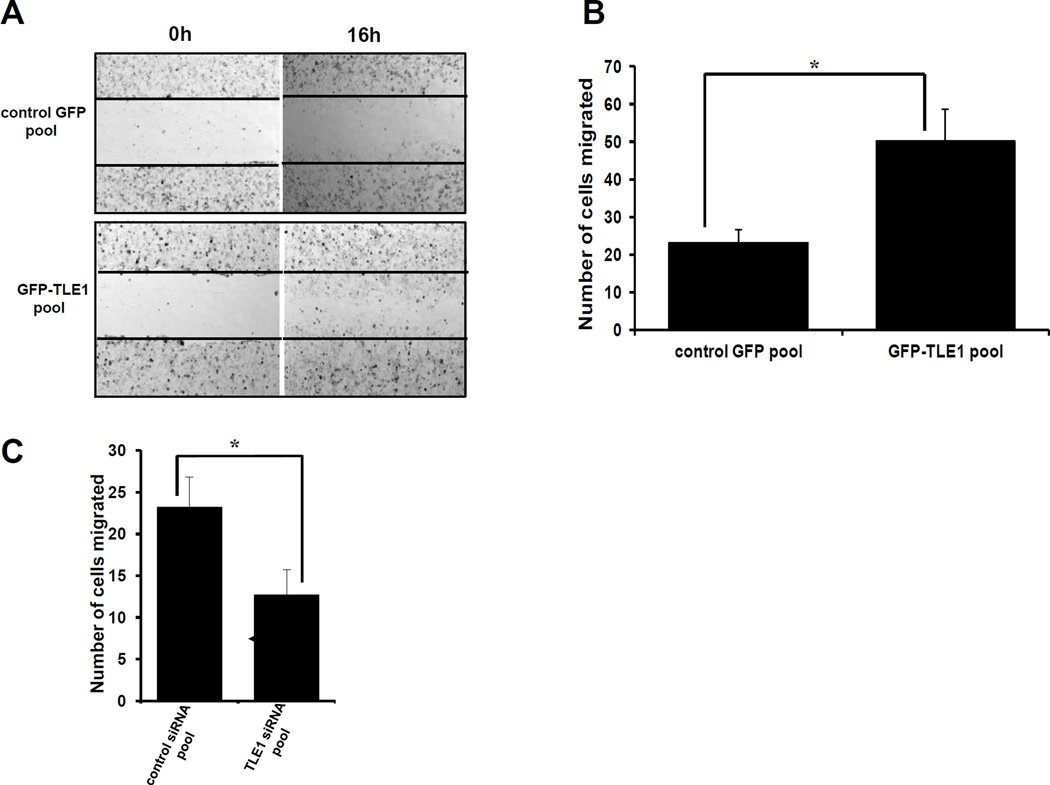
A critical hallmark and an early marker of EMT is the loss of the tumor suppressor epithelial marker E-cadherin expression. As a primary and direct target of EMT-promoting signals and due its important role in tumorigenicity (15–17), E-cadherin is a candidate gene targeted by the epigenetic regulator TLE1. To investigate the potential regulation of E-cadherin by TLE1, we examined whether TLE1 regulates E-cadherin expression at the transcriptional level via real-time PCR and promoter assays in A549 cells. In line with the observed reduction of E-cadherin protein, the GFP-TLE1 pool exhibited a significantly reduced level of E-cadherin mRNA transcript (Fig. 3A) as compared to control GFP cells. Consistently, the exogenous TLE1 expressing cells showed repressed activity of the transfected E-cadherin promoter luciferase construct (Fig. 3B). To confirm the transcriptional regulation of E-cadherin expression by TLE1, we also investigated the effect of removal of endogenous TLE1 on E-cadherin mRNA transcript and promoter activity. Knockdown of endogenous TLE1 in A549 cells resulted in higher levels of E-cadherin transcript (Fig. 3C) and increased activity of the transfected E-cadherin promoter reporter construct (Fig. 3D). Taken together, these results indicate that TLE1 suppresses E-cadherin expression at the transcriptional level.
Fig. 3.
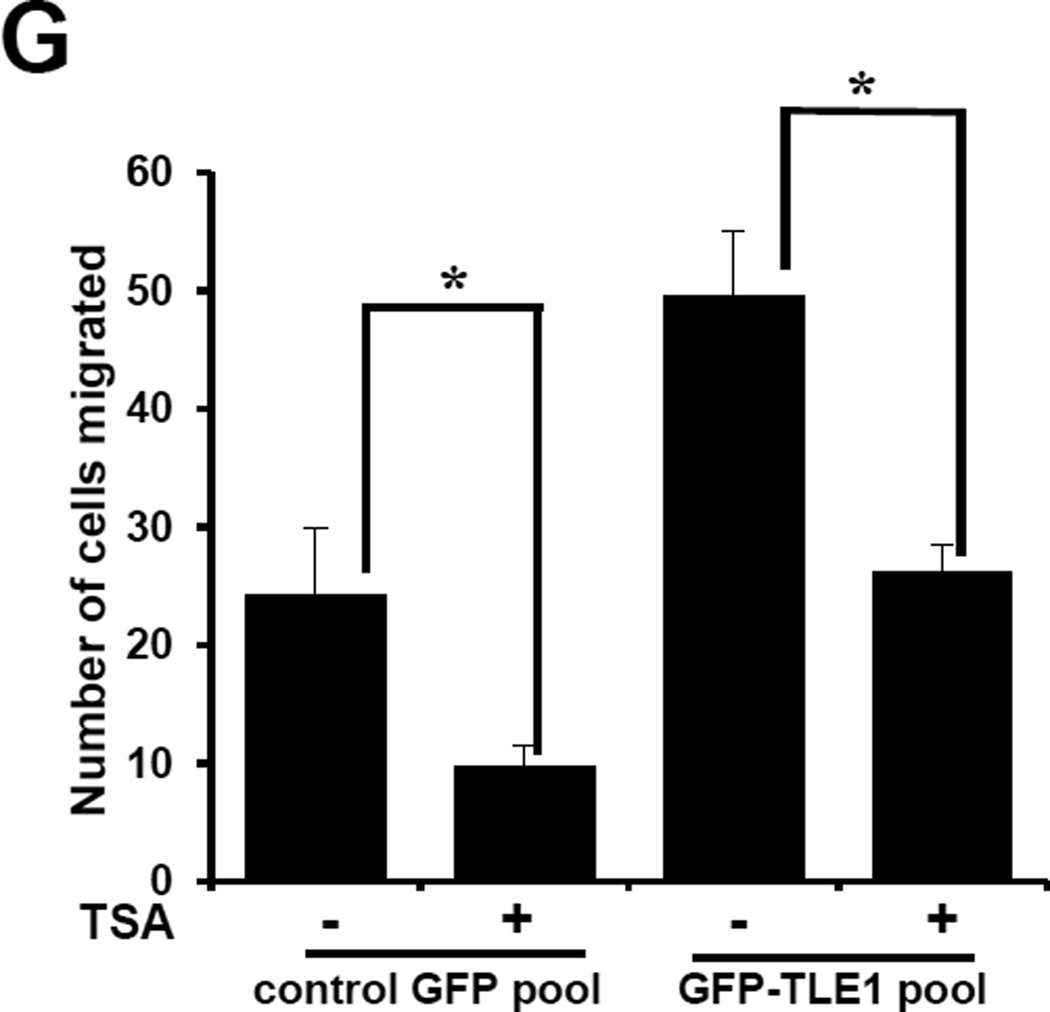
TLE1-induced E-cadherin repression occurs at the transcriptional level and is inhibited by the HDACi TSA. A. Stable control GFP and GFP-TLE1 cells were subjected to total RNA isolation, and E-cadherin mRNA expression levels were quantified by reverse transcription and real time PCR analysis using specific E-cadherin primers. B. Stable control GFP and GFP-TLE1 cells were subjected to E-cadherin promoter luciferase activity assays as described in materials and methods. C and D. A549 cells transfected with control or TLE1 siRNAs pool were subjected to RNA isolation and Real time PCR analysis (C) to assess for E-cadherin mRNA levels and E-cadherin promoter luciferase assay (D) to quantify E-cadherin promoter activity. E, F, and G. Control GFP and GFP-TLE1 treated or untreated with 300 nM TSA for 24h were subjected to RNA isolation and Real time PCR analysis to assess for E-cadherin mRNA levels (E), E-cadherin promoter luciferase assay (F) to assay for E-cadherin promoter activity, and Boyden Chamber migration assay (G). In A to G, three independent experiments were performed in triplicates, * indicates p<0.05 by Student’s t test.
Histone deacetylase (HDAC) inhibition attenuates TLE1-induced E-cadherin suppression and cell motility
Previous studies indicate that E cadherin repression by known EMT regulatory transcription factors depends on recruited HDAC activity (18). Considering that TLE1 interacts with and recruits HDAC to repress its target gene(s) (4,5,19), we investigated whether the observed repression of E-cadherin expression by TLE1 is dependent on HDAC function. To address this possibility, the control GFP and GFP-TLE1 pool of cells were treated with or without the HDAC inhibitor, Trichostatin A (TSA), and examined for E-cadherin expression. Consistent with previous finding (20), treatment of control A549 cells with TSA resulted in induction of TLE1 expression (Figs. 3E and 3F). Importantly, HDAC inhibition also attenuated the TLE1-induced E-cadherin suppression in GFP-TLE1 cells (Figs. 3E and 3F). These results indicate that histone deacetylation in part plays a role in the suppression of E-cadherin by TLE1. We then examined the impact of histone deacetylation on the EMT promoting effect of TLE1. To this end, we tested whether HDAC inhibitors can block TLE1-induced motility in A549 cells by subjecting untreated and TSA treated control GFP and GFP-TLE1 cells to migration assays. As shown in Fig. 3G, TSA treatment decreased the basal migratory potential of control cells. Importantly, TSA also attenuated the induced motility of exogenous TLE1 expressing cells. These studies, in conjunction with the observed TSA effect on TLE1-mediated E-cadherin suppression, suggest that the TLE1-induced EMT is mediated in part by HDAC activity.
TLE1 interacts in vivo with the E-cadherin promoter and recruits HDACs
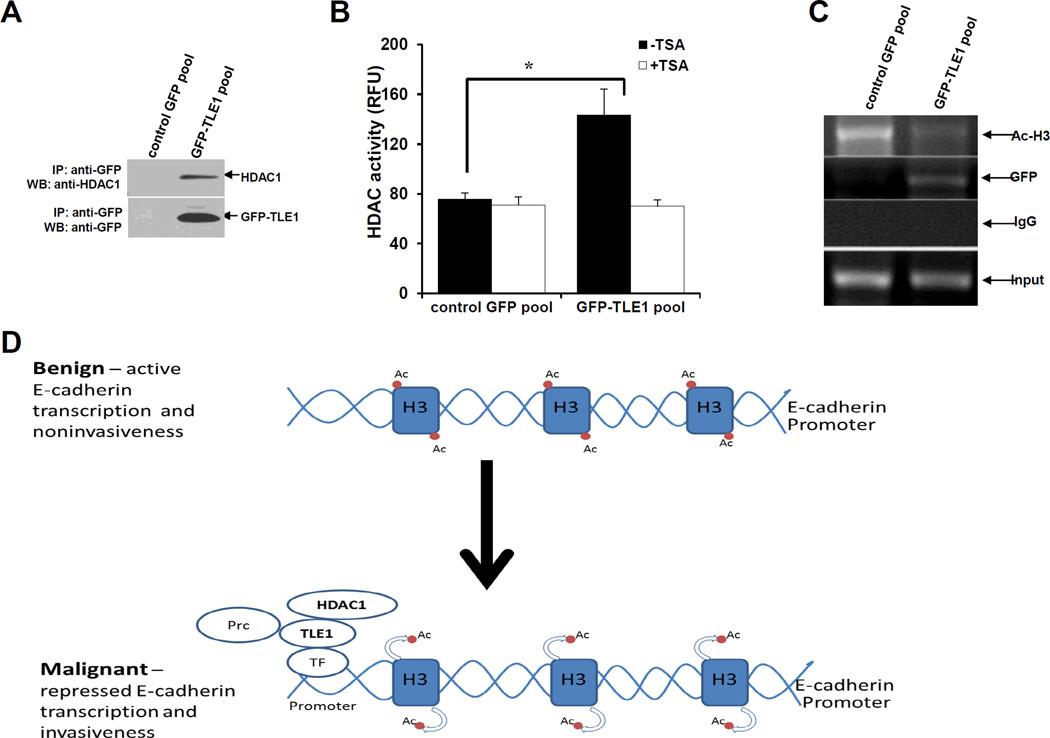
To further examine the role of HDAC in TLE1-mediated E-suppression, we first confirmed the interaction of TLE1 with HDAC1 in A549 cells by co-immunoprecipitation (Fig. 4A). Consistent with previous findings (4,5,19), the immunoprecipitate of GFP-TLE1 was associated with endogenous HDAC1. Consistently, the GFP-TLE1 immunoprecipitated fraction contained a significantly higher level of HDAC activity as compared to the control GFP (Fig. 4B). The observed enhanced HDAC activity in the GFP-TLE1 immunoprecipitate was inhibited by TSA (Fig. 4B). HDAC recruitment to the E-cadherin promoter by known EMT regulatory transcription factors such as SNAIL has been previously shown as a mechanism to repress E-cadherin transcription (18). To assess the possibility that the corepressor TLE1 facilitates the recruitment of HDAC to the E-cadherin promoter, we examined the interaction of TLE1 with the E-cadherin promoter using ChIP assay with the highly specific antibodies against GFP and acetylated histone H3 in the control GFP and GFP-TLE1 cells. As shown in Fig. 5C, the GFP-TLE1 protein was shown to interact with the endogenous E-cadherin promoter and this interaction was specific for the GFP-TLE1 due to the absence of the E-cadherin promoter sequences in the control GFP immunoprecipitate. In line with the HDAC recruitment function of TLE1, we observed that the level of acetylated histone H3 at the E-cadherin promoter is significantly reduced in GFP-TLE1 cells as compared to control GFP cells (Fig. 4C). Taken together, these findings indicate that TLE1 recruits HDAC to the E-cadherin promoter to repress transcription.
Fig. 4.
TLE1 interacts with HDAC1 and recruits HDAC activity to the endogenous E-cadherin promoter. A. Control GFP and GFP-TLE1 cells were harvested and cell extracts were prepared, immunoprecipitated (IP) with agarose-immobilized anti-GFP, and immunoblotted with anti-HDAC1 and anti-GFP antibodies. B. Control GFP and GFP-TLE1 cells treated or untreated with 300 nM TSA for 24h were subjected to immunoprecipitation (IP) with anti-GFP antibodies, and the HDAC activity in the anti-GFP immunoprecipitated fractions was determined as described in materials and methods. C. Control GFP and GFP-TLE1 cells were analysed by ChIP assays with anti-GFP, anti-acetyl-histone H3 (Ac-H3), and control IgG as detailed in the materials and methods. D. A model depicting the mechanism of TLE1-induced E-cadherin repression. In benign cells wherein active E-cadherin transcription is occurring, the E-cadherin promoter is not occupied by TLE1 and exhibits basal Histone 3 acetylation. In aggressive lung cancer, the increased expression of TLE1 will lead to its binding to the promoter of E-cadherin (likely through DNA binding transcriptional regulatory factors (TF)) followed by recruitment of HDAC1, deacetylation of Histone H3, and repression of E-cadherin transcription resulting to increased invasiveness. The inability of HDAC inhibitors to completely block TLE1-mediated E-cadherin suppression indicates TLE1 may also recruit additional chromatin remodelling proteins such as the histone methylating Polycomb repressive complex (Prc) (6). In B, three independent experiments were performed in triplicates, * indicates p<0.05 by Student’s t test.
Conclusion
In summary, we have uncovered a novel function of the corepressor TLE1 in EMT by suppressing the expression of the tumor suppressor E-cadherin. Although the exact mechanism(s) by which TLE1 is recruited to the E-cadherin promoter remains to be examined (likely through known EMT regulatory DNA binding transcriptional factors), the present findings indicate that TLE1 is a functional component of the E-cadherin corepressor complex in part to recruit HDAC1 (Fig. 4D). With numerous evidence indicating that defects in E-cadherin expression and the resulting EMT can promote tumor progression (15–17), the observed ability of TLE1 to suppress E-cadherin expression and to potentiate EMT may underlie its previously reported oncogenic function in lung cancer (14). Furthermore, our findings indicate that suppression of TLE1 could provide a novel therapeutic strategy to specifically block lung cancer aggressiveness and metastasis.
Highlights.
Exogenous TLE1 in A549 cells enhances cell migration and suppresses E-cadherin.
Knockdown of TLE1 in A549 cells reduces cell motility and upregulates E-cadherin.
TLE1 suppresses E-cadherin expression at the transcriptional level.
TLE1 recruits HDAC1 to the E-cadherin promoter.
HDAC inhibition attenuates TLE1-induced E-cadherin downregulation and motility.
Acknowledgments
This work was supported by Louisiana Cancer Research Consortium (LCRC) Start up Grant (HB), NIH RCMI G12RR026250-03 Grant (to Xavier University of Louisiana), and NIH 1R15CA158677-01A1 Grant (HB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen G, Courey AJ. Groucho/TLE family proteins and transcription repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 2.Gasperowicz M, Otto F. Mammalian Groucho homologs: redundancy or specificity? J. Cell Biochem. 2005;95:670–687. doi: 10.1002/jcb.20476. [DOI] [PubMed] [Google Scholar]
- 3.Palaparti A, Baratz AA, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 1999;96:6797–6801. doi: 10.1073/pnas.96.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscarlet M, Hermann R, Lo R, Tang Y, Joachim K, Stifani S. Cofactor-activated phosphorylation is required for inhibition of cortical neuron differentiation by Groucho/TLE1. PLoS One. 4:e8107. doi: 10.1371/journal.pone.0008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flowers EB, Poole RJ, Tursun B, Bashllari E, Pe’er I, Hobert O. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegens. Development. 2010;137:1799–17805. doi: 10.1242/dev.046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonderegger CK, Vogt PK. Binding of the corepressor TLE1 to Qin enhances Qin-mediated transformation of chicken embryo fibroblasts. Oncogene. 2003;22:1749–1757. doi: 10.1038/sj.onc.1206308. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Chen HM, Jaramillo E, Wang L, D’Mello SR. Histone deacetylase-related protein inhibits AES-mediated neuronal cell death by direct interaction. J. Neuroscience Res. 2008;86:2423–2431. doi: 10.1002/jnr.21680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo SW, Lee H, Lee HI, Kim HS. The role of TLE1 in synovial sarcoma. J. Orthop. Res. 2011;29:1131–1136. doi: 10.1002/jor.21318. [DOI] [PubMed] [Google Scholar]
- 12.Jan Y, Matter M, Pai JT, Chen YL, Pilch J, Komatsu M, Ong E, Fukuda M, Ruoslahti E. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell. 2004;116:751–762. doi: 10.1016/s0092-8674(04)00204-1. [DOI] [PubMed] [Google Scholar]
- 13.Yao X, Jennings S, Ireland SK, Pham T, Temple B, Davis M, Chen R, Davenport I, Biliran H. The anoikis effector bit1 displays tumor suppressive function in lung cancer cells. PLoS One. 2014 Jul 8;9(7):e101564. doi: 10.1371/journal.pone.0101564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen T, van Tuyl M, Iyengar P, Jothy S, Post M, Tsao MS, Lobe CG. Grg1 acts as a lung-specific oncogene in a transgenic mouse model. Cancer Res. 2006;66:1294–1301. doi: 10.1158/0008-5472.CAN-05-1634. [DOI] [PubMed] [Google Scholar]
- 15.Nollet F, Berx G, van Roy F. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol. Cell Biol. Res. Commun. 1999;2:77–85. doi: 10.1006/mcbr.1999.0155. [DOI] [PubMed] [Google Scholar]
- 16.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 17.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 18.Peinado H, Ballestar E, Esteller M, Cano A. Snail Mediates E-Cadherin Repression by the Recruitment of the Sin3A/Histone Deacetylase 1 (HDAC1)/HDAC2 Complex. Mol Cell Biol. 2004;24(1):306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su L, Sampaio AV, Jones KB, Pacheco M, Goytain A, Lin S, Poulin N, Lin Yi, Rossi FM, Kast J, Capecchi MR, Michael U, Nielsen TO. Deconstruction of the SS18-SSX Fusion Oncoprotein Complex: Insights into Disease Etiology and Therapeutics. Cancer Cell. 2012;21(3):333–347. doi: 10.1016/j.ccr.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagathihalli NS, Massion PP, Gonzalez AL, Lu P, Datta PK. Smoking Induces Epithelial-to-Mesenchymal Transition in Non–Small Cell Lung Cancer through HDAC-Mediated Downregulation of E-Cadherin. Mol. Cancer Ther. 2012;11:2362–2372. doi: 10.1158/1535-7163.MCT-12-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]