Abstract
The widely used anticoagulant Coumadin (R/S-warfarin) undergoes oxidation by cytochromes P450 into hydroxywarfarins that subsequently become conjugated for excretion in urine. Hydroxywarfarins may modulate warfarin metabolism transcriptionally or through direct inhibition of cytochromes P450 and thus, UGT action toward hydroxywarfarin elimination may impact levels of the parent drugs and patient responses. Nevertheless, relatively little is known about conjugation by UDP-glucuronosyltransferases in warfarin metabolism. Herein, we identified probable conjugation sites, kinetic mechanisms and hepatic UGT isoforms involved in microsomal glucuronidation of R- and S-7-hydroxywarfarin. Both compounds underwent glucuronidation at C4 and C7 hydroxyl groups based on elution properties and spectral characteristics. Their formation demonstrated regio- and enantioselectivity by UGTs and resulted in either Michaelis-Menten or substrate inhibition kinetics. Glucuronidation at the C7 hydroxyl group occurred more readily than at the C4 group, and the reaction was overall more efficient for R-7-hydroxywarfarin due to higher affinity and rates of turnover. The use of these mechanisms and parameters to model in vivo clearance demonstrated that contributions of substrate inhibition would lead to underestimation of metabolic clearance than that predicted by Michaelis-Menten kinetics. Lastly, these processes were driven by multiple UGTs indicating redundancy in glucuronidation pathways and ultimately metabolic clearance of R- and S-7-hydroxywarfarin.
Keywords: Glucuronidation, UGT (UDP-Glucuronosyl transferase), Metabolism, Warfarin, 7-Hydroxywarfarin, in vitro
1. Introduction
Coumadin (R/S-warfarin) is a commonly prescribed anticoagulant for more than 20 million Americans to treat atrial fibrillation, mechanical heart valves, venous thromboembolism, and other coagulopathies; however, optimal dosing requires adequate consideration of metabolic pathways that determine levels of the active drugs R-warfarin and especially S-warfarin [1, 2]. Warfarin is essentially inactivated through oxidation by several cytochromes P450 (P450 or CYP for specific isoforms) into 6-, 7-, 8-, 10- and 4′-hydroxywarfarin [3, 4]. While hydroxywarfarins are commonly reported in patient plasma plasma [5–7], there is indirect evidence for extensive conjugation of hydroxywarfarins, most notably to glucuronides, in patient urine samples [3, 8]. Glucuronidation involves the addition of a glucuronic acid moiety from the cofactor UDPGA catalyzed by the superfamily of conjugative enzymes UDP-glucuronosyltransferases (UGTs) [9]. UGT metabolic processes rank second in clinical relevance for the top 200 drugs on the market after those by cytochromes P450 [10].
Recent studies have implicated two possible mechanisms for hydroxywarfarins to modulate warfarin metabolism and thus impact patient responses to the drug. First, these warfarin metabolites may interact with receptors to control expression of many genes involved in drug metabolic clearance including enzymes and transporters. Hydroxywarfarins can bind to the pregnane X receptor (PXR) and induce expression of CYP2C9 and CYP3A4 and to a less extent the transporter P-glycoprotein 1 (also known as multidrug resistance protein 1 (MDR1) based on gene reporter assays with transformed HepG2 cells and treatment of primary human hepatocytes [11]. Second, hydroxywarfarins could inhibit warfarin metabolism by cytochromes P450. We have shown that hydroxywarfarins, including 7-hydryoxywarfarin metabolite, one of the most abundant metabolites in human plasma [5, 7], can potently inhibit CYP2C9, the major pathway for S-warfarin metabolism [12]. Therefore, the second phase of metabolism by UGTs may alter the levels of hydroxywarfarin levels in vivo and ultimately impacts the metabolism of the parent drug warfarin. For example, studies in mice have shown that R-warfarin clearance likely involves a coupled process between P450 and UGT metabolism of the drug [13]. Further studies are clearly needed to advance an understanding of the potential clinical relevance of UGT metabolic pathways in warfarin metabolic clearance.
Recent efforts have begun to identify possible glucuronidation pathways for hydroxywarfarins and the UGTs responsible for them. Recombinant UGT1A1, 1A3, 1A8, 1A9, and 1A10 display regioselective activity toward racemic warfarin and hydroxywarfarins [14], although only 6-, 7-, and 8-hydroxywarfarin underwent appreciable metabolism. The enantioselectivity of these reactions were investigated in a subsequent study using enantiomers of the parent drugs and hydroxywarfarins and an emphasis on recombinant UGT1A1 and 1A10. In addition, high correlations were reported between with bilirubin glucuronidation (UGT1A1) and UGT1A1 protein content for 6- and 7-hydroxywarfarin, whereas the poorest correlations occurred with UGT2B7 and 2B15 [15].
Despite these advances, there remain significant gaps in our understanding of hepatic glucuronidation pathways in humans. Neither of studies identified the structure of glucuronide metabolites generated in those reactions [14, 15]. The parent drug, warfarin, possesses a C4 hydroxyl group, and upon P450 oxidation, another hydroxyl group becomes available for modification. Both of these sites are possible targets for glucuronidation as shown for 7-hydroxywarfarin in Figure 1. Studies with rat hepatocytes suggest both sites could be accessible for glucuronidation and/or sulfonation leading to the formation of mono- and di-conjugates [16]. Knowledge of the actual products of conjugative reactions is necessary to guide the synthesis of authentic standards for direct quantification of glucuronide metabolites and facilitate metabolite-profiling approaches for use as biomarkers [4]. Moreover, identification of the structures for the respective glucuronides would enable the mapping of specific metabolic pathways for hydroxywarfarins as contributors to overall drug clearance. Lastly, the reliance on recombinant UGTs for in vitro metabolic mechanism and parameters fails to reflect the complexity of a hepatic microsomal system in which phospholipid and UGT dimerization impact UGT activity (reviewed in [17]). Moreover, the typical normalization of recombinant activity to total protein concentration does not allow a correction for differences among preparations and corresponding metabolic rates. This strategy makes it impossible to directly compare metabolic clearance rates among different recombinant UGT preparations.
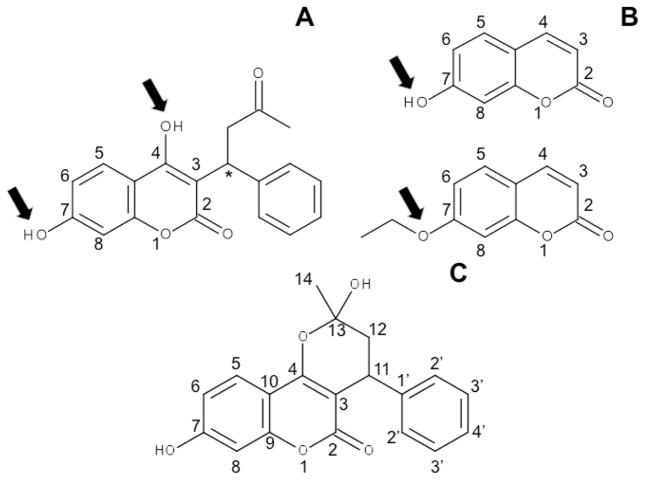
Figure 1. Structures of coumarin derivatives possessing a 7-hydroxyl group.
Panel A, The 7-hydroxywarfarin metabolite of warfarin is pictured with arrows indicating the hydroxyl groups on the coumarin that serve as potential sites of glucuronidation. The asterisk shows the chiral center for the compound resulting in R and S enantiomers of the drug metabolite. Panel B, 7-Hydroxycoumarin (top) and 7-ethoxycoumarin (bottom) are shown with an arrow highlighting the 7-hydroxyl group, which when modified, leads to a selective decrease in compound fluorescence but not absorbance. Panel C, 7-Hydroxywarfarin forms diastereomeric cyclic hemiacetals when present in organic solvents, like methanol, as observed during characterization of products synthesized through green chemistry.
In this study, we identified probable conjugation sites, kinetic mechanisms and hepatic UGT isoforms involved in microsomal glucuronidation of R- and S-7-hydroxywarfarin. For these studies, we synthesized R- and S-7-hydroxywarfarin substrates enantioselectively using novel green chemistry techniques. Experiments revealed for the first time the formation of two glucuronide metabolites and their corresponding spectral properties were used to identify whether glucuronidation occurred at the C4 or C7 hydroxyl group (Figure 1). We then measured steady-state kinetics for R- and S-7-hydroxywarfarin using human liver microsomes pooled from 150 donors (HLM150) as a model for the metabolic capacity of the average human liver [18]. The corresponding kinetic profiles mostly deviated from the relationship reported by Michaelis-Menten [19]. Consequently, we compiled the data and globally fit it to six possible mechanisms involving either a single binding site (Michaelis-Menten) or two binding sites and identified the most statistically probable one using DynaFit software [20] as described recently [21]. Inhibitor phenotyping experiments were carried out to determine relative roles for UGT1A1, 1A4, 1A6, and 1A9 in microsomal 7-hdyroxywarfairn metabolism. Lastly, we assessed the potential impact of these mechanisms and parameters on the prediction of in vivo hepatic metabolic clearance of R- and S-7-hydroxywarfarin.
2. Methods
2.1 Materials
All chemicals used in this study were ACS grade or higher. The following chemicals were purchased from Sigma-Aldrich (St. Louis, MO): 4,7-dihydroxycoumarin, E-4-phenylbut-3-en-2-one, (R,R)- and (S,S)-1,2-diphenylethylenediamine, tetrahydrofuran, glacial acetic acid, ethyl acetate, CD3OD, ethanol, UDP-glucuronic acid (UDPGA), perchloric acid, serotonin (inhibitor), hecogenin (inhibitor), bilirubin (inhibitor), and mefenamic acid (inhibitor). R/S-7-Hydroxywarfarin and R/S-warfarin (internal standard) were purchased from Toronto Research Chemicals (Toronto, Canada). HPLC Grade methanol, alamethicin, magnesium chloride, and saccharolactone were purchased from Fisher Scientific (Wilmington, MA). Pooled human liver microsomes were purchased from BD Biosciences (San Jose, CA).
2.2 Synthesis of R- and S-7-hydroxywarfarin through green chemistry
The enantiomers of 7-hydroxywarfarin were prepared by adapting the green chemistry procedure as described by others [22, 23]. 4,7-Dihydroxycoumarin (107 mg, 0.600 mmol), E-4-phenylbut-3-en-2-one (90 mg, 0.616 mmol), (R,R)- or (S,S)-1,2-diphenylethylenediamine (12 mg, 0.057 mmol), 1 mL of tetrahydrofuran and 285 μL of glacial acetic acid were stirred in a sealed 10 mL round-bottomed flask at the ambient temperature until a white solid precipitated (typically this took 2–4 days). The R,R catalyst produces the R configuration at C-11 in 7-hydroxywarfarin, while the S,S catalyst yields the S configuration (Figure 1, Panel C). The precipitate resulting from either reaction was collected by vacuum filtration, rinsed with water, and allowed to dry. The yield for each reaction was typically around 70% for the white solid. Recrystallization from ethanol/water provided each product with an enantiomeric excess greater than 99% based on chromatographic analyses.
We compared the products to R- and S-7-hydroxywarfarin standards that were prepared by using a chiral preparative chromatography of the racemic commercial compound as described previously [15]. In this study, recrystallized compounds were injected onto a Waters Breeze HPLC system and resolved using a 2.1 mm × 150 mm, 5.0 μM Astec Chirobiotic V column (Supelco) operated at room temperature (22.0 °C) using an isocratic method with 20% methanol: 80% 0.1% acetic acid/water at a flow rate of 1.2 mL/min. Elution of the compounds was monitored by absorbance at 275 and 325 nm and fluorescence (excitation: 325 nm; emission: 420 nm) and compared against individual R- and S-7-hydroxywarfarin standards.
The structure of the compounds were further analyzed by 1H and 13C NMR as well as high resolution MS. The 1H (400 MHz) and 13C (100 MHz) NMR spectra were acquired on a JEOL ESC 400 spectrometer with CD3OD as solvent; therefore, hydroxyl protons are exchanged with deuterium and are not observed. Assignment of chemical shifts was aided by comparison to published data for warfarin itself [24, 25], as well as use of DEPT-90, DEPT-135, and HMQC. Specific assignments for hydrogens or carbons refer to the numbering on Figure 1, Panel C. Solvent conditions for these studies favored the ring closure of warfarin to the hemiacetal and resulting two diastereomers (Figure 1, Panel C). Data for13C NMR experiments are expressed in ppm relative to the solvent absorbance at 49.15 ppm. All data for the NMR studies are summarized in Tables S2 and S3 in Supplemental Data. Melting points were obtained on an electrothermal apparatus and are uncorrected. The mp range was 223–228 °C with decomposition, which is slightly higher than reported previously (208–210 °C) [26]. Mass spectra for the synthesized compounds were acquired on a Waters/Micromass ESI-QTOF-MS spectrometer. The high resolution mass spectral molecular ion for C19H16O5 was calculated to be 324.33, which matched the experimentally-determined m/z.
2.3 HPLC analysis of 7-hydroxywarfarin glucuronidation reaction
Unlike our previous studies [14], we developed a new high performance liquid chromatography (HPLC) method for analyzing glucuronides. The samples were injected onto a Waters Breeze HPLC system and resolved using a 4.6 × 150 mm Zorbax Eclipse 3.5 μM XDB-C18 column (Agilent) heated to 45 °C using an isocratic method with 40 % methanol: 60% 0.1% acetic acid/water at a flow rate of 1.2 mL/min. Elution of the compounds was monitored for 10 min by absorbance at 275 and 325 nm and fluorescence (excitation: 325 nm; emission: 420 nm). For our reactions, peak areas of the two metabolites were normalized to R/S-warfarin and then quantitated relative to the standard response for 7-hydroxywarfarin due to the lack of authentic glucuronide standards. It has been shown previously that the addition of the glucuronic acid moiety does not alter the extinction coefficient from that of the unreacted substrate [27].
2.4 Impact of modification at 7-hydroxyl group on spectral properties and mass
Glucuronidation of 7-hydroxywarfarin can occur at C4 or C7 hydroxyl groups; we hypothesized that these modifications would differentially affect visible spectral properties based on the critical role the 7-hydroxyl group plays in fluorescence, but not absorbance, of coumarin derivatives. This property has been utilized for measuring the increase in fluorescence upon O-dealkylation of 7-ethoxycoumarin by P450s [28]. We tested our hypothesis by assessing the relative fluorescence and absorbance properties of 7-hydroxycoumarin and 7-ethoxycoumarin by HPLC. Specifically, samples of 50 μM 7-hydroxycoumarin and 7-ethoxycoumarin were prepared in water from 10 mM stocks in methanol and injected onto a Waters Breeze HPLC system, compounds were resolved employing the same method described for analyzing the microsomal reaction toward 7-hydroxywarfarin in Section 2.3. Elution of the compounds was then monitored by absorbance (370 nm) and fluorescence (excitation: 370 nm, emission: 450 nm) and the relative areas of the peaks compared.
As a complement to those efforts, we analyzed the fractions by mass spectrometry to determine the parent masses and fragmentation patterns for individual metabolites. Samples were injected onto an 6590 Agilent triple quadrupole mass analyzer using an isocratic flow of 0.4 mL/min 70% acetonitrile/0.1% formic acid. Ion spectra were acquired in full scan monde monitoring the m/z range of 50 – 1000 amu. Subsequently product ion spectra were generated from the theoretical precursor ion m/z 501 monitoring the m/z range 50–600 amu at various collision energies.
2.5 Steady-state kinetic assays for R- and S-7-hydroxywarfarin glucuronidation by human liver microsomes
We initially carried out control experiments to guide the design of steady-state studies on the metabolism of R- and S-7-hydroxywarfarin. Specifically, we ensured linearity of response as a function of protein concentration and time. Based on those findings, the kinetic reactions included 0.5 mg/mL HLM150 incubated with 0, 20, 50, 100, 150, 250, 300, 500, 750, 1000, 1250, 1500, 1750, and 2000 μM R- and S-7-hydroxywarfarin in the presence of 50 mM Tris-HCl pH 7.4 (under reaction conditions), 25 ug/mL alamethacin, 8 mM MgCl2, and 5 mM saccharolactone for 15 min at 37 °C. The reactions were initiated with 2 mM UDPGA and quenched at 1 hr with equal volume of 0.4 N perchloric acid. The reactions were centrifuged at 2500 RPM for 15 min at 4 °C and the glucuronide metabolites quantified by HPLC as described in Section 2.3. Initial rates at each concentration were determined based on the completion of four to ten experimental replicates.
2.6 Analysis of steady-state kinetic profiles for R- and S-7-hydroxywarfarin glucuronidation
Kinetic profiles were not hyperbolic, but demonstrated a decrease in rate at higher substrate concentrations. Thus, the data were initially fit and analyzed with respect to the hyperbolic Michaelis-Menten and uncompetitive substrate inhibition equations using GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA). A statistical comparison of the quality of the resulting fits using Akaike Information Criterion revealed a strong preference for the uncompetitive substrate inhibition mechanism; however, there are possibilities other than an uncompetitive mechanism that can explain substrate inhibition. Consequently, we further investigated the mechanism and parameters for equilibrium constants and reaction rates using DynaFit version 3.28 (BioKin, Ltd, Watertown, MA) [20].
For the reanalysis of the data, we globally fit kinetic profiles to six different kinetic mechanisms and identified the most statistically probable one using DynaFit version 3.28 [20]. Specifically, we fit the data to single binding site mechanism reported by Michaelis-Menten [19] and five different two-site binding mechanisms including those describing substrate inhibition and cooperativity (Figure S1 and Table S1 in Supplemental Data), as described previously [21]. These latter mechanisms possess a binary (ES) complex, ternary (ESS) complex, or both complexes, whereby they are active with the same or differing maximal rates. Binding events presented in the model are ordered with respect to stoichiometry, but the occupancy of each individual binding site was indistinguishable in this assay. The most probable mechanism and corresponding parameters for R- and S-7-hydroxywarfarin were identified using the advanced tools of numerical analysis and applied statistics, as implemented in the software DynaFit. After globally fitting reactions to these mechanisms, we generated a corresponding Akaike information criterion (AICc) to identify statistically the most plausible mechanism based on the quality of the fits [20]. The presentation of data and the corresponding fit to the appropriate mechanism was achieved through the use of GraphPad Prism version 5.0.
2.7 Inhibitor phenotypying of R- and S-7-hydroxywarfarin glucuronidation by UGTs present in human liver microsomes
For these experiments, 0.5 mg/mL protein (HLM150) was incubated with R- and S-7-hydroxywarfarin (100 μM or 1000 μM), UDPGA (2 mM), and a chemical inhibitor specific for an individual UGT in 50 mM Tris-HCl buffer, pH 7.4. The inhibitors used in these experiments were bilirubin (25 μM) for mainly UGT1A1 [29], hecogenin (10 μM) for UGT1A4 [30], serotonin (200 μM) for UGT1A6 [31], and mefenamic acid (25 μM) for UGT1A9 and UGT2B7 [32]. The inhibitors were initially dried down with R- or S-7-hydroxywarfarin before reconstituting the reactions. By contrast, bilirubin was prepared fresh daily in amber vials as a 2 mM stock solution in DMSO. The inhibitors were incubated with R- and S-7-hydroxywarfarin and HLM for 15 min prior to initiation of the reaction. Upon addition of UDPGA to start the reaction, the samples were incubated at 37 °C for 1 hr and analyzed as described for steady-state experiments. The statistical significance of the specific inhibition by each compound compared to the respective control reaction was calculated using a one-way analysis of variance (ANOVA) test as implemented by GraphPad Prism software, version 5.0 (San Diego, CA). Final values for inhibitor potency reflected a minimum of four experimental replicates.
2.8 Modeling R- and S-7-hydroxywarfarin metabolic clearance
We obtained a perspective on the potential clinical importance of these studies by modeling metabolic clearance with in vitro kinetic parameters [33, 34]. We fit our data to mechanistic equations describing the relationship between 7-hydroxywarfarin enantiomer concentration and its clearance (rate/[7-hydroxywarfarin]). The metabolic clearance of 7-hydroxywarfarin based on the traditional model was predicted using the parameters derived from fitting the HLM150 data to the Michaelis-Menten equation and applying the results to Equation 1 as follows:
| Equation 1 |
where ν is the initial rate of the reaction, Vmax the maximal rate of the reaction, and Km the Michaelis constant. For comparison, the impact of substrate inhibition on the metabolic clearance of 7-hydroxywarfarin by glucuronidation was predicted using Equation 2 and parameters derived from fitting the HLM150 kinetic data to a derivation of the equation describing substrate (uncompetitive) inhibition as follows:
| Equation 2 |
where ν is the initial rate of the reaction, Vmax the maximal rate of the reaction, Km the Michaelis constant, and Ks the inhibition constant for occupancy of the second site.
3. Results
3.1 Identification of steady-state conditions and potential glucuronides resulting from R- and S-7-hydroxywarfarin metabolism
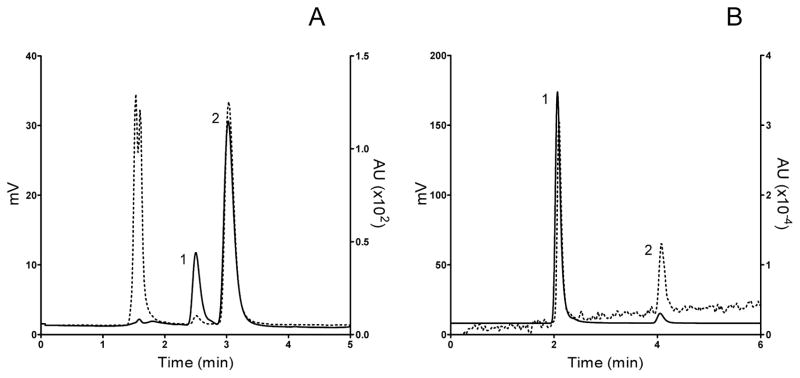
Initially, we determined conditions demonstrating linear product formation as a function of time and enzyme concentration. Over the course of a four hr incubation, microsomal reactions for both R- and S-7-hydroxywarfarins yielded two compounds, whose appearance seemed consistent with products of the reaction and occurred simultaneously minimizing the likelihood for a secondary glucuronidation reaction (Figure 2, Panel A). Consequently, we labeled them metabolites 1 and 2 based on elution order during HPLC analyses. The amounts of these metabolites increased as a function of time and linearly correlated with enzyme concentration, as shown in supplemental data (Figure S2 in Supplemental Data). A 60 min time point was chosen to avoid excessive substrate depletion and thus remain within steady-state conditions. Both metabolites eluted before 7-hydroxywarfarin during HPLC analyses, indicating a higher polarity as expected for glucuronide metabolites. Finally, the introduction of UGT-specific inhibitors blocked formation of the compounds (Section 3.4).
Figure 2. Modification of hydroxyl group alters the relative signal of 7-hydroxycoumarin derivatives.
HPLC traces of compounds are overlaid to demonstrate impact of possible modification of the 7-hydroxyl group on the coumarin ring on relative fluorescence and absorbance properties. Solid line, fluorescence; dotted line, absorbance. Panel A, 50 μM steady state reaction of glucuronide metabolite 1 (1) and glucuronide metabolite 2 (2) (fluorescence excitation at 325 nm and emission 420 nm; absorbance at 325 nm). These compounds were later identified as 7-OHWAR-4-GLUC and 7-OHWAR-7-GLUC, respectively, in the Discussion. Panel B, 50 μM 7-hydroxycoumarin (1) and 7-ethoxycoumarin (2) (fluorescence excitation at 370 nm and emission 450 nm; absorbance at 370 nm).
Glucuronidation can only occur at two locations of 7-hydroxywarfarin yielding molecules with different spectral properties, and thus we leveraged those differences to predict the structures for metabolites 1 and 2. The formation of a bond with the C7 hydroxyl group on a coumarin ring does not significantly impact absorbance yet causes a large drop in fluorescence. This property is leveraged in P450 catalytic assays whereby O-dealkylation of substrate to form 7-hydroxycoumarins result in a significant increase in fluorescence as a marker for product formation [28]. Therefore, we hypothesized that glucuronidation of the C7 hydroxyl group and not the one at C4 will decrease 7-hydroxywarfarin fluorescence selectively, and thus provide evidence for the regiospecificity of glucuronidation. As a control, we investigated this effect for 7-hydroxy- and 7-ethoxycoumarin under HPLC conditions used in this study (Figure 2, Panel B, Table 1). At equal molar concentrations, the spectral ratio between fluorescence and absorbance for these compounds was almost 10-fold higher value for 7-hydroxycoumarin relative to 7-ethoxycoumarin, indicating selective suppression of coumarin fluorescence upon modification of the hydroxyl group at C7. Analysis of the 7-hydroxywarfarin metabolites demonstrated that glucuronidation decreased compound fluorescence regardless of the site of modification; however, the effect was more severe for the second metabolite than the first (Figure 2, Panel A, Table 1). The magnitude of the difference in the spectral ratio between metabolites 1 and 2 for both UGT substrates was about 10-fold as observed for the coumarin model compounds.
Table 1.
Spectral properties for 7-hydroxycoumarin derivatives
| Compound | Amount (μM) | Spectral Properties
|
||
|---|---|---|---|---|
| Absorbancea | Fluorescenceb | Fluorescence | ||
| Absorbance | ||||
| 7-hydroxycoumarin | 50 | 1.48×103 | 1.24×106 | 837 |
| 7-ethoxycoumarin | 50 | 1.30×103 | 9.60×104 | 73.8 |
|
| ||||
| R-7-hydroxywarfarin | 10 | 5.64×104 | 4.75×106 | 84.2 |
| metabolite 1d | *c | 3.78×103 | 9.68×104 | 25.6 |
| metabolite 2e | *c | 1.37×105 | 3.69×105 | 2.70 |
|
| ||||
| S-7-hydroxywarfarin | 10 | 5.29×104 | 4.48×106 | 84.7 |
| metabolite 1d | *c | 2.45×103 | 4.83×104 | 19.7 |
| metabolite 2e | *c | 7.89×104 | 2.25×105 | 2.84 |
Area of absorbance peak measured at 370 nm and 325 nm for coumarin and hydroxywarfarin derivatives respectively
Area of fluorescence emission peak measured following excitation at 370 nm and 325 nm for coumarin and hydroxywarfarin derivatives respectively
Product areas observed for the reaction containing 500 μM substrate.
Later identified as 7-HOWAR-4-GLUC (see Discussion).
Later identified as 7-HOWAR-7-GLUC (see Discussion).
As a complement to kinetic, chromatographic, and spectral data, we subjected the metabolites form the four hr time course experiment to mass spectral analysis. These studies would not likely be able to identify the site of glucuronidation as reported by others [14, 15]; however, it would be possible to distinguish whether the metabolites were either a mono- or di-glucuronide based on mass. The calculated mass for the mono-glucuronide is 500.45 and that for the di-glucuronide is 676.56. The full mass spectrum for both metabolites showed signals consistent with the parent mass of a mono-glucuronide in positive and negative ion mode (including sodium adducts) (Figure S3 in Supplemental Data), and subsequent MS/MS experiments revealed product ions of m/z 443, 655, 267, 179, and 147, which match those reportedly previously by others for the mono-glucuronide [14, 15].
3.2 Substrate inhibition during microsomal glucuronidation of R- and S-7-hydroxywarfarin
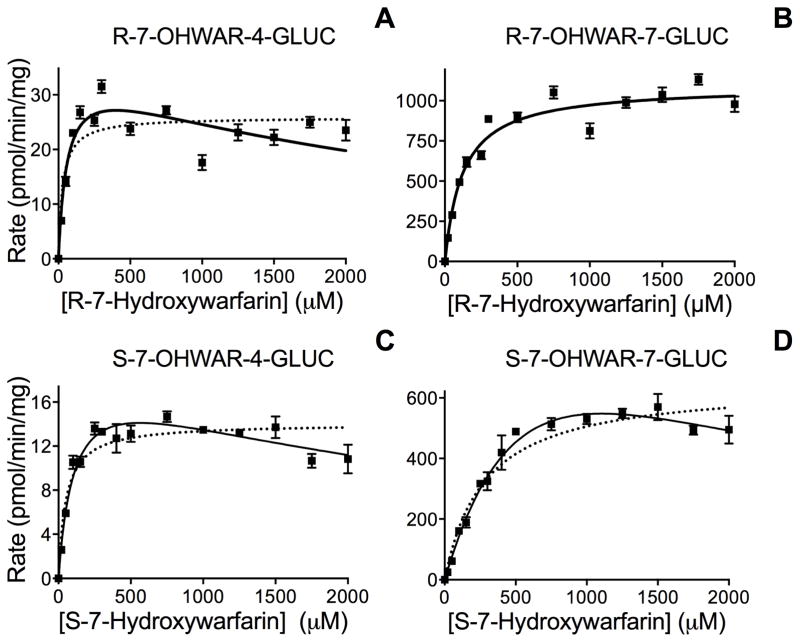
Microsomal glucuronidation reactions revealed that after an initial increase, the initial rate actually significantly decreased at higher substrate concentrations (Figure 3). An initial fit of the data revealed a strong statistical preference for the uncompetitive substrate inhibition mechanism over that for the Michaelis-Menten mechanism for both S-7-hydroxywarfarin metabolites, later identified as S-7-hydroxywarfarin β-D-4-glucuronide (S-7-OHWAR-4-GLUC) and S-7-hydroxywarfarin β-D-7-glucuronide (S-7-OHWAR-7-GLUC), respectively (see Discussion). The first metabolite (R-7-OHWAR-4-GLUC) for R-7-hydroxywarfarin was also formed through a substrate inhibition mechanism, while generation of the second one (R-7-OHWAR-7-GLUC) involved Michaelis-Menten kinetics (Table 2). Nevertheless, uncompetitive inhibition is only one of many possible kinetic mechanisms resulting in substrate inhibition.
Figure 3. Steady-state kinetics for glucuronidation of R- and S-7-hydroxywarfarin by HLM150.
For the kinetic reactions, 0.5 mg/mL protein for HLM150 was incubated with 0 to 2000 μM R- and S-7-hydroxywarfarin for 60 min at 37 °C. Panel A and B represent the kinetic profiles for S-7-hydroxywarfarin glucuronidation into 7-OHWAR-4-GLUC and 7-OHWAR-7-GLUC, respectively. Panel C and D represent the glucuronidation of R-7-hydroxywarfarin steady-state kinetics for glucuronidation at the C4 and C7 hydroxyl groups, respectively. The reported values represent the average from 4–10 replicates for each individual experiment. Solid line depicts fit of data to the preferred mechanism indicated in Table 3, i.e. Michaelis-Menten for the major metabolic pathway for R-7-hydroxywarfarin and substrate inhibition for the other reactions. For comparative purposes, dashed line depicts fit of data to Michaelis-Menten equation when substrate inhibition was the preferred mechanism.
Table 2.
Preliminary analysis of kinetic mechanism describing R- and S-7-hydroxywarfarin glucuronidationa
| Substrate | Metabolitesb | Model | Model Preference | Parameters for Preferred Model | ||
|---|---|---|---|---|---|---|
| Vmax1c | Km (μM) | Ki (μM) | ||||
| R-7-HOWAR | 7-HOWAR-4-GLUC | Substrate Inhibition | 70.7 % | 35.4 (30.4–40.4) | 60.8 (36.9–84.8) | 2640 (1360–3920) |
| 7-HOWAR-7-GLUC | Michaelis-Menten | 75.2 % | 1090 (1040–1140) | 122 (100–145) | - | |
| S-7-OHWAR | 7-HOWAR-4-GLUC | Substrate Inhibition | >99.99 % | 19.7 (17.19–22.2) | 111 (77.7–145) | 2840 (1650–4030) |
| 7-HOWAR-7-GLUC | Substrate Inhibition | 99.7 % | 1470 (790–2150) | 929 (362–1500) | 1310 (259–2370) | |
The nonsymmetrical 95 % confidence intervals for parameters are shown in parentheses.
Glucuronidation resulted in conjugation of the hydroxyl group at either the C4 or C7 position resulting in the formation of 7-HOWAR-4-GLUC or 7-HOWAR-7-GLUC, respectively, as described in the Discussion.
Units are nmol/min/mg protein
We employed DynaFit [20], a more powerful software package, and re-analyzed the data comparing the suitability of data fits to the Michaelis-Menten (single site) mechanism or multiple two-site mechanisms in which the UGT responsible for the reaction possesses catalytic and cooperative binding sites (Figure S1 and Table S1 in Supplemental Data). Based on Akaike Information Criterion, the most probable mechanism for both S-hydroxywarfarin metabolites was a two-site ES active model in which catalytically active ES complex forms as a function of S-hydroxywarfarin concentration. The most probable mechanisms for R-hydroxywarfarin glucuronidation into two metabolites involved different mechanisms of action. The first metabolite (R-7-OHWAR-4-GLUC) was preferentially formed through a two-site cooperative mechanism in which catalytically active ES and ESS complexes formed as a function of R-7-hydroxywarfarin, whereas generation of the second metabolite (R-7-OHWAR-7-GLUC) was more likely through a single-site mechanism following Michaelis-Menten kinetics. The parameters and corresponding 95 % confidence intervals for all reactions are summarized in Table 4. Similarly, both pathways for glucuronidation of S-7-hydroxywarfarin were best explained through a two-site cooperative mechanism in which catalytically active ES and ESS complexes formed as a function of S-7-hydroxywarfarin.
3.3 Impact of substrate inhibition on metabolic clearance of R- and S-7-hydroxywarfarin
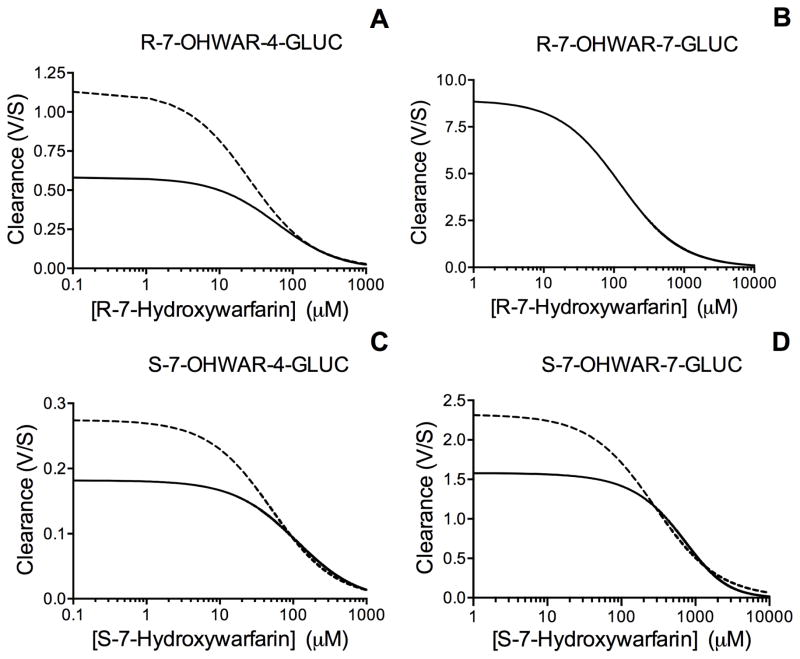
We employed the kinetic parameters from fits of the microsomal data to either the Michaelis-Menten or substrate inhibition mechanisms to assess their effects on 7-hydroxywarfarin metabolic clearance. The parameters for the traditional uncompetitive equation (Table 2) were used to simulate substrate inhibition as reflected in Equation 2 (solid line in Figure 4, Panels A, C, and D). The dashed line indicates a hypothetical plot for metabolite clearance through a process dependent on the Michaelis-Menten mechanism for comparative purposes. The resulting log plot demonstrates that substrate inhibition results in significantly lower clearance rates at low substrate concentrations such that the clearance of the metabolites is overestimated. At higher 7-hydroxywarfarin levels, the difference between the clearance models becomes insignificant. The exception to this observation is R-7-OHWAR-7-GLUC, the major metabolite derived form the reaction with R-7-hydroxywarfarin.
Figure 4. R- and S-7-Hydroxywarfarin clearance involving either Michaelis-Menten (dashed line) or substrate inhibition (solid line) kinetics.
GraphPad Prism software (San Diego, CA) was used to simulate clearance curves for R- and S-7-hydroxywarfarin using equations adapted by Houston and Kenworthy [34] as described in Materials and Methods. Panels A and B indicate the clearance of R-7-hydroxywarfarin as a solid line for both metabolites, i.e. R-7-OHWAR-4-GLUC and R-7-OHWAR-7-GLUC. Panels C and D indicate the clearance of S-7-hydroxywarfarin as a solid line for both metabolites, i.e. S-7-OHWAR-4-GLUC and S-7-OHWAR-7-GLUC. The dashed line in Panels A, C, and D shows the predicted clearance of the compounds if the metabolic process conformed to the Michaelis-Menten mechanism to highlight the overestimation of 7-hydroxywarfarin clearance by traditional Michaelis-Menten kinetic models.
3.4 Multiple microsomal UGT enzymes contribute to R-and S-7-hydroxywarfarin glucuronidation
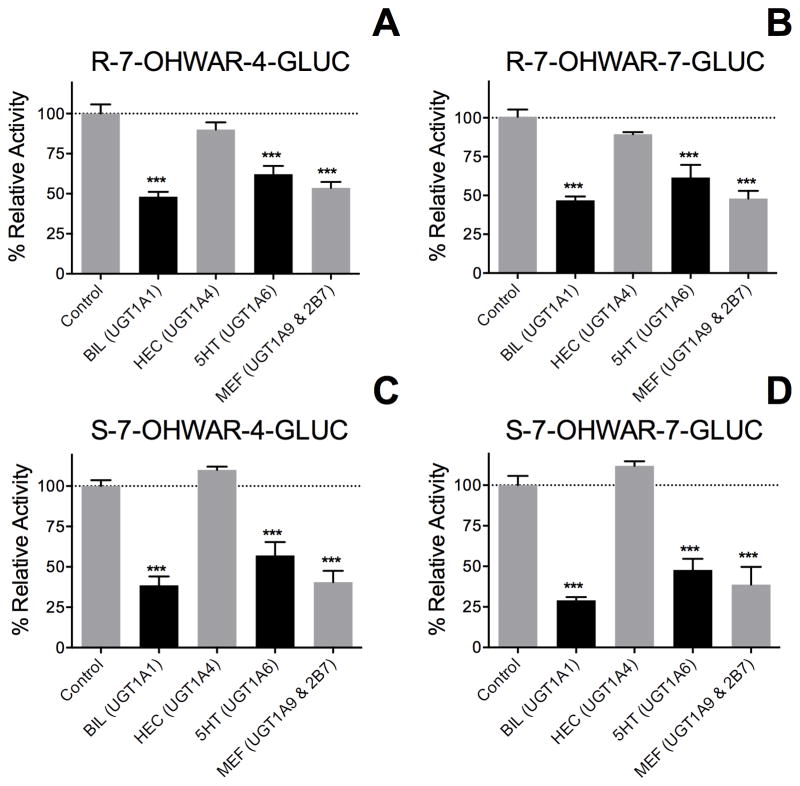
Glucuronidation of 7-hydroxywarfarin to either metabolite by human liver microsomes was decreased in the presence of relatively specific inhibitors toward individual UGTs. The presence of bilirubin, serotonin, and mefenamic acid but not hecogenin inhibited activity toward 100 μM substrate by about 50 % (Figure 5). The potency of this effect was lost at a higher substrate concentration (1000 μM, data not shown). These patterns were not impacted by chirality of the substrates and were conserved for both metabolites, such that the metabolic pathways may be the same. Nevertheless, the effects of these inhibitors on reactions implicate roles for hepatic UGT1A1 and 1A6, as well as 1A9 and/or UGT2B7, and rule out UGT1A4 in glucuronidation of R- and S-7-hydroxywarfarin.
Figure 5. Inhibitor phenotyping of steady-state glucuronidation of R- and S-7-hydroxywarfarin by HLM150.
Reactions carried out with 0.25 mg/mL protein, 100 μM 7-hydroxywarfarin, and 2 mM UDPGA in the presence of specific inhibitors for specific UGT isoforms at 37 °C and pH 7.4. Panels A and B depict selective inhibition of R-7-hydroxywarfarin to R-7-OHWAR-4-GLUC and R-7-OHWAR-7-GLUC, respectively. Panels C and D depict selective inhibition of S-7-hydroxywarfarin to S-7-OHWAR-4-GLUC and S-7-OHWAR-7-GLUC, respectively. Reaction activities were normalized to the control rate and reported as a percent, e.g. 100 % in the absence of inhibition. The reported activities represent the average from a minimum of 4 individual experiments and are shown as the mean ± standard deviation. Asterisks indicate statistical significance of inhibition compared to control (*** p < 0.001).
4. Discussion
The first kinetic analysis of hepatic microsomal metabolism of R- and S-7-hydroxywarfarin revealed important features of the reaction. Both compounds underwent glucuronidation at the C4 and C7 hydroxyl group. In the absence of authentic standards, we were able to putatively identify these metabolites based on elution properties and fluoresce and mass spectral characteristics. Their formation by microsomal UGTs demonstrated regio- and enantioselectivity and resulted in either Michaelis-Menten or substrate inhibition kinetics. Glucuronidation at the C7 hydroxyl group occurred more readily than at the C4 hydroxyl group, and the reaction was overall more efficient for R-7-hydroxywarfarin due to higher affinity and rates of turnover for the substrate. The use of those mechanisms and parameters to model in vivo clearance of the 7-hydroxywarfarins demonstrated that the contributions of substrate inhibition would lead to underestimation of their metabolic clearance at low 7-hydroxywarfarin levels than that predicted by the traditional Michaelis-Menten kinetics. Lastly, these processes were driven by multiple UGTs indicating redundancy in glucuronidation pathways and ultimately metabolic clearance of R- and S-7-hydroxywarfarin.
Preliminary experiments revealed that hepatic UGTs target both hydroxyl groups of R- and S-7-hydroxywarfarin for glucuronidation. Based on HPLC analyses, two new peaks formed over the course of the reaction simultaneously and eluted before the substrates. This observation is consistent with the creation of polar glucuronides, which were labeled metabolites 1 and 2. It is possible that the same glucuronide conjugate at the C7 position, yet differ in whether the compound exists in open and closed structures [25, 35], as shown in Figure 1, Panels A and C, due to the presence of organic solvent (methanol) in the mobile phase. This experimental artifact is not likely, given the absence of this effect with 7-hydroxywarfarin substrate (or other warfarin derivative). Moreover, the ratio between the peak areas was not the same as expected for an equilibrium process and. Alternatively, these UGT products may reflect the creation of two monoglucuronides or a mono- and di-glucuronide. The formation of a di-glucuronide would require the initial accumulation of the monoglucuronide to drive the reaction, yet no lag in the time course was observed. Moreover, the addition of a second glucuronic acid moiety to the monoglucuronide would significantly increase the polarity of the molecule making metabolite 1 the only probable di-glucuronide; however, the fluorescence properties of metabolite 1 was consistent with modification only at the C4 hydroxyl group. Lastly, mass spectral analysis of the metabolite yielded identical masses and fragmentation patterns consistent with mono-glucuronides of hydroxywarfarins as reported previously reported by others [14, 15]. Taken together, these findings suggest that both R- and S-7-hydroxywarfarin undergoes glucuronidation into two monoglucuronides by UGT action present in liver microsomes.
Glucuronidation of the C4 or C7 hydroxyl groups would result in metabolites with different spectral properties and thus provide a basis for their identification in the absence of authentic standards. Coumarin rings are inherently fluorescent. That property is significantly increased when a free C7 hydroxyl group is present on the molecule, but the effect is selective. Absorbance properties are minimally affected by the presence of a free or modified 7-hydroxyl group, as demonstrated by our HPLC analysis of 7-hydroxy- and 7-ethoxycoumarin. The same principle can be applied to predicting the structure of the 7-hydroxywarfarin glucuronides. Modification of the C7 hydroxyl group would lead to a much lower fluorescence signal relative to absorbance than that expected for the C4 hydroxyl group. Analysis of the glucuronidation reaction showed that metabolite 2 yielded a much lower relative fluorescent signal than metabolite 1. In fact, the magnitude of this difference (~10-fold) is similar to that observed between 7-hydroxy- and 7-ethoxycoumarin. Consequently, the two monoglucuronides, metabolites 1 and 2, generated from the hepatic microsomal reaction toward R- and S-7-hydroxywarfarin are likely 7-hydroxywarfarin β-D-4-glucuronide (7-OHWAR-4-GLUC) and 7-hydroxywarfarin β-D-7-glucuronide (7-OHWAR-7-GLUC).
Our strategy for assigning possible structures to these glucuronides provides an opportunity to speculate on the identity of metabolites generated by recombinant UGTs. In previous studies [14, 15], recombinant hepatic UGT1A1 and extrahepatic UGT1A10 metabolized R-7-hydroxywarfarin into distinctly different products based on chromatographic properties and patterns of mass spectral signals. This information allowed the authors to conclude that the enzymes generated monoglucuronides in the reaction but the specific site of modification could not be determined. Nevertheless, those monoglucuronides were resolved chromatographically using the same HPLC column and mobile phase, as employed in our study, and thus, the elution order of the metabolites would be conserved between the studies. Analysis of the recombinant enzyme reactions showed that the UGT1A10 product eluted before the UGT1A1 product like metabolites 1 and 2 in our study. Based on this elution order, UGT1A10 glucuronidates R-7-hydroxywarfarin to form 7-OHWAR-4-GLUC, while the UGT1A1 reaction produces 7-OHWAR-7-GLUC. Through this re-analysis of data employing recombinant enzymes [14, 15], we advance an understanding of the stereoselectivity of possible hepatic (UGT1A1) and extrahepatic (UGT1A10) metabolic pathways for R- and S-7-hydroxywarfarin catalyzed by individual UGTs.
The major microsomal pathway for hepatic glucuronidation of R- and S-warfarin involves preferential conjugation at the C7 and not the C4 hydroxyl group (Figure 1). The C7 hydroxyl group is located on the edge of a flat coumarin ring distal from the chiral center possessing bulky substituents. This arrangement makes the C7 hydroxyl group more accessible to glucuronidation and likely explains the regiospecificity of the reaction. The chiral center does influence the reaction mechanism and corresponding parameters though. Metabolism of R-7-hydroxywarfarin yields a hyperbolic kinetic profile as predicted by Michaelis-Menten kinetic for a single active site enzyme [19]. By contrast, metabolism of S-7-hydroxywarfarin demonstrated a kinetic profile consistent with substrate inhibition. These types of non-hyperbolic kinetic profile may occur when initial steady-state conditions are not met during the course of the experiment. We eliminated those possibilities by measuring initial rates in the linear response range for product formation as a function of time and protein concentration. Moreover, product accumulation was not significant during the course of the reaction (< 20 % of substrate concentration), suggesting that the possibility of UDP inhibition of the reaction was minimal [36]. This validation of the kinetic profile confirms the observation of substrate inhibition for S-7-hydroxywarfarin glucuronidation but not the mechanism underlying its occurrence.
The corresponding metabolic mechanism rests on the meaning of the second binding event (Kss). The simplest and most common explanation is the presence of two binding sites for the enzyme. The first binding event results in a binary substrate-enzyme complex that follows traditional Michaelis-Menten kinetics; however, at higher substrate concentrations, a second binding event occurs, whereby the activity of the enzyme decreases or is eliminated altogether. This proposed mechanism has been used to explain multiple UGT reactions [37] including UGT1A10 metabolism of 8-hydroxywarfarins [15]. Alternatively, at higher 7-hydroxywarfarin concentrations, substrate may bind a different UGT present in the microsomal fraction. The formation of that complex could alter protein-protein interactions among the UGTs that govern activity as reported for UGT1A1, 1A6, 1A9, and 2B7 (reviewed in [38]). In this case, a less active UGT complex may supplant a more productive one at higher substrate concentrations. The distinction between these possibilities would require further study. Regardless, R-7-hydroxywarfarin was more efficiently metabolized than the S enantiomer to form the 7-OHWAR-7-GLUC due to higher substrate affinities and rates of turnover.
Hepatic glucuronidation at the C4 hydroxyl group was a minor pathway for both 7-hydroxywarfarins. Presumably, the bulky substituents on the nearby chiral center sterically occlude access to the site for glucuronidation to occur (Figure 1). The orientation of the substituents did impact metabolism as evidenced by a much higher turnover rate for R-7-hydroxywarfarin relative to the S enantiomer. The overall kinetic profile in both cases replicated the substrate inhibition observed for glucuronidation of S-7-hydroxywarfarin at the C7 hydroxyl group. In following, high substrate concentrations induce a shift in observed enzyme activity for these pathways reflecting the contribution of two binding events for the same enzyme or different enzymes that collectively contribute to formation of the 7-OHWAR-4-GLUC. In either case, these metabolic pathways are far more inefficient than glucuronidation at the C7 hydroxyl group for R- and S-7-hydroxywarfarin, such that the latter pathways dominate the metabolic clearance of the warfarin metabolite.
We modeled the overall in vivo metabolic clearance of R- and S-7-hydroxywarfarin taking into consideration the specific mechanisms as described by others [33, 34]. The major metabolic pathway for R-7-hydroxywarfarin conformed to the traditional Michaelis-Menten model for clearance. However, the other metabolic pathways involved substrate inhibition mechanism that predicted a more efficient clearance of the compounds at lower concentrations. In other words, the traditional model would underestimate 7-hydroxywarfarin metabolic clearance in those cases. An important driver in this process is the concentration or plasma level of 7-hydroxywarfarin in warfarin patients, which have been reported to range from 6 to 50 nM for R-7-hydroxywarfarin and 188 to 1140 nM for S-7-hydroxywarfarin [5, 7, 39]. The amount accessible for glucuronidation is actually 20-fold lower than these reported ranges, because only 4.49 % of R-7-hydroxywarfarin and 4.27 % of S-7-hydroxywarfarin are not bound to plasma proteins [40]. Consequently, plasma levels of 7-hydoxywarfarin available for glucuronidation are sub-nanomolar for R-7-hydroxywarfarin and mid to low nanomolar for S-7-hydroxywarfarin. These levels are at least a 1000-fold lower than the binding constants for the reaction reported in this study, and thus they would not be sufficient to saturate the capacity of the liver to glucuronidate R- and S-7-hydroxywarfarin under in vivo conditions. Furthermore, the process of glucuronidation is not efficiently coupled with the formation of 7-hydroxywarfarin by cytochromes P450, because hydroxywarfarin metabolites accumulate in the plasma. Their elimination may ultimately require glucuronidation (or possibly sulfonation) and in fact, nearly all hydroxywarfarins including those in this study are excreted in the urine as conjugates based on the analysis of a limited number of patient samples [8, 41].
Inhibitor phenotyping studies described herein demonstrated that none of the hepatic UGTs dominated 7-hydroxywarfarin metabolism. Multiple UGT1A enzymes as well as the possibility of UGT2B7 contributed to glucuronidation of both hydroxywarfarin enantiomers and the corresponding two glucuronides metabolites. For comparative purposes, only kinetic parameters for recombinant UGT1A1 have been reported for glucuronidation at the C7 hydroxyl group [15]. The Km for the UGT1A1 reaction toward R-7-hydroxywarfarin (130 μM) was comparable to the one observed for the microsomal reaction (121 μM); however, the values were very different for the S-7-hydroxywarfarin, i.e. 230 μM versus 927 μM, respectively. It is conceivable then that UGT1A1 may play an important role in the metabolic clearance of the R-7-hydroxywarfarin, but not the S enantiomer. Nevertheless, the demonstration of multiple UGTs metabolizing 7-hydroxywarfarin is plausible. These UGTs show significant activity toward 4-methylumbelliferone [30], i.e. 4-methyl-7-hydroxycoumarin, which is isosteric for the portion of 7-hydroxywarfarin mainly targeted for glucuronidation. Moreover, others have shown that microsomal glucuronidation of R- and S-7-hydroxywarfarin correlates with the activities of multiple UGT1A and 2B enzymes UGTs [15]. The contributions of multiple UGTs to these metabolic reactions indicate significant redundancy in glucuronidation pathways such that variations in individual UGT activities may be compensated by other UGT activities. This mechanism could explain failure of genome-wide association studies [42–44] to identify any UGT polymorphisms being linked to warfarin dose-responses in patients, such that the clinical relevance of glucuronidation remains unclear.
In sum, we report for the first time the mechanism and parameters for microsomal glucuronidation of R- and S-7-hydroxywarfairn by hepatic UGTs. These findings link together the metabolic pathways determining the conversion of R- and S-warfarin to inactivate metabolites that are ultimately excreted in the urine. These details are critical to assess which metabolic pathways are clinically relevant and understand their mechanism of interaction with other pathways. The glucuronidation of R- and S-7-hydroxywarfarin occurs through multiple pathways carried out by multiple UGTs in the liver. This redundancy precludes the dominance of a single UGT, but does not minimize the potential importance of glucuronidation in the overall clearance of the drug enantiomers. Their clinical relevance rests on the ability of UGT activities to influence the initial oxidative processes mediated by P450s that render the active parent drugs into hydroxywarfarins with little to no anticoagulant properties [3, 4, 11] or capacity to inhibit P450 reactions through feedback inhibition [12]. Further research is necessary to assess the clinical importance of the coupling between those processes. Knowledge of glucuronidation pathways and the identification of putative glucuronide metabolites reported herein provide a sound basis for addressing these gaps in our understanding of warfarin metabolism and bolster efforts to utilize those kinds of metabolites as predictive biomarkers for patient dose-responses to warfarin [4].
Supplementary Material
Table 3.
Preferred model mechanism and parameters for steady-state glucuronidation of R- and S-7-hydroxywarfarin by HLM150a
| Substrate | Metabolitesb | Modelc | Parameters for Preferred Model | |||
|---|---|---|---|---|---|---|
| Vmax1d | Vmax2d | Ks (μM) | Kss (μM) | |||
| R-7-HOWAR | 7-HOWAR-4-GLUC | Substrate Inhibition | 58.0 (49.5–66.1) | 19.0 (16.3–21.4) | 145 (112–183) | 145 (112–183) |
| 7-HOWAR-7-GLUC | Michaelis-Menten | 1090 (1050–1140) | - | 121 (101–144) | - | |
| S-7-OHWAR | 7-HOWAR-4-GLUC | Substrate Inhibition | 19.7 (17.6–22.4) | - | 110 (83.0–149) | 2850 (1950–4400) |
| 7-HOWAR-7-GLUC | Substrate Inhibition | 1470 (1020–2700) | - | 927 (559–1960) | 1310 (538–2810) | |
The nonsymmetrical 95 % confidence intervals for parameters are shown in parentheses.
Glucuronidation resulted in conjugation of the hydroxyl group at either the C4 or C7 position resulting in the formation of 7-HOWAR-4-GLUC or 7-HOWAR-7-GLUC, respectively, as described in the Discussion.
See Table 3 for details for the specific metabolic mechanism describing formation of the respective glucuronides metabolites.
Units are nmol/min/mg protein.
Highlights.
Synthesis of R- and S-7-hydroxywarfarin through green chemistry
Two novel 7-hydroxywarfarin glucuronides generated by microsomal hepatic UGTs
Prediction of glucuronide metabolite structures based on spectral properties
Glucuronidation involved Michaelis-Menten and substrate inhibition kinetics
Multiple UGTs in HLMs contribute to R- and S-7-hydroxywarfarin metabolism
Acknowledgments
Financial support for undergraduate research was provided in part by the Summer Undergraduate Research Fellowship program sponsored by the Biochemistry and Molecular Biology Department at the University of Arkansas for Medical Sciences (CPP and DLP), the UAMS Summer Undergraduate Research Program to Increase Diversity in Research under award number R25HL108825 from the NIH National Heart, Lung and Blood Institute (NHLBI) (DLP), and the Arkansas INBRE program, supported by grant funding from the NIH National Institute of General Medical Sciences (NIGMS) (P2003429, formerly P20RR016460) (CPP). In addition, financial support was provided to GPM through a Grant-in-Aid from the SouthWest Affiliate of the American Heart Association (13GRNT16960043) and a pilot study grants provided by the NIH (UL1 TR000039) awarded to the University of Arkansas for Medical Sciences awarded to GB and GPM. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the University of Arkansas for Medical Sciences, the American Heart Association, or National Institutes of Health.
TEG gratefully acknowledges funding from the Hendrix College Odyssey and Rwanda Presidential Scholars Programs, as well as from John and Laura Byrd. Funding was provided by the U.S. National Science Foundation for the NMR spectrometer (NSF MRI grant to Hendrix College, Award No. 1040470). Nick Heathscott, Doug Hammon, Jake Leffert, Kenyon Plummer, Robert Rurangwa, and Aline Umuhire-Juru contributed to the early stages of the research at Hendrix College.
Abbreviations
- UGT
UDP glucuronosyl transferase
- UDPGA
UDP-glucuronic acid
- OHWAR
hydroxywarfarin
- 7-OHWAR-4-GLUC
7-hydroxywarfarin β-D-4-glucuronide
- 7-OHWAR-7-GLUC
7-hydroxywarfarin β-D-7-glucuronide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ansell J, Hirsh J, Hylek E, Jacobson A, Crowther M, Palareti G. Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2008;133:160S–198S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 2.Breckenridge A, Orme M, Wesseling H, Lewis RJ, Gibbons R. Pharmacokinetics and pharmacodynamics of the enantiomers of warfarin in man. Clin Pharmacol Ther. 1974;15:424–430. doi: 10.1002/cpt1974154424. [DOI] [PubMed] [Google Scholar]
- 3.Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 4.Jones DR, Miller GP. Assays and applications in warfarin metabolism: what we know, how we know it and what we need to know. Expert Opin Drug Metab Toxicol. 2011;7:857–874. doi: 10.1517/17425255.2011.576247. [DOI] [PubMed] [Google Scholar]
- 5.Chan E, McLachlan AJ, Pegg M, MacKay AD, Cole RB, Rowland M. Disposition of warfarin enantiomers and metabolites in patients during multiple dosing with rac-warfarin. Br J Clin Pharmacol. 1994;37:563–569. doi: 10.1111/j.1365-2125.1994.tb04305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ufer M, Kammerer B, Kirchheiner J, Rane A, Svensson JO. Determination of phenprocoumon, warfarin and their monohydroxylated metabolites in human plasma and urine by liquid chromatography-mass spectrometry after solid-phase extraction. J Chromatogr B. 2004;809:217–226. doi: 10.1016/j.jchromb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Jones DR, Boysen G, Miller GP. Novel multi-mode ultra performance liquid chromatography-tandem mass spectrometry assay for profiling enantiomeric hydroxywarfarins and warfarin in human plasma. J Chromatogr B. 2011;879:1056–1062. doi: 10.1016/j.jchromb.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Miller GP, Jones DR, Sullivan SZ, Mazur A, Owen SN, Mitchell NC, Radominska-Pandya A, Moran JH. Assessing cytochrome P450 and UDP-glucuronosyltransferase contributions to warfarin metabolism in humans. Chem Res Toxicol. 2009;22:1239–1245. doi: 10.1021/tx900031z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones D, Moran JH, Miller GP. Warfarin and UDP-glucuronosyltransferases: writing a new chapter of metabolism. Drug Metab Rev. 2010;42:53–59. doi: 10.3109/03602530903209395. [DOI] [PubMed] [Google Scholar]
- 10.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug-Drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCI/AUC) ratios. Drug Metab Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 11.Rulcova A, Prokopova I, Krausova L, Bitman M, Vrzal R, Dvorak Z, Blahos J, Pavek P. Stereoselective interactions of warfarin enantiomers with the pregnane X nuclear receptor in gene regulation of major drug-metabolizing cytochrome P450 enzymes. J Thromb Haemost. 2010;8:2708–2717. doi: 10.1111/j.1538-7836.2010.04036.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones DR, Kim SY, Guderyon M, Yun CH, Moran JH, Miller GP. Hydroxywarfarin Metabolites Potently Inhibit CYP2C9 Metabolism of S-Warfarin. Chem Res Toxicol. 2010;23:939–945. doi: 10.1021/tx1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Weller P, Farrell E, Cheung P, Fitch B, Clark D, Wu S-y, Wang J, Liao G, Zhang Z, Allard J, Cheng J, Nguyen A, Jiang S, Shafer S, Usuka J, Masjedizadeh M, Peltz G. In silico pharmacogenetics of warfarin metabolism. Nat Biotech. 2006;24:531–536. doi: 10.1038/nbt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielinska A, Lichti CF, Bratton S, Mitchell NC, Gallus-Zawada A, Le VH, Finel M, Miller GP, Radominska-Pandya A, Moran JH. Glucuronidation of Monohydroxylated Warfarin Metabolites by Human Liver Microsomes and Human Recombinant UDP-Glucuronosyltransferases. J Pharmacol Exp Ther. 2008;324:139–148. doi: 10.1124/jpet.107.129858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bratton SM, Mosher CM, Khallouki F, Finel M, Court MH, Moran JH, Radominska-Pandya A. Analysis of R- and S-Hydroxywarfarin Glucuronidation Catalyzed by Human Liver Microsomes and Recombinant UDP-Glucuronosyltransferases. J Pharmacol Exp Ther. 2012;340:46–55. doi: 10.1124/jpet.111.184721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansing R, Chao E, Kaminsky L. Phase II metabolism of warfarin in primary culture of adult rat hepatocytes. Mol Pharmacol. 1992;41:209–215. [PubMed] [Google Scholar]
- 17.Ishii Y, Takeda S, Yamada H, Oguri K. Functional protein-protein interaction of drug metabolizing enzymes. Front Biosci. 2005;10:887–895. doi: 10.2741/1583. [DOI] [PubMed] [Google Scholar]
- 18.Crespi C, Francis E, Patten C. Optimizing Donor Number for Consistent Pooled Human Liver Microsomes. BD Biosciences – Discovery Labware; Billerica, MA: 2009. [Google Scholar]
- 19.Michaelis L, Menten ML. Die Kinetik der Invertinwirkung. Biochem Z. 1913;49:333–369. [Google Scholar]
- 20.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV protease. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 21.Hartman JH, Knott K, Miller GP. CYP2E1 hydroxylation of aniline involves negative cooperativity. Biochem Pharmacol. 2014;87:523–533. doi: 10.1016/j.bcp.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Yen C, Preston P, Chin J. Substrate-Directed Stereoselectivity in Vicinal Diamine-Catalyzed Synthesis of Warfarin. Organic Lett. 2006;8:5239–5242. doi: 10.1021/ol062000v. [DOI] [PubMed] [Google Scholar]
- 23.Wong TC, Sultana CM, Vosburg DA, Green A. Enantioselective Synthesis of Warfarin for the Undergraduate Organic Laboratory. J Chem Educ. 2010;87:194–195. [Google Scholar]
- 24.Valente E, Porter WR, Trager WF. Conformations of selected 3-substituted 4-hydroxycoumarins in solution by nuclear magnetic resonance. Warfarin and phenprocoumon. J Med Chem. 1978;21:231–234. doi: 10.1021/jm00200a019. [DOI] [PubMed] [Google Scholar]
- 25.Giannini D, Chan KK, Roberts JD. Carbon-13 nuclear magnetic resonance spectroscopy. Structure of the anticoagulant warfarin and related compounds in solution. Proc Natl Acad Sci U S A. 1974;71:4221–4223. doi: 10.1073/pnas.71.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermodson M, Barker WM, Link KP. Studies on the 4-hydroxycoumarins. Synthesis of the metabolites and some other derivatives of warfarin. J Med Chem. 1971;14:167–169. doi: 10.1021/jm00284a022. [DOI] [PubMed] [Google Scholar]
- 27.Doerge DR, Chang HC, Churchwell MI, Holder CL. Analysis of Soy Isoflavone Conjugation In Vitro and in Human Blood Using Liquid Chromatography-Mass Spectrometry. Drug Metab Dispos. 2000;28:298–307. [PubMed] [Google Scholar]
- 28.Greenlee WF, Poland A. An improved assay of 7-ethoxycoumarin O-deethylase activity: induction of hepatic enzyme activity in C57BL/6J and DBA/2J mice by phenobarbital, 3-methylcholanthrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Pharmacol Exp Ther. 1978;205:596–605. [PubMed] [Google Scholar]
- 29.Bosma PJ, Seppen J, Goldhoorn B, Bakker C, Oude Elferink RP, Chowdhury JR, Chowdhury NR, Jansen PL. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J Biol Chem. 1994;269:17960–17964. [PubMed] [Google Scholar]
- 30.Uchaipichat V, Mackenzie PI, Guo XH, Gardner-Stephen D, Galetin A, Houston JB, Miners JO. Human UDP-Glucuronosyl Transferases: Isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclophenac and probenecid. Drug Metab Dispos. 2004;32:413–423. doi: 10.1124/dmd.32.4.413. [DOI] [PubMed] [Google Scholar]
- 31.Krishnaswamy S, Duan SX, von Moltke LL, Greenblatt DJ, Court MH. Validation of Serotonin (5-Hydroxtryptamine) as an in Vitro Substrate Probe for Human UDP-Glucuronosyltransferase (UGT) 1A6. Drug Metab Dispos. 2003;31:133–139. doi: 10.1124/dmd.31.1.133. [DOI] [PubMed] [Google Scholar]
- 32.Gaganis P, Miners JO, Knights KM. Glucuronidation of fenamates: Kinetic studies using human kidney cortical microsomes and recombinant UDP-glucuronosyltransferase (UGT) 1A9 and 2B7. Biochem Pharmacol. 2007;73:1683–1691. doi: 10.1016/j.bcp.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Houston J. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem Pharmacol. 1994;47:1469–1479. doi: 10.1016/0006-2952(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 34.Houston JB, Kenworthy KE. In vitro-in vivo scaling of CYP kinetic data not consistent with the classical Michaelis-Menton model. Drug Metab Dispos. 2000;28:246–254. [PubMed] [Google Scholar]
- 35.Valente E, Lingafelter EC, Porter WR, Trager WF. Structure of warfarin in solution. J Med Chem. 1977;20:1489–1493. doi: 10.1021/jm00221a025. [DOI] [PubMed] [Google Scholar]
- 36.Luukkanen L, Taskinen J, Kurkela M, Kostiainen R, Hirvonen J, Finel M. Kinetic Characterization of the 1A Subfamily of Recombinant Human UDP-Glucuronosyl Transferases. Drug Metab Dispos. 2005;33:1017–1026. doi: 10.1124/dmd.105.004093. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, Miners J. Enzyme Kinetics of Uridine Diphosphate Glucuronosyltransferases (UGTs) In: Nagar S, Argikar UA, Tweedie DJ, editors. Enzyme Kinetics in Drug Metabolism. Vol. 1113. Humana Press; 2014. pp. 203–228. [DOI] [PubMed] [Google Scholar]
- 38.Ishii Y, Takeda S, Yamada H. Modulation of UDP-glucuronosyltransferase activity by protein-protein association. Drug Metab Rev. 2010;42:145–158. doi: 10.3109/03602530903208579. [DOI] [PubMed] [Google Scholar]
- 39.Zuo Z, Wo SK, Lo CMY, Zhou L, Cheng G, You JHS. Simultaneous measurement of S-warfarin, R-warfarin, S-7-hydroxywarfarin and R-7-hydroxywarfarin in human plasma by liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal. 2010;52:305–310. doi: 10.1016/j.jpba.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Chan E, McLachlan AJ, Rowland M. Warfarin metabolites: stereochemical aspects of protein binding and displacement by phenylbutazone. Chirality. 1993;5:610–615. doi: 10.1002/chir.530050808. [DOI] [PubMed] [Google Scholar]
- 41.Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 42.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen AL, Al-Zubiedi S, Zhang JE, Keniry A, Hanson A, Hughes DA, Eker D, Stevens L, Hawkins K, Toh CH, Kamali F, Daly AK, Fitzmaurice D, Coffey A, Williamson PR, Park BK, Deloukas P, Pirmohamed M. Genetic and environmental factors determining clinical outcomes and cost of warfarin therapy: a prospective study. Pharmacogenet Genomics. 2009;19:800–812. doi: 10.1097/FPC.0b013e3283317ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.