Abstract
Identifying driver genes in cancer remains a crucial bottleneck in therapeutic development and basic understanding of the disease. We developed Helios, a novel algorithm that integrates genomic data from primary tumors with data from functional RNAi screens to pinpoint driver genes within large recurrently amplified regions of DNA. Applying Helios to breast cancer data identified a set of candidate drivers highly enriched with known drivers (p-value < e−14). 9/10 top scoring Helios genes are known drivers of breast cancer and in vitro validation of 12 novel candidates predicted by Helios found 10 conferred enhanced anchorage independent growth, demonstrating Helios’s exquisite sensitivity and specificity. We extensively characterized RSF-1, a driver identified by Helios whose amplification correlates with poor prognosis, and found increased tumorigenesis and metastasis in mouse models. We have demonstrated a powerful approach for identifying novel driver genes and how it can yield important insights into cancer.
Introduction
Cancer genome data collected by projects such as the TCGA or ICGC is defining the landscape of genetic alterations that underlie cancer. Tumor cells may harbor thousands of genetic lesions including point mutations, somatic copy-number alterations, and translocations that localize to hundreds or even thousands of genes. However, most affected genes are so-called passengers and their alteration does not confer any type of advantage to tumors (Vogelstein et al., 2013). A pivotal challenge in cancer genomics is to identify the small subset of altered genes (so-called drivers) that directly contribute to tumor fitness and progression.
Exome sequencing studies helped identify driver genes (Curtis et al., 2012; Stephens et al., 2012), however the majority of point mutations display low population frequencies, with only a handful altered in greater than 5% of patients (Stephens et al., 2012). In breast cancer, only 6 genes have point mutations in >5% of samples, and of these, only PIK3CA (36% frequency) is currently targeted therapeutically (TCGA, 2012). Instead, the most recurrent genetic lesions in breast cancer are somatic copy number alterations (SCNAs), often driven by inactivation of DNA repair genes such as BRCA1/2. Indeed, HER2, one of the most therapeutically targeted drivers in breast cancer, is primarily dysregulated by copy-number amplification.
The ability to discern drivers from copy-number alteration promises to dramatically expand the set of therapeutic targets in this disease. However, this potential is crucially hindered by the difficulty of driver discovery (Yuan et al., 2012). The crux of the difficulty is that in all but a few instances, these lesions contain dozens of genes and no previously characterized drivers (Albertson et al., 2003). A recent study analyzing multiple tumor types reported that over 70% of 140 recurrently altered regions did not contain a known oncogene or tumor suppressor (Zack et al., 2013). As a result, most recent driver discovery efforts have focused on point mutations, which directly indicate the target genes by virtue of their precise location (Kandoth et al., 2013; Lohr et al., 2012; Wong et al., 2011), and less progress has been made with respect to SCNAs. However, the increased frequency of recurring SCNAs relative to mutations (87 SCNA regions vs. 6 mutated genes with >5% population frequency) (Figure 1A) highlights the need for methods to pinpoint drivers within these regions.
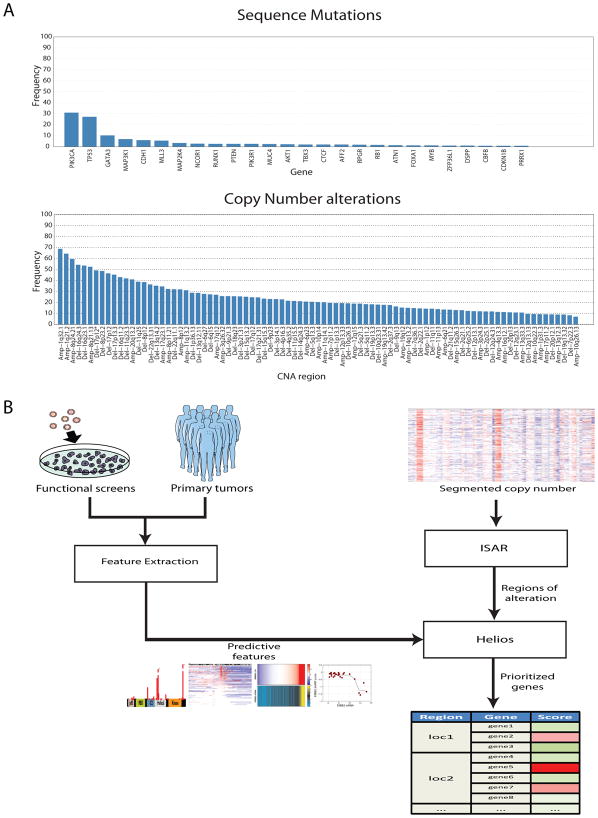
Figure 1. Helios integrates data from primary tumor and functional screens.
(A) Frequency of alteration in the TCGA breast cancer dataset of (top) genes with recurrent point mutations and (bottom) regions of recurrent copy number alteration. Significant genes and regions were downloaded from the DBroad Genome Data Analysis Center, selecting the TCGA pipeline algorithms GISTIC2 (v. 4.2012021700.0.0) and MutSig (v. 4.2011112800.0.0) (B) A schematic of our pipeline for the identification of candidate driver genes. The method first uses ISAR to identify regions of focal SCNAs. To pinpoint drivers within those regions, it extracts features from genetic, genomic and functional data, which are integrated into a single probabilistic score by Helios.
Genome-wide pooled-RNAi screening is an alternative approach to driver gene discovery. In these studies, a shRNA library is transduced into cancer cell lines and the growth effect of each individual gene knock-down is assessed for each cell line (Cheung et al., 2011; Marcotte et al., 2012; Silva et al., 2008). While such studies can provide gene-level resolution, they are currently limited by the high degree of noise, the potential for off-target effects of shRNAs and by the artificiality of the in-vitro screening system (Kaelin, 2012). Moreover, cell-lines are not fully representative of primary tumor biology as these lack tissue structure and microenvironment, which are key to cellular behavior (Bissell and Hines, 2011).
Given the largely orthogonal strengths and weaknesses of descriptive analysis of primary cancer genomes and in-vitro genome-wide functional screening, we hypothesized that integrating the two data types into a single approach would result in increased resolution and accuracy for driver gene discovery. Therefore, we developed Helios (Figure 1B), a novel algorithm that incorporates primary tumor SCNA, point mutation, gene expression, and RNAi screens into a single candidate driver score. Helios runs in two steps, first identifying regions of focal SCNAs and then identifying driver genes within each region by integrating functional screens and other data using a Bayesian transfer-learning framework.
Helios displayed a remarkable capacity to pinpoint bona fide cancer drivers when the algorithm was used to analyze the SCNA landscape of breast cancer. In a systematic evaluation of Helios’s performance, we selected 12 novel driver candidates identified by Helios, based on their frequency of occurrence, for experimental investigation. We found 10/12 candidate genes induced increased anchorage independent growth when over-expressed in vitro. Thus, Helios demonstrated an unprecedented sensitivity and specificity in identifying genes that promote oncogenic capabilities. Helios doubled the number of SCNA drivers identified in breast cancer and substantially increased our understanding of the breast cancer SCNA landscape.
Results
ISAR expands the list of significantly amplified regions in breast cancer
The first step for identifying SCNA-drivers is identification of significantly altered regions. There are multiple algorithms that successfully perform this task (Mermel et al., 2011; Walter et al., 2011), GISTIC2 being the most widely used among these. We noted a number of oncogenes (e.g. BCL2) that were not detected as falling within a significantly altered region by GISTIC2 in the TCGA breast cancer data (TCGA, 2012). By visual inspection of chromosome 18, we noted that while BCL2 does not appear significantly amplified based on its absolute copy-level, its copy-number is nevertheless significantly higher than the adjacent chromosomal regions (Supplementary Figure 1). Most SCNA detection algorithms, including GISTIC2, compute a null distribution across the entire genome to estimate the significance of alterations. However, the alteration rate can strongly differ across different genomic regions, due to features such as DNA secondary structure and DNA hypomethylation (De and Michor 2011).
Therefore, we developed ISAR (Identification of Significantly Altered Regions), an algorithm that accounts for local differences in SCNA rate due to these and other forces. By computing the significance locally, the algorithm is capable of identifying both global alteration events, as well as subtle events, such as a focal amplification within largely deleted regions, that would be missed if the background distribution for the whole genome were employed (See Methods). We applied ISAR to 785 breast cancer samples (TCGA, 2012) and identified 83 significantly amplified regions (see Supplementary table 1), compared to the 30 regions originally reported by the TCGA consortium. ISAR captures all significant regions captured by GISTIC2 and many additional regions. Among the new regions we find many bona-fide or likely oncogenes, including MYB, BCL2, CDK4, ESR1, FGFR2, FGFR3 and FGFR4. Identified regions contained an average of 14 genes resulting in a total of 1226 significantly amplified genes across all 83 regions.
Helios: An Integrative Approach to Pinpoint Drivers
Helios seeks to exploit additional properties —e.g. recurrent domain-specific point mutations or depletion in a lethality shRNA screen—to implicate likely driver genes targeted by the SCNA. Helios considers the entire significantly altered region, but prioritizes the genes within this region by incorporating cues from additional genetic and genomic data to estimate the probability that each is a driver (Figure 2A). It is a statistically rigorous framework for combining multiple signals that might lack power individually into a single score for the likelihood that each gene’s amplification specifically increases tumor fitness. Here, we integrate features derived from exome-sequencing, shRNA screening and gene-expression, but due to the flexibility of our framework, these could readily be removed, modified or extended for subsequent studies.
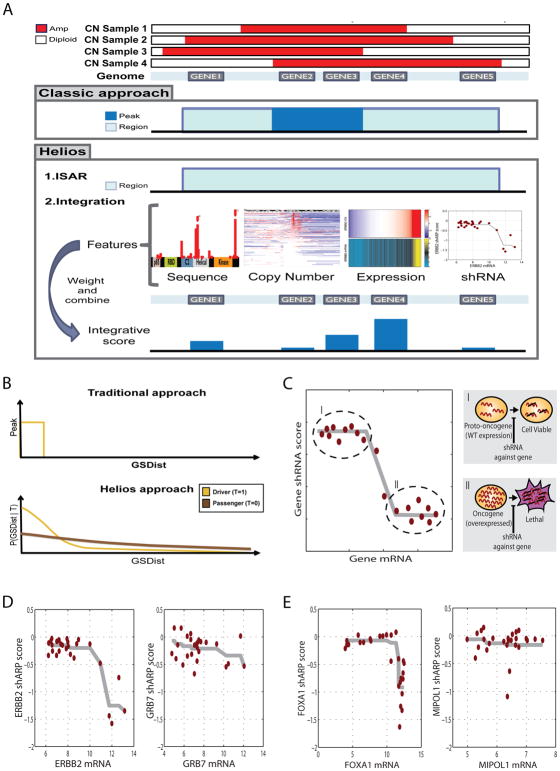
Figure 2. Helios features.
(A) Diagram of the classic and Helios approach. While the classic approach relies solely on copy number, both to identify significantly altered regions and to further narrow down those region to a minimal region of maximal alteration, Helios identifies regions in the same fashion, but then integrates features extracted from different data sources to compute the probability of each gene being a target of the region. (B) Diagram of the copy number model of the Helios Algorithm. The classic approach (top) calculate a hard threshold on the delta to the most altered marker (GSDist, X axis) to define the peak region (Y axis). Helios (bottom) instead calculates the probability (Y axis) of displaying a GSDist value (X axis) for both driver and passenger genes (yellow or brown curves respectively). (C) Our oncogene addiction score uses monotonic regression to measure the association between gene dosage (X axis) and shRNA dropout (Y axis), aiming to differentiate the proto-ongonenic state (I) of the driver, which is expressed at wild type levels, and the oncogenic state (II), which is characterized by high expression and high dependency on the gene for survival. (D) Monotonic regression of the shRNA dropout (Y axis) based on the gene dosage (X axis) for the two top scoring genes for oncogene addiction in the 17q12 region. (E) Monotonic regression of the shRNA dropout (Y axis) based on the gene dosage (X axis) for the two top scoring genes for oncogene addiction in the 14q13 region.
Helios uses a set of features to classify genes as either drivers or passengers, based on inference within a hierarchical Bayesian mixture model (see Methods, Supplementary Figure 2). Standard classification approaches rely on an initial list of examples—drivers and passengers—to train the model. Unfortunately, the list of known oncogenic drivers is relatively small and strongly biased towards kinases and extreme phenotypes that facilitate discovery. Instead, Helios begins with the assumption that a driver gene is more likely to be near the most frequently amplified segment (defined as peak) of the ISAR region. This is used to initialize the algorithm by providing an estimated list of drivers to start from. Helios then iterates between 2 stages until convergence:
Learning the parameters to distinguish passengers and drivers on the basis of their SCNA profile and on the additional genomic data
Re-computing the probability that each gene is a driver using the parameters determined in step 1
Helios uses a transfer learning approach (Widmer and Rätsch, 2011) whereby drivers with clearer signal (e.g. at the peak of their region) are used to extract informative features to improve performance in cases with less obvious signal. Helios automatically learns the weights of features directly from the data by leveraging information among features. In each iteration, Helios learns a better classification of drivers and passengers, which in turn is used to learn better parameters, until convergence (see Supplementary Methods). Helios utilizes a mixture of two copy-number distributions– one for drivers and one for passengers, thus avoiding the problematic selection of a hard threshold for defining aberrant regions (Figure 2B). Additionally, Helios permits final models where more than one gene in a region is identified as a likely driver, or where no probable driver genes are identified.
Finally, Helios can readily incorporate additional features, including complex features generated by combinations of multiple data sources. It automatically learns the contribution and importance of each feature directly from the data, making it easily extendable and adaptable to other cancer types. For example, here, we integrate data from functional screens based on the concept of oncogene addiction (Weinstein and Joe, 2008) by deriving a composite statistic reflecting the extent to which shRNA-depletion in a genome-wide screen correlated with over-expression of the gene at baseline. A similar idea has recently been used to discover the novel oncogene HNF1B (Shao, Tsherniak et al. 2013). Our oncogene addiction score allows for both linear and non-linear relations between gene expression and lethality (See Figure 2B–D, Methods). This ability to combine multiple weaker pieces of evidence from heterogeneous data types into a single score enables Helios to effectively pinpoint the driver gene from within the recurrently altered region.
Helios identifies candidate drivers of breast cancer
We used Helios to integrate TCGA data from 785 primary breast cancer tumors, including DNA copy number, gene expression and sequence mutations (TCGA, 2012), with data from 27 breast cancer cell lines including gene expression, copy number and shRNA depletion in a genome-wide shRNA screen (Barretina et al., 2012; Marcotte et al., 2012).
Using stringent criteria, we defined 64 candidate drivers by selecting only the top gene in each region and applying a threshold of Helios score > 0.5 (see Supplementary Table 2). Some significant SCNA regions did not contain a high scoring protein-coding gene; these amplifications potentially target non-coding RNA or other genomic features. For example, all protein-coding genes were low-scoring in an amplified region containing the known oncomir mir21 (O’Day and Lal, 2010). While approximately 20% of the regions contained more than one high scoring gene, we limited our initial analysis to the highest scoring gene in each region.
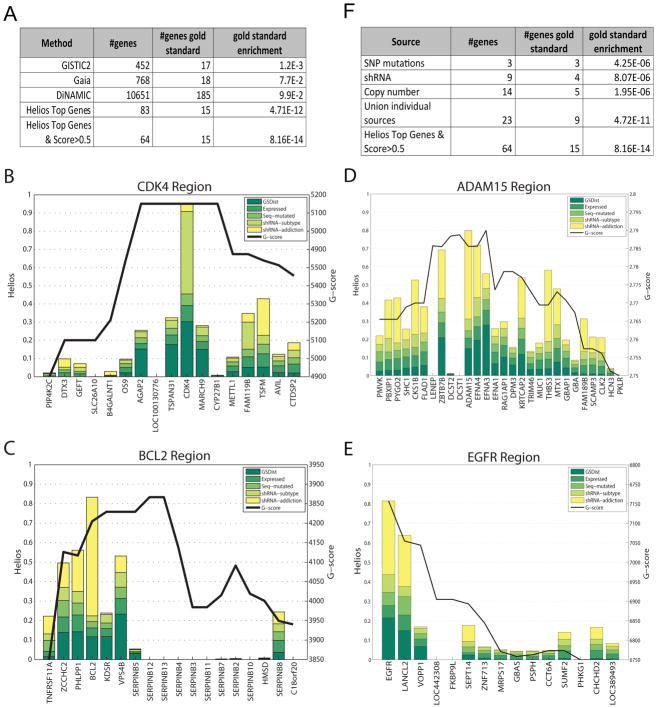
To evaluate the sensitivity of our approach, we combined several publically available resources to create a comprehensive set of breast cancer oncogenes ((Beroukhim et al., 2010; Consortium, 2013; Frankild and Jensen), (Supplementary methods)). Among the 10 top scoring Helios genes, 9 were included in this set (FOXA1, PIK3CA, CCND1, CDK4, MYB, ERBB2, IGF1R, BCL2, ESR1), while only 5 of these appear in regions that are significant based on GISTIC2. Moreover, the entire list of 64 Helios candidates was significantly enriched for our compiled set of breast cancer drivers (16/64, p-value < 4e−15), a large improvement over the set of all genes in amplified regions identified by GISTIC2 (17/452, p-value > e−3) (TCGA, 2012) (Figure 3A). The performance of the method was also compared against two other algorithms, GAIA (Morganella et al., 2011) and DiNAMIC (Walter et al., 2011), outperforming both of them (18/768, p-value > e−3 and 185/10651, p-value > e−3 respectively). This demonstrates the significant improvement of our integrative approach over the state of the art.
Figure 3. Helios analysis of Breast Cancer.
(A) A comparison of enrichment for a literature- compiled set of breast cancer drivers between our Helios genes defined as the top gene in each region with a score greater than 0.5) and three state of the art methods. (B),(C),(D) and (E) display the result of the Helios analysis for the 12p14, 18q21, 1q21 and 7p12 regions respectively. Genes in the ISAR regions are displayed in the X axis and the Helios score is represented by bars colored proportionally to the contribution of each feature (a logistic regression approximation is employed to approximate the contribution of each feature). The ISAR score is displayed as a black line. (F) A comparison between Helios and the results from the analysis of the data sources individually, testing for enrichment based on our literature compiled set of breast cancer drivers. See Supplementary Figure 3A–D for information about convergence and stability of the results.
Helios’s integration across multiple data sources is key to its ability to be both specific and sensitive. Sequence mutations are gene-specific, but only few drivers harbor such mutations recurrently. SCNAs typically cover a large number of genes, making it hard to identify the target of the amplification based on copy number alone. For instance, CDK4 shares exactly the same copy number profile with its five closest neighbors, but the lethality displayed by CDK4 in the shRNA screen raises its Helios score (Figure 3B). More strikingly, BCL2 is only the sixth gene in its region in terms of copy number alteration frequency, but its dramatic oncogene addiction score raises its Helios score well above all others in the region (Figure 3C). In many cases (e.g. EGFR or ADAM15, Figure 3D–E), it is not any single feature, but a combination of features that identifies the top-scoring gene in the region. Figure 3F shows how Helios outperforms the simple use of the data sources independently to identify drivers. Even if all of the candidates obtained by each data source are joined together naively, Helios provides significantly better sensitivity (15 versus 9 detected driver genes) and specificity (hypergeometric enrichment p-value of driver genes 8.16E−14 versus 4.72E−11).
Candidate selection for systematic in vitro validation of Helios-predicted genes
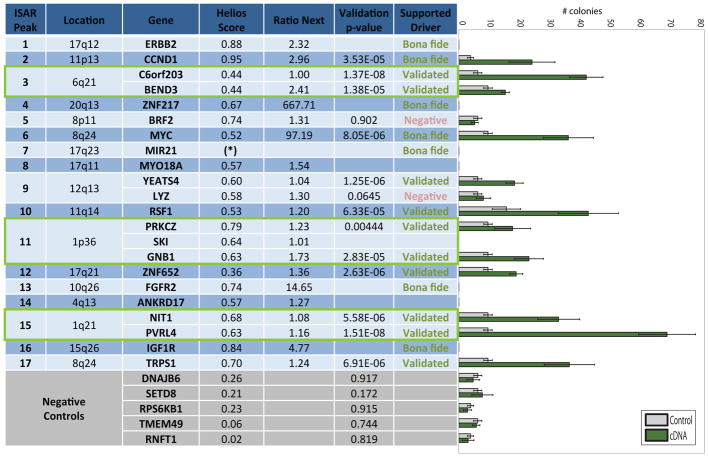
Helios is designed to rank genes within an amplified region based on their likely driver capacity. Contrary to most prior work that prioritized kinases for experimental validation, for an unbiased evaluation of Helios, we chose a systematic score driven approach to validation. To perform an unbiased and comprehensive assessment, over a wide range of Helios scores, we used the independent ISAR score > 5.5 to select regions and used Helios to pinpoint the most likely driver within each region. Thus we sought to assess how often could Helios pinpoint the correct driver for each of the 17 most frequently and significantly amplified regions.
In 7 of the 17 regions, the top Helios gene was a bona-fide breast cancer oncogene (ERBB2, CCND1, ZNF217, MYC, miR-21, FGFR2 and IGF1R) and these oncogenes scored well above the next best scoring gene. For example, MYC’s Helios score was 100 times greater than the 2nd best gene in the region (Figure 4, “Ratio-next” column). There was no known breast cancer oncogene present among 10 additional regions and therefore we decided to perform in vitro validation for the top scoring Helios genes in each of these regions. Since an amplified region can harbor more than one oncogene, we selected multiple genes if more than one was significantly scoring (4/10 regions). We failed to clone over-expression vectors for three genes, resulting in a final selection of 12 predicted oncogenes for validation. The selected genes encompassed a wide range of functional roles including chromatin remodeling, transcription factors, cell surface and cell adhesion proteins and metabolic enzymes.
Figure 4. Helios validations.
Results of the systematic in-vitro validation of Helios candidates including the selected genes and 5 genes selected as negative controls (highlighted in grey). The ‘Ratio Next’ column indicates the ratio between the Helios score of the candidate gene and the score of the next best scoring gene in the region. The ‘Validation p-value’ displays the statistical significance of the change in colony size between the 6 empty vector controls and the 6 repeats of the cDNA overexpressing the candidate driver gene. This p-value was computed using a right-tailed unpaired two-sample t-test. The ‘Supported Driver’ column indicates if the gene has been positively validated by the in-vitro assay or a known driver based on previous literature. The rightmost panel shows the box plots of the colony numbers for each gene in the validation experiment, where grey indicates the control and green the cDNA overexpressing the candidate driver gene. The colony assay was not performed for several genes that we failed to clone (MYO18A, SKI), or were bona fide drivers at the top of their peak (ERBB2, ZNF217, FGFR2, ANKRD17, IGF1R). Additionally, no gene scored above 0.3 in the 17q23 region, suggesting that the target was another regulatory element, in this case the bona fide onco-microRNA MIR21. The three green boxes highlight amplified regions in which we confirmed more than one driver. The colony data that supports this figure is available in Supplementary Figures 4B (candidate drivers) and 4C (negative controls).
One of the hallmarks of transformation that is commonly used to investigate putative epithelial oncogenes is the ability to promote attachment independent growth of a non-transformed cell line (Hanahan and Weinberg, 2011). This capacity likely reflects the cumulative impact of multiple signals such as increased resistance to stress, increased cellular growth rates and changes in metabolism (Davison et al., 2013). As a result, many driver alterations in cancer may potentially impact attachment independent growth through multiple mechanisms. Therefore, we based our candidate validation strategy on assaying this phenotype.
Experimental in vitro validation confirms Helios-predicted genes
For each of the 12 candidate genes, we evaluated the ability of a clone of MCF-10A cells (human mammary epithelium) with intrinsic low attachment independent growth ability, (see methods) to form colonies in semi-solid media when the putative oncogene was experimentally upregulated. These cells were transduced with viral vectors over-expressing the putative driver and evaluated for growth in soft agar. CCND1 and MYC were used as positive controls, and for negative controls, we selected 5 genes from significantly amplified ISAR regions (ISAR > 5.5) that did not have a high Helios score (score < 0.3). The agar assays for each gene was tested with a minimum of 6 replicates and statistical significance was evaluated by unpaired two-sample t-test between the 6 test and 6 control plates.
10/12 tested genes (C6ORF23, BEND3, YEATS4, RSF-1, PRKCZ, GNB1, ZNF652, NIT1, PVRL4 and TRPS1) were able to significantly increase MCF10A anchorage independent activity with a p-value of 0.005 or below (Figure 4). None of the negative controls demonstrated an increase in colony formation. This provides in vitro evidence that Helios is highly specific in identifying genes that provide a selective advantage for breast cancer cells. Note that a negative result for BRF2 (demonstrated to be an oncogene in lung cancer (Lockwood et al., 2010)) does not conclusively rule it out as a driver gene, since attachment independent growth is not the only hallmark of cancer and the assays were performed in a single genetic background.
Overall, Helios demonstrated unprecedented accuracy in identifying genes that promote oncogenic capabilities. Helios correctly scored 13/14 drivers at the top of their respected region (93%). Moreover, 10/12 empirically tested genes validated (83%), thus we identified 9 new genes that promote tumorigenic capabilities in breast cancer (excluding PVRL4 which was recently published (Pavlova et al., 2013)). Additionally, since the genes were selected based on the region’s significance, rather than their Helios score, a wide range of Helios scores were tested (between 0.36 to 0.79), increasing our confidence in the candidates identified in other regions. Based on this performance, we expanded our list of likely drivers based on Helios predictions with more permissive criteria (Supplementary Table 3).
Importantly, Helios identified multiple high scoring (likelihood>0.5) genes for over 20% of the regions. Indeed, we validated three regions with multiple genes and each gene independently induced colony formation in vitro (Figure 4, green boxes), indicating that an amplicon often targets more than one gene. In summary, while previously only 7/17 of the most frequently altered regions in breast cancer harbored a known oncogene, following our validation 14/17 regions can be assigned a driver with substantial confidence.
RSF-1 Promotes Colony Growth In Vitro
Among the 10 validated candidates, RSF-1 is an especially compelling putative driver because it is recurrently amplified in several cancers (Chen et al., 2011; Fang et al., 2011; Li et al., 2012; Liu et al., 2012; Shih Ie et al., 2005). Additionally, an amplicon containing RSF-1 was recently associated with a breast cancer subtype bearing one of the worst clinical prognoses (Curtis et al., 2012). Although high expression levels of RSF-1 has been associated with poor prognosis in several malignancies (Hu et al., 2012; Li et al., 2012; Liu et al., 2012; Sheu et al., 2013), its involvement in breast cancer pathogenesis has not yet been explicitly demonstrated. Therefore, we chose to follow-up our analysis of RSF-1 with further in-vitro and in-vivo experiments.
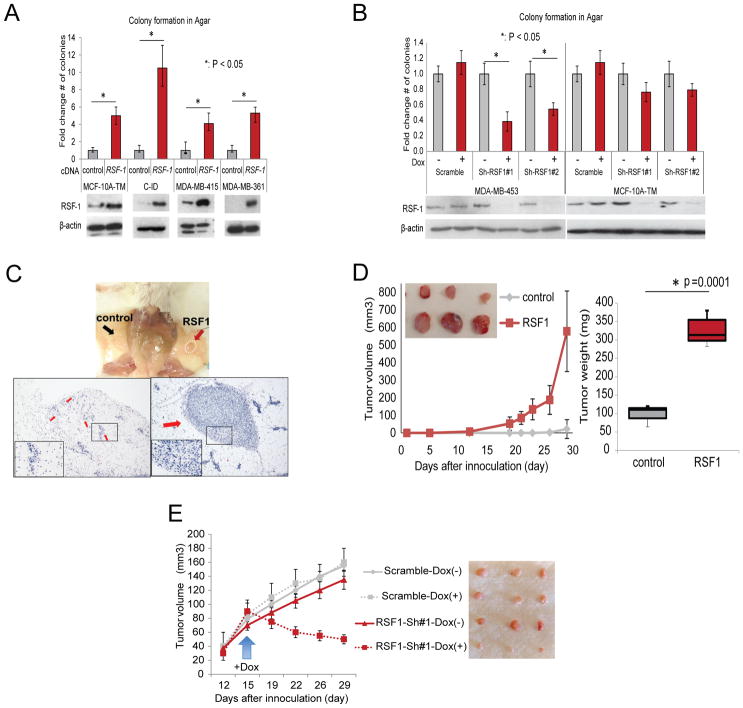
We selected four additional mammary epithelial cell lines non-amplified for RSF-1. The human MCF-10A-Triple Modified (a MCF-10A variant sensitized to transformation called here MCF-10A-TM (Pires et al., 2013)), MDA-MB-415 and MDA-MB-361; and the mouse Comma-ID (C-ID) (Campbell et al., 1988). We also selected one cell line (MDA-MB-453, human) with amplified and over-expressed RSF-1 (Supplementary Figure 5A). Overexpression of RSF-1 in all non-amplified cell lines increased the ability to form colonies in semisolid media (Figure 5A). To assay RSF-1 oncogene addiction, we selected two doxycycline (Dox) inducible shRNA-miRs that efficiently silenced RSF-1 and assayed colony formation of the RSF-1 amplified MDA-MB-453 line. As expected, silencing of RSF-1 significantly reduced the number of colonies formed (Figure 5B). To demonstrate that the loss of tumorigenic potential is not an off-target effect, we restored RSF1 expression in these cells by overexpressing the RSF1 cDNA (Supplementary Figure 5C). Restoring RSF1 levels rescued the ability of MDA-MB-453 to form colonies in agar despite the expression of RSF1 shRNAs.
Figure 5. High expression levels of RSF-1 promote tumorigenesis.
(A) Overexpression of RSF-1 in multiple cell lines enhances its ability to form colonies in agar. (B) downregulation of RSF-1 using dox inducible shRNAs in a cell line with amplification of the locus (MDA-MB-453) reduced its ability to form colonies in agar. Overexpression of RSF-1 in (C) MCF-10A-TM and (D) CID cells enhanced their tumorigenic potential in vivo. The MCF-10A-TM model generated small tumor masses, thus H&E images are also provided. Number of tumors formed for each model is available in Supplementary Figure 5B. (E) Silencing of RSF-1 in MDA-MB-453 attenuated its tumorigenic potential when orthotopically transplanted in SCID mice.
RSF-1 Promotes Growth in Xenograft Models
Next, we conducted experiments to assay RSF-1 in vivo. MCF-10A-TM and C-ID were orthotopically transplanted into the fat pad of immunocompromised (SCID) mice with and without prior transduction of an RSF-1 over-expression vector. We then tracked the development of tumors and compared growth between controls and those over-expressing RSF-1.
MCF-10A cells are not tumorigenic, and overexpression of RSF-1 did not transform them. While some transplanted MCF-10A-TM cells remained in the fat pad, these did not produce tumor. However MCF-10A-TM overexpressing RSF-1 was able to establish small primary tumor outgrowths (Figure 5C and Supplementary Figure 5B). C-ID overexpressing RSF-1 cells generated palpable masses as early as 2 weeks after transplantation—significantly earlier than control mice, which lacked detectable tumor burden after 1 month. (P=0.0001) (Figure 5D and Supplementary Figure 5B).
Finally, we also transplanted RSF-1 amplified MDA-MB-453 cells and an MDA-MB-453 variant bearing a doxycycline inducible RSF-1-ShRNA into the fat pad of SCID mice. As expected, in the absence of Dox all MDA-MB-453 variants generated tumors that grew at a comparable rate. However, supplementing the mice with Dox reduced the tumorigenic growth specifically in the tumors carrying the RSF-1 shRNA (Figure 5E). This data provides evidence that RSF-1 can contribute to tumor progression in vivo and that inhibition of RSF-1 expression can cause tumor regression.
RSF-1 Promotes invasion in Xenograft Models
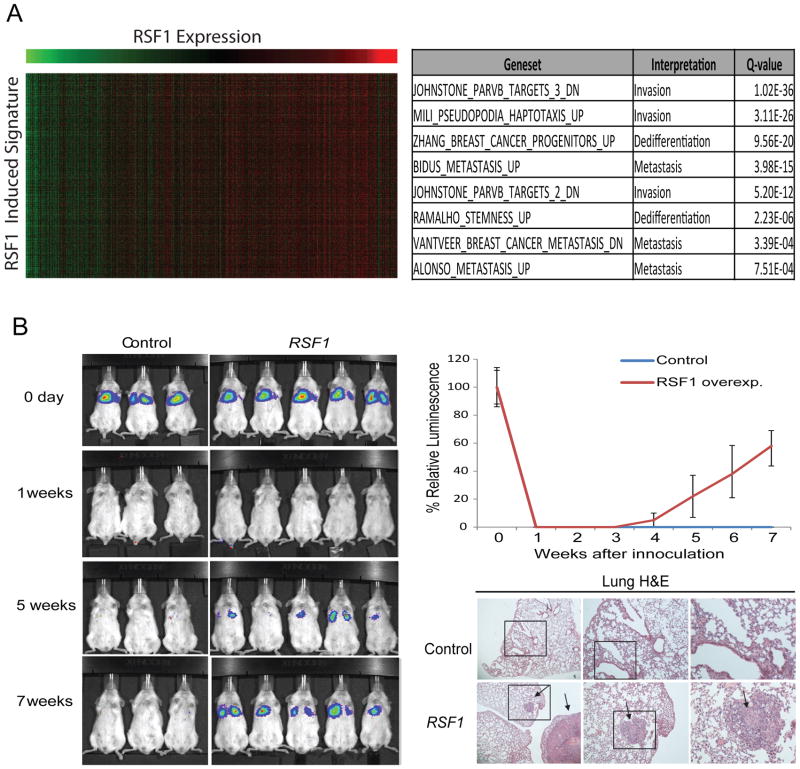
To further characterize the role of RSF-1 in breast cancer, we analyzed the TCGA gene expression data and identified gene-expression signatures associated with RSF-1 expression levels ((Akavia et al., 2010; Danussi et al., 2013), (Supplementary Methods). Genes associated with RSF-1 in this procedure are putative downstream targets of RSF-1 activity. We performed gene set enrichment in these signatures using the MSigDB database (Subramanian et al., 2005) and found enrichment for gene sets involved in invasion, metastasis, and de-differentiation (Figure 6A, Supplementary Figure 6A).
Figure 6. RSF-1 alteration promotes metastasis.
(A) The analysis of the expression changes related to RSF-1 overexpression in basal primary tumors revealed a signature enriched for invasiveness, migration and dedifferentiation (table at right). The heat map at left shows genes in the signature as rows and samples as columns and the color indicates the relative expression (green-low and red-high) and demonstrates the tight correlation of the signature genes across patients. Similar results were observed for luminal primary tumors (Supplementary Figure 6A). See Supplementary Figure 6B-C for analysis of downregulated genes. (B) Comparison of lung metastasis formation in SCID mice subjected to tail vein injection of MCF-10A-TM cells expressing a luciferase reporter and either an RSF-1 over-expression vector or a control vector. H&E of sectioned lungs from mice injected with control and RSF-1 overexpressing cells is also shown. The arrows indicate the presence of metastatic outgrowths in the lungs.
Therefore, we hypothesized that RSF-1 overexpression may promote metastatic potential in-vivo. To test this, we performed intravenous tail injection of MCF-10A-TM cells expressing a luciferase reporter into SCID mice. When cells are injected intravenously in the tail of recipient mice, the cells travel through the circulatory system and are deposited in the lungs, where the majority of the cells die due to the absence of a supportive microenvironment (Yang et al., 2012). Both control and RSF-1 overexpressing cells were rapidly cleared and no signal was detected one week after the injection. Importantly, after 7 weeks all the mice injected with cells overexpressing RSF-1 showed luciferase signal in the lungs indicating the formation of lung metastases while luciferase signal was never recovered in mice injected with control cells (Figure 6B). This demonstrates that RSF-1 over-expression promotes increased invasive capacity in the lungs and therefore a pro-metastatic state in breast cancer cells.
In summary, we have shown that over-expression of RSF-1 confers increased anchorage independent growth in vitro and promotes the formation of lung metastases in mouse models. Additionally, we have identified a transcriptional signature associated with RSF-1 amplification in primary tumors that was enriched for genes related to metastasis and invasion. The identification of RSF-1 as an oncogene that increases metastasic potential provides an explanation for the steep mortality of a recently identified molecular subgroup of breast cancer al (Curtis et al., 2012).
Discussion
Cancer research has recently been driven by the hope that therapies targeting drivers will be especially effective in tumors harboring genetic alterations in the target. This approach relies on the oncogene addiction effect whereby cancer cells become dependent on the activity of their altered oncogenes, so that inhibiting them compromises cellular viability. This “personalized medicine” is the basis of some of the most effective therapies, e.g. those targeting ERBB2 amplification in breast cancer (Ashworth et al., 2011). The success of these therapies has fueled efforts to catalog the genomic alterations in numerous cancers with the hopes of discovering new therapeutically actionable mutations.
However, even as data from cancer genomes accumulates, the identification of actionable driver genes remains a crucial limitation to therapeutic development. We see at least two significant bottlenecks. First, only a small subset of established driver genes are druggable given the current pharmalogical state of the art (Collins and Workman, 2006). Second, even when a driver is druggable, it may occur in a very small fraction of patients, limiting its clinical utility. At present, there is an untapped resource of driver genes in SCNAs that have evaded discovery. Moreover, due to the high frequency of SCNA events, actionable drivers can impact more patients (Figure 1A). However, to date, this possibility has been crucially limited by the difficultly of distinguishing passengers and drivers in the majority of SCNAs.
Here, we have presented a major advance in addressing this challenge, using a method that integrates data from primary tumors with functional assays on cell lines to prioritize candidate drivers. The unparalleled sensitivity and specificity of Helios enabled us to execute the first reported systematic validation of an algorithm designed to identify tumor dependencies. Helios’s performance was confirmed by a success rate of 10/12 candidates in an anchorage independent growth assay, successfully characterizing several regions for which there was no previously implicated driver. Importantly, because we selected the genes for validation based on their amplification significance (ISAR score), rather than their Helios score, we expect that this success rate will extend to additional regions that have equally strong Helios scores. Moreover, many of these genes are amplified in additional epithelial cancers (e.g. C6orf203, NIT1, ZNF652) suggesting possible drivers in those cancers as well.
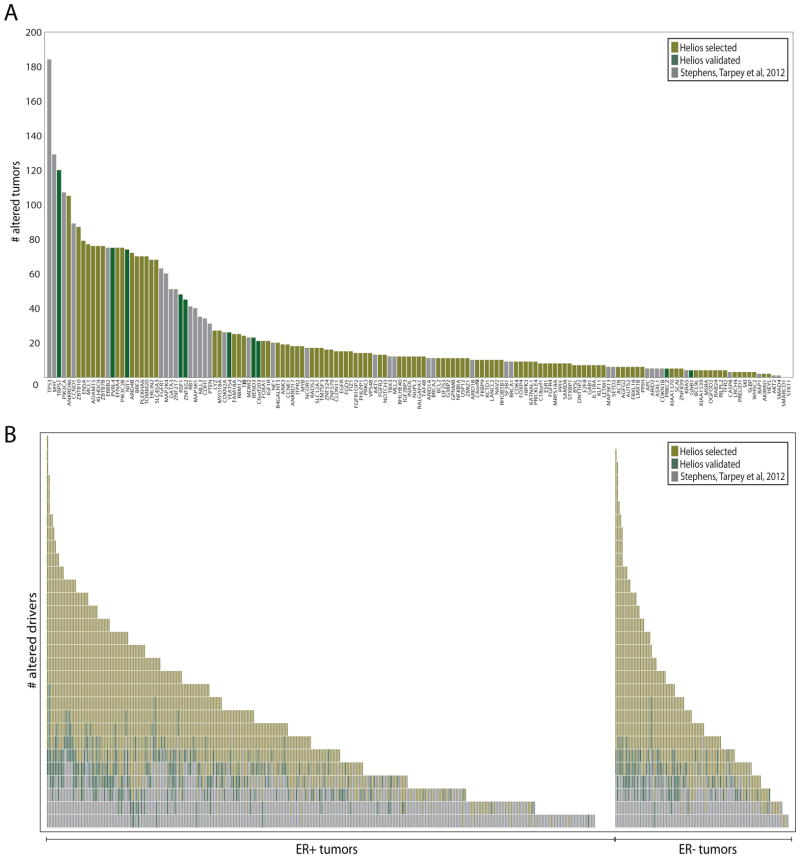
Using Helios, we have significantly expanded the landscape of high-confidence breast cancer drivers by more than two fold (Figure 7 and Supplementary Figure 7). Previous analyses of breast cancer cohorts (Stephens et al., 2012; TCGA, 2012) had identified 15 driver genes amplified in at least 5% of breast cancer tumors (both SCNA and sequence mutations). Our analysis has doubled this number to 29, substantially expanding the list of potential drug targets. Even more importantly, we have increased the number of drivers identified in each tumor, thus raising the possibility that at least one might be actionable in a given patient. A previous study (Figure 7B, grey boxes (Stephens et al., 2012)), could assign each tumor a median of 2 established drivers. Adding the Helios validated genes increases this number to a median of 3 drivers per tumor (Figure 7B, green boxes). Adding all predicted drivers with a high Helios score further expands this number to a median of 5 drivers in each tumor (Figure 7B, yellow boxes). Thus Helios has substantially expanded the set of high-confidence drivers in breast cancer.
Figure 7. The landscape of driver mutations in breast cancer.
For the driver genes described in (Stephens et al., 2012) (grey), Helios validated genes (green) and other Helios genes scoring > 5.5 (yellow) we compute (A) the number of tumors altered (copy number or sequence mutation) for each driver gene and (B) the number of driver genes altered (copy number or sequence mutation) per tumor. For this Figure we consider the 485 primary tumors in TCGA for which both copy number and DNA-Seq were available.
Helios uses a technique called transfer learning, whereby drivers with clearer signal (e.g. at the peak of their region) help learn informative features to improve performance in cases with less obvious signal. Helios learns the list of candidate drivers without using any prior list of driver genes and therefore it does not suffer from any bias that would hinder the discovery of novel biology. The algorithm uses all data in its learning process, transferring information across different genes, as well as between copy number and other features, until it converges into a final ranking of candidate driver genes. By leveraging information in this fashion, Helios is capable of learning how to weigh and combine features into a probabilistic score that represents the likelihood of the gene being the target of the recurrent alteration. This computational framework is independent of the features and tumor type and it can be applied to analyze additional cancers using a similar or even different set of features.
Genetic, genomic and functional data on cancers will continue to accumulate from large-scale projects in the coming years (Cheung et al., 2011; TCGA, 2008). Such datasets continue to accelerate drug development and to yield deep insights into oncogenesis. However, they also create new analytical challenges such as the need to pinpoint the alterations that promote cancer. Helios can be viewed as an accurate in silico screen for drivers. As such, it can be applied to additional cancer types and data types to accelerate the identification of cancer drivers.
Experimental Methods ISAR
ISAR is based on the G-score metric, a significance measure of the aberration for each marker, which was originally defined in GISTIC (Beroukhim, Getz et al. 2007). Specifically, the G-score for a marker m is the summation of the copy number across samples that surpass an aberration threshold θ. Given the copy number for N samples, the G-score for a marker m in the case of amplifications is:
| Eq. 1 |
Where CN(m,i) is the copy number of marker m in sample i and I is the indicator function. ISAR uses a local sliding window of constant size that moves along the chromosome, calculating the null distribution for each window. Once the distribution has been computed in all windows within a chromosome, each genomic marker is associated with several overlapping windows. The algorithm takes a conservative approach by selecting the least significant q-value among the values computed for all overlapping windows containing the marker (See Supplementary Methods for more detail).
Modeling copy number
We aim to model a distribution of SCNA that reflects the differences between driver and passenger genes, independently of the chromosomal region. However, in contrast to the subtle differences in SCNA within each altered region, the distribution of alterations differs dramatically between regions. Indeed, the median difference in G-score between genes in a region is significantly smaller (172) than the difference for genes across different regions (6405). Thus, without appropriate normalization, the G-score should not be used to prioritize drivers across regions. We aim to model whether the gene is among the most altered genes in its own region (and therefore more likely to be the driver of that region) and therefore define a metric that measures the difference in terms of G-score to the highest value in each region. For a single gene g, we define the GSDist score as:
| Eq. 2 |
The most altered gene(s) in a region will have GSDist=0, while any other gene will have a positive value that indicates the “distance” to the most frequently amplified gene in the region. Note that traditional approaches would use a threshold on this metric to make a hard decision on whether genes in the altered region are peak genes (Figure 2B). Instead Helios models this metric using two exponential distributions (one for drivers and one for passengers):
| Eq. 3 |
Driver genes have a GSDist distribution that exponentially decreases from zero with small variance, whereas passenger genes are modeled by a uniform distribution, which is approximated by an exponential distribution with large variance (See Supplementary Methods for more detail).
Features used in the Helios algorithm
We use MutSig (Banerji, Cibulskis et al. 2012) to compute the statistical significance of the recurrence of point mutations.
Helios uses features extracted from RNA-Seq based gene expression in two different ways: (1) To identify genes that are not expressed and therefore unlikely to be drivers. (2) We expect the oncogenic activity of an amplified driver gene to be reflected in the gene’s mRNA dosage (Akavia et al., 2010).
The oncogene addiction score for a hairpin is defined as the log-likelihood of the monotonic regression that predicts the lethality based on the gene mRNA. We use the PAVA algorithm (Brunk, 1955) to estimate the best fit for the regression (See Supplementary Methods for more detail).
Helios Algorithm
Helios uses a hierarchical Bayesian mixture model to distinguish drivers from passengers among the genes present in significantly altered regions. The unsupervised Bayesian algorithm discriminates driver genes (T=1) by integrating the copy number alteration information (SCNA), with cues from different data sources (X). The hierarchical framework naturally separates these two components using the following model:
| Eq. 4 |
This model separates the modeling of copy number (P(SCNA|T=t)) from other sources of information (P(T=t|X)), focusing on predicting the observed copy number landscape (P(SCNA)). The algorithm iteratively fits a model for each part: P(SCNA|T=t) and P(T=t|X) and updates the estimations for each gene (T) taking both parts into account. The algorithm is executed until the model converges into a stable solution that incorporates all the information into a single probability score for each gene.
Figure S2A shows the graphical model for Helios, where N genes are classified by combining the information from different data sources X and SCNA. w represents the parameters that control the integration of X, while λ parameterizes the influence of SCNA. In this model, when the values Tn for the genes are given, the parameters for the different sources (W) and copy number (λ) are independent. This property makes it possible to fit the model efficiently using the Expectation Maximization (EM) algorithm (See Supplementary Methods for more details).
Data Sets Used for Helios
We used the following public datasets:
Primary tumor data from the TCGA Project (TCGA 2012): copy number Affymetrix 6.0 SNP arrays (n=785), Illumina HiSeq RNA sequencing (n=732) and whole-exome sequencing (n=507).
Cell line shRNA screens (n=29) collected by Marcotte et al. (Marcotte, Brown et al. 2012).
Cell line data from the Cancer Cell Line Encyclopedia (Barretina, Caponigro et al. 2012) for the cell lines screened with shRNA: copy number Affymetrix 6.0 SNP arrays (n=27) and messenger RNA Affymetrix U133 plus 2.0 arrays (n=27)
Data Sets Used to generate Gold Standard Set
To assess performance, a gold standard set of 330 genes was compiled from the following sources:
The set of known amplified oncogenes from Beroukhim et al. (Beroukhim, Mermel et al. 2010)
The set of genes related to Breast cancer according to the University of Copenhagen DISEASES database (Frankild and Jensen) with score greater than 2.5. We filtered out genes categorized as tumor suppressors according to Uniprot.
See supplementary Methods for more information
Cell culture and reagents
To generate cell lines overexpressing a gene, cells were plated at 60% of confluence in a 6 well plate and after 24 hours infected with virus expressing the different plasmids containing the different genes. Media containing virus was replaced in 12h for fresh media. After that cells were re-infected for other 12h. Cells were grown in fresh media for 24h and selected with the appropriate drug. Alternatively, to generate MDA-MB 453 deficient in RSF1, cells where infected with virus expressing doxycycline-inducibles pTRIPz shRNA against RSF1 and selected with the puromycin (2ug/mL).
See Supplementary methods for Cell lines, DNA constructs and gene cloning strategy.
Validation of Helios predictions was based on the ability of MCF-10A to form colonies in semi-solid media when the putative oncogene was experimentally upregulated. Because low passage MCF-10A are very resistant to transformation, to increase the sensitivity of our assay, we selected a passage with intrinsic low attachment independent growth ability (5–15 colonies per 5,000 plated cells) that demonstrated robust higher growth ability when bona-fide breast oncogenes were overexpressed (Supplementary Figure 4A).
Colony formation assay in semisolid media was performed in 6 well plates. First, a layer of 2 mls of 0.6% agar (Fisher #9002-18-0) in regular MCF-10A media was placed at the bottom of each well and allowed gelification. Then, layer of 2mls of 0.3% agar containing 5,000 cells was seeded on top of the bottom agar layer and allow gelification. Finally, 1 ml of regular MCF-10A media was placed covering the agar. The colonies were allowed to form for 1 month. After this period 2 mls of MTT solution (Sigma #M5655) at 0.5mgr/ml was used to stain the colonies. A minimum of 6 replicas per gene were plated. To ensure comparability, transformation assays for each gene are compared to empty-vector controls performed together on the same day. The number of colonies was independently evaluated by two researchers. All the different MCF-10A clones carrying controls and genes of interest were maintained growing exponentially for 48 hours (plates were at 50–70 confluency) before being plated in agar to homogenize assay conditions.
Tumorigenicity in mice
Animal maintenance and experiments were performed in accordance with the animal care guidelines and protocols approved by Columbia University animal care unit. For Comma-1D cell line, 21 days old female NOD SCID immunocompromised mice NOD.CB17-Prkdcs SCID mice (Harlan) mice were injected with 5×105 cells, resuspended in PBS, into a fat mammary gland. For MDA-453 cell line, eight-weeks old female NOD SCID immunocompromised mice NOD.CB17-Prkdcs SCID mice (Harlan) mice where injected with 5×106 cells, resuspended in 1:2 Matrigel (BD Biosciences) plus normal growth media, into a fat pad mammary gland. Doxicyclin was added to drinking water at a final concentration of 2.0 mg/mL. Tumor growth was monitored twice a week with callipers at the site of injection. Animals were sacrificed as soon as tumor size reached 1.5 cm diameter.
In the experimental metastasis assays, eight-weeks old female NOD SCID immunocompromised NOD.CB17-Prkdcs SCID mice (Harlan) where injected with 5×106 cells, resuspended in PBS, via the tail vein. To measure the luciferase intensity of injected cells, 2.25 ug ml-1 luciferin was injected intravenously through the tail and luciferase activity was assessed 5 minutes after luciferin injection using a IVIS Spectrum Pre-clinical in vivo Imaging System (PerkinElmer, IVISSPE) machine. The presence of established metastases was confirmed by euthanizing the mice.
Supplementary Material
Figure S1. 18q21 copy number region, related to Figure 1. Heatmap of the copy number profile of the TCGA Breast cancer cohort for the 18q arm (A) and a zoom-in of the 18q21 region (B). ISAR score and regions are displayed at the bottom.
Figure S2. Helios modeling approach, related to Figure 2. (A) Graphical model for the Helios Algorithm. N input genes are classified as either driver gene (Tn=1) or passenger gene (Tn=0) by combining the information from CNA and different data sources X. The model needs to fit both the parameters that control the integration of X (w) and the parameters that influence the CNA (λ). (B) Bayesian Network for data integration in the Helios Algorithm. Helios uses a Bayesian network with sigmoid nodes to integrate different data sources. The current version of Helios contains two input nodes (shRNA-addiction and shRNA-subtype) that integrate genome-wide vulnerability assays and that are merged into a single node (shRNA-node), an input node for sequence mutations (DNASeq) and an input node for gene expression (RNASeq). The output of this network gives the expected classification of a gene as target (T=1) or passenger (T=0) when CNA information is not taken into account.
Figure S3. Helios analysis of breast cancer, related to Figure 3. (A) Likelihood of the Helios model for each iteration of the algorithm (B) Parameters for the GSDist distributions (λ0 and λ1) for each iteration of the Helios algorithm. (C) ISAR peak stability versus S-score. The percentage of runs in which a peak was called across subsample runs is plotted against the S-score. (D) Histogram of Pearson correlation of Helios scores across subsample runs of the algorithm. The average correlation between Helios scores across executions (0.96) is highlighted with a discontinuous line. (E) Contribution of each feature to the Helios score. GSEA enrichment of gold standard genes both in the final run and in runs where each individual data source was excluded.
Figure S4. Helios validation assay, related to Figure 4. Most representative repeat for each of the genes assayed for anchorage independent growth: (A) controls, (B) putative drivers selected by HELIOS and (C) Randomly selected genes with high ISAR score but low HELIOS score. (D) QRT-PCR data showing efficient overexpression in MCF-10A for each of the genes studied. The data are represented as the difference in Delta-Ct (gene of interest-β/actin) between overexpressing and parental cells and then normalized to cells transduced with the control vector.
Figure S5. Overexpression of RSF1 promotes tumorigenesis, related to Figure 5. (A) The composition demonstrates that copy number drives the expression of RSF1 in cell lines. The left panels shows the correlation between CNV and mRNA levels and the right panels between CNV and protein levels. (B) The table shows the number of animal orthotopically transplanted with three different cell lines (MCF-10A, MCF-10ATM and C-ID) and its ability to form tumors when they were engineered to overexpress RSF1. (C) Experimentally rescued RSF-1 expression in MDA-MB-453 cells expressing shRNAs against RSF-1 restored their colony formation ability.
Figure S6. RSF1 expression signatures, related to Figure 6. (A) The analysis of the overexpression changes related to RSF1 overexpression in luminal primary tumors revealed a signature enriched for invasiveness, migration, EMT and stem cell. (B–C) The analysis of the underexpression related to RSF1 overexpression in luminal (B) and basal (C) primary tumors reveals signatures enriched for metabolic changes, dedifferentiation and metastasis.
Figure S7. The landscape of driver mutations in breast cancer, related to Figure 7. For the driver genes described in (Stephens et al., 2012) (grey), Helios validated genes (green) and other Helios selected genes (yellow) the figure displays the alterations in the 485 primary tumors in TCGA for which both copy number and DNA-Seq were available. Each row represents a gene while each column represent a tumor. Both point mutations and copy number changes are considered for this plot.
Supplementary Table 1. Regions of recurrent amplification detected by ISAR, related to Figure 1. Significantly amplified regions identified by ISAR in the TCGA Breast cancer cohort.
Supplementary Table 2. Helios score for the genes in the regions of recurrent amplification detected by ISAR, related to Figure 4.
Supplementary Table 3. Candidate driver genes selected by Helios, related to Figure 7. 119 genes selected by from Helios analysis of breast cancer.
Supplementary Table 4. Primers used for DNA constructs, related to Supplementary section 10.2
Supplementary Table 5. Forward and reverse sequences for determination of RNA levels, related to Supplementary section 10.3
Acknowledgments
The authors would like to thank Sourav Bandyopadhyay, Oren Litvin, Richard Marcotte, Ben Neel, Ramon Parsons and Sagi Shapira for valuable comments. This research was supported by National Institutes of Health grant number (R01CA164729), National Centers for Biomedical Computing Grant 1U54CA121852-01A1 and D.P. holds a Packard Fellowship for Science and Engineering.
Footnotes
Author Contributions
FSG, JMS, DP conceived the study. FSG, DP designed ISAR and HELIOS. FSG designed and implemented ISAR and HELIOS. FSG, DK, BJ preprocessed data. FSG, DK, UDA, DP performed statistical analysis of Helios results. FSG, PV, DK, JMS, DP performed biological interpretation of Helios and selected genes for validation. PV, JMS designed biological validation experiments. PV, JM, VC, LSC, RDB, DLN, JMS performed biological validation of Helios genes. PV, JM, VC and JMS performed characterization experiments for RSF1. FSG, PV, DK, JMS, DP wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An Integrated Approach to Uncover Drivers of Cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Ashworth A, Lord Christopher J, Reis-Filho Jorge S. Genetic Interactions in Cancer Progression and Treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk HD. Maximum Likelihood Estimates of Monotone Parameters. The Annals of Mathematical Statistics. 1955;26:607–616. [Google Scholar]
- Chen TJ, Huang SC, Huang HY, Wei YC, Li CF. Rsf-1/HBXAP overexpression is associated with disease-specific survival of patients with gallbladder carcinoma. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2011;119:808–814. doi: 10.1111/j.1600-0463.2011.02808.x. [DOI] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TU. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Research. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danussi C, Akavia UD, Niola F, Jovic A, Lasorella A, Pe’er D, Iavarone A. RHPN2 Drives Mesenchymal Transformation in Malignant Glioma by Triggering RhoA Activation. Cancer research. 2013 doi: 10.1158/0008-5472.CAN-13-1168-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison CA, Durbin SM, Thau MR, Zellmer VR, Chapman SE, Diener J, Wathen C, Leevy WM, Schafer ZT. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Cancer research. 2013;73:3704–3715. doi: 10.1158/0008-5472.CAN-12-2482. [DOI] [PubMed] [Google Scholar]
- Fang FM, Li CF, Huang HY, Lai MT, Chen CM, Chiu IW, Wang TL, Tsai FJ, Shih Ie M, Sheu JJ. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. The American journal of pathology. 2011;178:2407–2415. doi: 10.1016/j.ajpath.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankild S, Jensen LJ. DISEASES database. University of Copenhagen; ( http://diseases.jensenlab.org) [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hu BS, Yu HF, Zhao G, Zha TZ. High RSF-1 expression correlates with poor prognosis in patients with gastric adenocarcinoma. International journal of clinical and experimental pathology. 2012;5:668–673. [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Use and Abuse of RNAi to Study Mammalian Gene Function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Dong Q, Wang E. Rsf-1 is overexpressed in non-small cell lung cancers and regulates cyclinD1 expression and ERK activity. Biochemical and biophysical research communications. 2012;420:6–10. doi: 10.1016/j.bbrc.2012.02.095. [DOI] [PubMed] [Google Scholar]
- Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:1485–1491. doi: 10.1007/s13277-012-0399-y. [DOI] [PubMed] [Google Scholar]
- Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, Campbell J, Williams AC, Hwang D, Zhu C-Q, et al. Integrative Genomic Analyses Identify BRF2 as a Novel Lineage-Specific Oncogene in Lung Squamous Cell Carcinoma. PLoS Med. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JLY, et al. Essential Gene Profiles in Breast, Pancreatic, and Ovarian Cancer Cells. Cancer Discovery. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel C, Schumacher S, Hill B, Meyerson M, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganella S, Pagnotta SM, Ceccarelli M. Finding recurrent copy number alterations preserving within-sample homogeneity. Bioinformatics. 2011;27:2949–2956. doi: 10.1093/bioinformatics/btr488. [DOI] [PubMed] [Google Scholar]
- O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast cancer research: BCR. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Pallasch C, Elia AE, Braun CJ, Westbrook TF, Hemann M, Elledge SJ, Staudt L. A role for PVRL4-driven cell cell interactions in tumorigenesis. eLife. 2013;2 doi: 10.7554/eLife.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires MM, Hopkins BD, Saal LH, Parsons RE. Alterations of EGFR, p53 and PTEN that mimic changes found in basal-like breast cancer promote transformation of human mammary epithelial cells. Cancer biology & therapy. 2013;14:246–253. doi: 10.4161/cbt.23297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu JJ, Choi JH, Guan B, Tsai FJ, Hua CH, Lai MT, Wang TL, Shih Ie M. Rsf-1, a chromatin remodelling protein, interacts with cyclin E1 and promotes tumour development. The Journal of pathology. 2013;229:559–568. doi: 10.1002/path.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Ie M, Sheu JJ, Santillan A, Nakayama K, Yen MJ, Bristow RE, Vang R, Parmigiani G, Kurman RJ, Trope CG, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA . Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter V, Nobel AB, Wright FA. DiNAMIC: a method to identify recurrent DNA copy number aberrations in tumors. Bioinformatics. 2011;27:678–685. doi: 10.1093/bioinformatics/btq717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. Oncogene Addiction. Cancer research. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- Widmer C, Rätsch G. Transfer Learning in Computational Biology. Paper presented at: Journal of Machine Learning Research - Proceedings Track 2011 [Google Scholar]
- Wong WC, Kim D, Carter H, Diekhans M, Ryan MC, Karchin R. CHASM and SNVBox: toolkit for detecting biologically important single nucleotide mutations in cancer. Bioinformatics. 2011;27:2147–2148. doi: 10.1093/bioinformatics/btr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang JJ, Huang XY. Mouse models for tumor metastasis. Methods in molecular biology. 2012;928:221–228. doi: 10.1007/978-1-62703-008-3_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Zhang J, Zhang S, Yu G, Wang Y. Comparative Analysis of Methods for Identifying Recurrent Copy Number Alterations in Cancer. PLoS ONE. 2012;7:e52516. doi: 10.1371/journal.pone.0052516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 18q21 copy number region, related to Figure 1. Heatmap of the copy number profile of the TCGA Breast cancer cohort for the 18q arm (A) and a zoom-in of the 18q21 region (B). ISAR score and regions are displayed at the bottom.
Figure S2. Helios modeling approach, related to Figure 2. (A) Graphical model for the Helios Algorithm. N input genes are classified as either driver gene (Tn=1) or passenger gene (Tn=0) by combining the information from CNA and different data sources X. The model needs to fit both the parameters that control the integration of X (w) and the parameters that influence the CNA (λ). (B) Bayesian Network for data integration in the Helios Algorithm. Helios uses a Bayesian network with sigmoid nodes to integrate different data sources. The current version of Helios contains two input nodes (shRNA-addiction and shRNA-subtype) that integrate genome-wide vulnerability assays and that are merged into a single node (shRNA-node), an input node for sequence mutations (DNASeq) and an input node for gene expression (RNASeq). The output of this network gives the expected classification of a gene as target (T=1) or passenger (T=0) when CNA information is not taken into account.
Figure S3. Helios analysis of breast cancer, related to Figure 3. (A) Likelihood of the Helios model for each iteration of the algorithm (B) Parameters for the GSDist distributions (λ0 and λ1) for each iteration of the Helios algorithm. (C) ISAR peak stability versus S-score. The percentage of runs in which a peak was called across subsample runs is plotted against the S-score. (D) Histogram of Pearson correlation of Helios scores across subsample runs of the algorithm. The average correlation between Helios scores across executions (0.96) is highlighted with a discontinuous line. (E) Contribution of each feature to the Helios score. GSEA enrichment of gold standard genes both in the final run and in runs where each individual data source was excluded.
Figure S4. Helios validation assay, related to Figure 4. Most representative repeat for each of the genes assayed for anchorage independent growth: (A) controls, (B) putative drivers selected by HELIOS and (C) Randomly selected genes with high ISAR score but low HELIOS score. (D) QRT-PCR data showing efficient overexpression in MCF-10A for each of the genes studied. The data are represented as the difference in Delta-Ct (gene of interest-β/actin) between overexpressing and parental cells and then normalized to cells transduced with the control vector.
Figure S5. Overexpression of RSF1 promotes tumorigenesis, related to Figure 5. (A) The composition demonstrates that copy number drives the expression of RSF1 in cell lines. The left panels shows the correlation between CNV and mRNA levels and the right panels between CNV and protein levels. (B) The table shows the number of animal orthotopically transplanted with three different cell lines (MCF-10A, MCF-10ATM and C-ID) and its ability to form tumors when they were engineered to overexpress RSF1. (C) Experimentally rescued RSF-1 expression in MDA-MB-453 cells expressing shRNAs against RSF-1 restored their colony formation ability.
Figure S6. RSF1 expression signatures, related to Figure 6. (A) The analysis of the overexpression changes related to RSF1 overexpression in luminal primary tumors revealed a signature enriched for invasiveness, migration, EMT and stem cell. (B–C) The analysis of the underexpression related to RSF1 overexpression in luminal (B) and basal (C) primary tumors reveals signatures enriched for metabolic changes, dedifferentiation and metastasis.
Figure S7. The landscape of driver mutations in breast cancer, related to Figure 7. For the driver genes described in (Stephens et al., 2012) (grey), Helios validated genes (green) and other Helios selected genes (yellow) the figure displays the alterations in the 485 primary tumors in TCGA for which both copy number and DNA-Seq were available. Each row represents a gene while each column represent a tumor. Both point mutations and copy number changes are considered for this plot.
Supplementary Table 1. Regions of recurrent amplification detected by ISAR, related to Figure 1. Significantly amplified regions identified by ISAR in the TCGA Breast cancer cohort.
Supplementary Table 2. Helios score for the genes in the regions of recurrent amplification detected by ISAR, related to Figure 4.
Supplementary Table 3. Candidate driver genes selected by Helios, related to Figure 7. 119 genes selected by from Helios analysis of breast cancer.
Supplementary Table 4. Primers used for DNA constructs, related to Supplementary section 10.2
Supplementary Table 5. Forward and reverse sequences for determination of RNA levels, related to Supplementary section 10.3