Abstract
Toll-like receptors (TLRs) play essential roles in the initiation and modulation of immune responses. TLR1/TLR2 heterodimers recognize tri-acylated bacterial lipopeptides, including the synthetic TLR1/2 lipopeptide, Pam3CSK4. Genetic variation in TLR1 is associated with outcomes in diseases in which regulatory T cells (Treg) play a role, including asthma and allergy. To determine whether genetic polymorphisms in TLR1 are associated with alterations in Treg suppression of effector T cells (Teff), we performed in vitro suppression assays in healthy individuals of varying haplotypes in TLR1. We show that functional genetic polymorphisms in TLR1 modify surface expression of TLR1 on T lymphocytes and confer enhanced Teff resistance to Treg suppression in the presence of Pam3CSK4. These effects are mediated in part by IL-6 and inhibited by blocking IL-6 signaling through STAT3. These findings suggest that TLR1 polymorphisms could influence immune-related disease through Teff resistance to Treg suppression.
Introduction
Toll-like receptors (TLRs) are a family of germline-encoded pattern recognition receptors essential to the detection of microbial components (1). Known for their role in innate immunity, TLRs detect a wide range of pathogen-associated molecular patterns (PAMPs). The genes encoding human TLRs are dispersed throughout the genome with the exception of TLR1, TLR6, and TLR10 which lie in a single locus on chromosome 4. TLR2 in combination with TLRs 1, 6, and possibly 10 recognize different PAMPs (2–4). Pertinent to our current study, the TLR1/2 heterodimer recognizes tri-acylated lipopeptides isolated from bacterial pathogens or produced synthetically (Pam3CSK4).
Common genetic variation in the TLR10/1/6 locus is associated with multiple disease states and functional changes in immune responsiveness. Recently in multiple genome wide association studies, the importance of the TLR10/1/6 locus has been highlighted by very strong associations with Helicobacter pylori seroprevalance (p=1.42×10−18) (5), self-reported allergy (p=5.3×10−21) (6), and allergic sensitization (p=5.2×10−11) (7). Multiple high frequency missense variants and varying LD structure across populations has made it difficult to pinpoint a single causative variant (8). However, two non-synonymous coding variants associated with clinical phenotypes rs4833095 (Asn248Ser) and rs5743618 (Ser602Ile) may play a role. In Caucasians, the minor alleles of these coding variants (rs5743618T and rs4833095C) have been associated with increased whole blood cytokine responses and increased TLR1 surface expression on monocytes due to enhanced trafficking to the cell surface rather than changes in messenger RNA or total cell protein levels (9–12). The rs5743618 (Ser602Ile) coding variant has also been shown to confer increased Pam3CSK4-induced NFκB reporter activity in transfected epithelial cell lines that do not express TLR1 (9, 13). We have also identified a SNP in the TLR1 promoter in high linkage disequilibrium (LD) with these SNPs in which the minor allele rs5743551G is highly associated with death in sepsis (9). To date, the consequences of these polymorphisms with respect to T cell function have yet to be explored.
Multiple different TLRs are expressed in T cells and are capable of modulating the function of various T cell subsets. CD4+CD25+FoxP3+ regulatory T cells are pivotal to the suppression of cellular immune responses. Important to these studies, TLR1/2 agonists have been shown in humans to impair Treg suppressive capacity (14, 15). Of interest, this effect was observed in only a subset of subjects implying inter-individual variation (14). In this study, we tested whether common genetic variation in TLR1 affects TLR1/2 agonist-induced changes in Treg and Teff function.
Materials and Methods
Study subjects
We obtained fresh peripheral blood, frozen PBMC, and DNA from healthy volunteers from whom written informed consent was obtained. This work was approved by the Benaroya Research Institute and University of Washington human subjects committees.
Genotyping
We genotyped DNA for three SNPs in TLR1: rs5743618, rs4833095, and rs5743551 by Taqman PCR-based allelic discrimination. We identified Caucasian individuals who either carried two copies of the haplotype for the three minor alleles (rs5743618T, rs5743551G, rs4833095C) and age and gender matched controls carrying two copies of the haplotype for the three major alleles (rs5743618G, rs5743551A, rs4833095T).
Treg isolation
Natural Treg (nTreg) from freshly isolated PBMC were sorted by flow cytometry for the CD4+ cells expressing the highest 3–5% of CD25 (16). The percent of FoxP3+ T Cells was 90% on average.
CFSE based suppression assay
CD4+CD25− T effector cells (Teff) were isolated from thawed frozen autologous or heterologous donors’ PBMC by negative selection with microbeads to CD4 and CD25 (Miltenyi Biotec) and CFSE labeled (16). nTregs (above) and Teff were co-cultured at a ratio of 1:2 with anti-CD3/anti-CD28 coated Dynabeads (Invitrogen) at a ratio of 1:10 (beads:Teff) in the presence of media, Pam3CSK4 (1μg/ml; Invivogen), 0114:B4 LPS (1μg/ml; Invivogen), PGN (100ng/ml; Sigma-Fluka) or exogenous IL-6 (50ng/mL; BD Pharmigen). On day 4, cells were stained for anti-CD4 and anti-CD25 (Biolegend) and analyzed by flow cytometry. Data were excluded from analysis when suppression was less than 10% in media treated cultures. For STAT3 inhibition, CD4+ T cells were incubated with phosphorylation inhibitor of STAT3 (Stattic V, Santa Cruz Biotechnology) at 1200ng/ml for one hour, then washed and cultured as above.
We calculated percent reduction in suppression in two stages (Supplemental Figure 1). First, percent suppression was calculated as [(%Teff alone proliferation - %Treg:Teff co-culture proliferation)/%Teff alone proliferation] × 100. We then determined the difference in percent suppression between co-cultures treated with PAMPs or media alone and expressed this as a percent of media alone: [(% suppression media-treated - % suppression PAMP-treated)/% suppression media treated] × 100.
TLR1 staining
PBMC were stained with human anti-CD4 (RPA-T4, Biolegend), and anti-TLR1 (GD2.F4, Biolegend), anti-TLR5 (624915, R&D Systems) or isotype control. For a subset, this was followed by intracellular staining using anti-FoxP3 (206D, Biolegend) and a FOXP3 Fix/Perm buffer set (Biolegend).
Cytokine Measurement
Cell culture supernatants (25μL) were collected at 48 hours of autologous co-culture of Treg:Teff or Teff alone cultures described above. Cytokines were measured by electrochemiluminesence multiplex immunoassay (Meso Scale Discovery, Rockville, MD).
Statistical Analysis
We used two-tailed unpaired t tests, two-tailed paired t tests, or two-tailed nonparametric (Spearman) correlation as indicated.
Results and Discussion
Minor allele haplotype is associated with enhanced surface expression of TLR1
We and others have previously shown the minor alleles of non-synonymous coding polymorphisms in TLR1 (rs5743618T and rs4833095C) are associated with altered cell surface expression of TLR1 on monocytes (9–11). We used flow cytometry to determine whether these TLR1 alleles also alter cell surface expression of TLR1 on T lymphocytes. We obtained PBMCs from Caucasian subjects carrying two copies of the TLR1 “minor” allele haplotype (rs5743618T, rs5743551G, rs4833095C) and control subjects carrying two copies of the “major” allele haplotype (rs5743618G, rs5743551A, rs4833095T). The study subjects were predominantly male (63%) and had a mean age of 36±14yrs. PBMC were stained for surface expression of CD4 and TLR1. Subjects homozygous for the TLR1 minor allele haplotype had a significantly higher TLR1 median fluorescence intensity (MFI) on CD4+ T cells (Figure 1A) than those homozygous for the TLR1 major allele haplotype (p=0.004). Cell surface expression of TLR5 did not differ by TLR1 haplotype (Figure 1B). When we further differentiated T cells by FoxP3 staining, we found that both CD4+FoxP3+ and CD4+FoxP3− T cells from subjects homozygous for the TLR1 minor allele haplotype had a significantly higher percentage of cells expressing TLR1 relative to the major allele haplotype (Supplemental Figure 2A, B). Thus, TLR1 variants associated with increased expression of TLR1 on peripheral blood monocytes are also associated with enhanced TLR1 surface expression on both Treg and Teff.
Figure 1. Variant TLR1 polymorphisms are associated with enhanced TLR1 surface expression on CD4+ T cells.
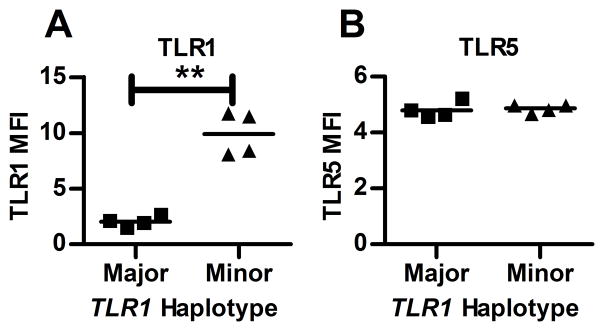
Frozen, thawed PBMC were stained with anti-CD4 and anti-TLR1 or anti-TLR5 for 4 subjects of the major and minor allele TLR1 haplotype. Data are depicted as median fluorescence intensity (MFI) after subtraction of MFI of isotype control. Subjects carrying two copies of the minor allele haplotype showed significantly increased surface expression of TLR1 (A) but not TLR5 (B) in the CD4+ T cell population with **p<0.01 by paired t test.
The minor allele haplotype of TLR1 is associated with greater Pam3CSK4 – induced impairment of Treg suppression of Teff
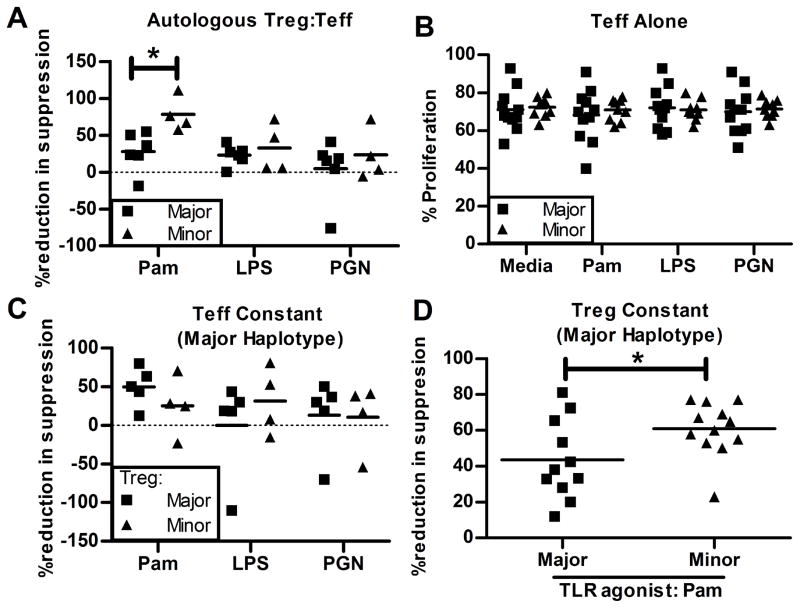
We tested the effects of treatment with Pam3CSK4, a TLR1/2 agonist, on Treg function in subjects of differing TLR1 haplotype. CD4+CD25HI Treg (mean FoxP3+ 90%) were isolated from each subject. Using an in vitro CFSE-based suppression assay, we compared the ability of Treg to suppress the proliferation of autologous CD4+CD25− Teff co-cultured with anti-CD3/anti-CD28 coated beads in the presence or absence of various TLR agonists for 96 hours. We found that treatment of co-cultures with Pam3CSK4 decreased the average suppression of Teff by Treg (mean of 20%) compared to co-cultures treated with media alone (mean of 41%; p = 0.01; data not shown) which is consistent with previous reports (14, 15). We then compared the magnitude of this Pam3CSK4-induced effect in cultures of cells from subjects with either the minor or major allele TLR1 haplotypes. We found that in the presence of Pam3CSK4, Treg suppression of Teff was impaired to a greater degree in cells from subjects harboring the TLR1 minor allele haplotype as compared to those with the major allele haplotype (Figure 2A, p=0.02). Evidence that the effect of the TLR1 haplotypes is specific to TLR1/2-mediated responses was provided by the finding that there were no haplotype-specific effects observed when co-cultures were treated with LPS (p=0.48), a TLR4 agonist, or peptidoglycan, a TLR2 agonist that does not require TLR1 (p=0.51)(2). Our data show that Pam3CSK4 impairs Treg suppression of Teff proliferation to a greater extent in subjects who harbor the TLR1 minor allele haplotype that confers higher TLR1 surface expression.
Figure 2. TLR1 minor allele haplotype is associated with greater Pam3CSK4-induced Teff resistance to Treg suppression.
(A) Freshly isolated nTreg were co-cultured at a ratio of 1:2 with CFSE-labeled CD4+CD25− Teff from frozen autologous PBMC. (B) CFSE-labeled CD4+CD25− Teff from frozen PBMC were cultured alone (no Tregs) and percent proliferation determined. (C) Freshly isolated nTreg from minor or major allele haplotype subjects were co-cultured at a ratio of 1:2 with CFSE-labeled CD4+CD25− Teff (major allele haplotype). (D) nTreg (major allele haplotype) were co-cultured at a ratio of 1:2 with CFSE-labeled CD4+CD25− Teff isolated from subjects of either the major or minor allele haplotype in the presence of media or Pam3CSK4. Cultures were performed in the presence of anti-CD3 and anti-CD28 beads for 4 days ± TLR agonists. Percent reduction in suppression represents the difference in percent suppression between co-cultures treated with TLRs or media alone and expressed as a percent of media alone. Each graph represents data from at least 4 independent experiments. Statistical significance was determined using an unpaired Students t-test. *, P<0.05.
Absence of Pam3CSK4 –induced effect on Teff suppression in absence of Tregs
The proliferation or activation state of effector T cells can change the sensitivity of Teff to Treg-mediated suppression. In order to assess whether Pam3CSK4 altered the proliferation of Teff in the absence of Tregs, we measured proliferation of Teff with anti-CD3/anti-CD28 coated beads in the presence of TLR agonists or media alone. There was no significant difference in proliferation of Teff between TLR1 haplotypes for media- or any of the TLR-treated cultures (Figure 2B). Thus, the genotypic differences in Pam3CSK4-induced modulation of Treg suppression of Teff are not due to alteration of autonomous Teff proliferation.
TLR1 minor allele haplotype is associated with Teff resistance
For subjects harboring the TLR1 minor allele haplotype, greater Pam3CSK4-induced reduction of Treg suppression could be attributed to impaired Treg suppression or increased Teff resistance to Treg suppression. To address these possibilities we performed allogeneic co-culture experiments where either the genotype of the Treg or Teff population was held constant as the major allele haplotype. We have previously demonstrated that Treg suppression is not different in autologous versus heterologous assays (17). When we incubated Teff isolated from a major allele haplotype subject with Tregs from subjects carrying either the minor or major allele haplotype in the presence of Pam3CSK4, we observed no significant association between TLR1 haplotype and Treg suppressive capacity (Figure 2C). This suggests that increased TLR1 surface expression on Tregs is not sufficient to observe the effect of TLR1 variants on Pam3CSK4-induced alteration of Treg suppression. In contrast, when we incubated Tregs isolated from a subject carrying the major haplotype with Teff from subjects carrying either the minor or major allele TLR1 haplotype, the haplotype-dependent Pam3CSK4-induced reduction in Treg suppression was observed (p=0.04, Figure 2D). These data suggest that increased stimulation of Teff conferred by the minor allele haplotype of TLR1 causes Teff resistance to Treg suppression.
Impaired Treg suppression correlates with higher IL-6 production and is reversed by STAT3 inhibition
Pro-inflammatory cytokine production, particularly IL-6, is associated with impaired regulatory T cell suppression (18). Teff resistance to Treg suppression has been implicated in the pathogenesis of several disease states including psoriasis, diabetes, and relapsing remitting multiple sclerosis and the IL-6 pathway has been shown to mediate this resistance (17, 19, 20). We reasoned that enhanced cell surface expression of TLR1 on T cells from subjects carrying the minor allele haplotype would result in increased Pam3CSK4-induced cytokine levels in the T cell co-cultures. We measured cytokine levels in supernatants collected after 48 hours of incubation from co-cultures of autologous Treg and Teff and the cultures of Teff alone stimulated with TLR agonists in the presence of anti-CD3/anti-CD28 coated beads. In supernatants from co-cultures of Teff with Treg there was a trend towards increased IL-6 production in subjects with the minor allele haplotype but this difference did not achieve statistical significance (Figure 3A). We found that Pam3CSK4-induced IL-6 levels were significantly higher in Pam3CSK4-treated Teff isolated from subjects carrying the minor allele haplotype (p=0.03, Figure 3B). IL-2 and TNF-α levels did not differ by TLR1 genotype for either cultures of Teff alone or co-cultures of Treg with Teff (Supplemental Figure 2C, D). These data support a model whereby Teff-driven differences in Pam3CSK4-induced IL-6 production participate in Teff resistance to Treg suppression. Further support for this model was demonstrated through the observed relationship between IL-6 production and the impairment in autologous Treg suppression of Teff. Although IL-6 levels were not significantly different by genotype from the co-culture assays, we observed that IL-6 levels in Teff:Treg co-cultures were strongly positively correlated with inhibition of Treg suppression (Spearman’s r=0.85, p=0.002, Figure 4A).
Figure 3. Higher IL-6 production is seen in co-cultures of cells from subjects harboring the TLR1 minor allele haplotype.
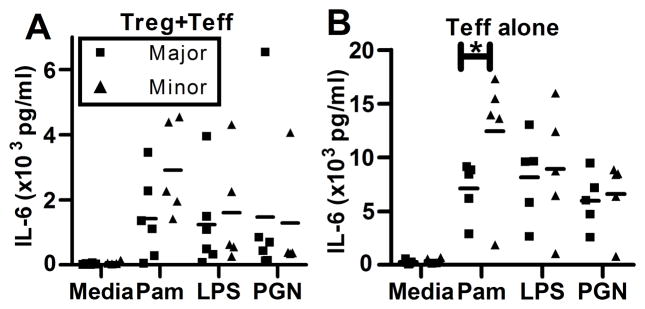
IL-6 was measured by multiplex immunoassay in 48 hour supernatants from autologous co-cultures of Treg:Teff cultured at a ratio of 1:2 (A) and Teff alone (B) in the presence of anti-CD3/anti-CD28 beads ± TLR agonists. Data are from 10 subjects, 5 of each genotype. Statistical analysis was performed using a paired t test. *, P<0.05.
Figure 4. Co-culture IL-6 production correlates with greater reduction in Treg suppression in the presence of Pam3CSK4.
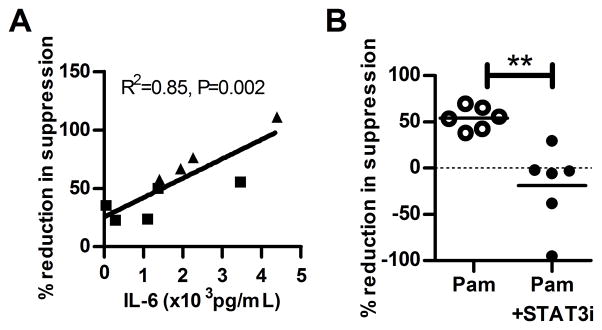
(A) The correlation (Spearman’s) between IL-6 production at 48 hours in autologous co-culture supernatants and percent reduction of suppression from the corresponding assays was performed. (B) Treg and Teff were incubated ± STAT3i (Stattic V), an inhibitor of STAT3 phosphorylation, at 1200ng/ml for one hour, then washed and co-cultured in a Treg suppression assay at a ratio of 1:2 in the presence of anti-CD3/anti-CD28 beads and media or Pam3CSK4. ‘%Reduction in suppression’ corresponds to the difference in % suppression between co-cultures treated with Pam3CSK4 or media alone expressed as a percent of media alone. The Pam3CSK4 + STAT3i culture is normalized to a media + STAT3i treated culture. Data shown are from 6 subjects, 3 of each genotype. The Pam3CSK4-induced reduction of suppression is blocked in the presence of inhibition of STAT3 phosphorylation (**p<0.01).
To determine if IL-6 is directly involved in the impairment of Treg suppression in our system, we tested whether inhibition of STAT3 phosphorylation, a key event in IL-6 intracellular signaling, abrogated Teff resistance. We found that STAT3 inhibition decreased the average Pam3-CSK4-mediated impairment in Treg suppression (Figure 4B) in co-cultures with Teff for subjects of both TLR1 haplotype (p=0.005). Addition of exogenous IL-6 to co-cultures increased major allele Teff resistance partially abrogating the difference between genotypes (Supplemental Figure 2E). In another published study, blockade of IL-6 independently reversed the effects of TLR1/2-mediated impaired Treg suppression (15). While other pro-inflammatory cytokines may be involved in modulating Teff resistance in this system, these data demonstrate that IL-6 signaling participates in the induction of Teff resistance to Treg suppression in response to TLR1/2 stimulation.
Work from our laboratory and others have highlighted a potential role for genetic variation in TLR1 in various immune-mediated and inflammatory diseases. Here, we have shown that a haplotype in TLR1 composed of the minor alleles identified in these clinical association studies is associated with increased Teff resistance to Treg suppression after stimulation with a TLR1/2 agonist. These findings provide a novel potential mechanism through which these TLR1 variants might affect clinical outcomes. For example, Mayerle et al. detected a strong association between the TLR10/1/6 genetic locus and seroprevalence for Helicobacter pylori (5). The most highly associated TLR1 SNP in this study, rs10004195, is in high LD with the two non-synonymous coding SNPs included in the haplotype in our study (rs4833095 r2=1 and rs5743618 r2=0.95). Our data suggests that Teff from subjects bearing the minor alleles at these loci could be resistant to Treg suppression after chronic exposure to TLR1/2 ligands that are abundant at these mucosal sites. Similar mechanisms could be responsible for associations with asthma through altered immune responses in the respiratory mucosa.
In summary, we have shown that Caucasian subjects harboring the minor allele TLR1 haplotype rs5743618T, rs5743551G, rs4833095C have greater cell surface expression of TLR1 on both CD4+FoxP3+ and CD4+FoxP3− T cells and greater impairment of Treg mediated suppression of Teff proliferation after treatment with the TLR1/2 agonist Pam3CSK4. This effect is due to Teff resistance and is largely mediated by IL-6 signaling, though other factors may also play a role. These studies highlight the potential importance of common genetic variation in TLR1 in mediating inter-individual differences in Treg effects on Teff and provide a new mechanism through which SNPs in TLR1 might alter susceptibility to disease states in which Treg:Teff interactions play a key role.
Supplementary Material
Acknowledgments
This work was supported by the Parker B Francis Fellowship Program (C.M.), National Heart, Lung, and Blood Institute grant K23 HL120896-01 (C.M.), R01 HL089807-01 (M.M.W), and National Institute of Allergy and Infectious Diseases grant U54 AI057141 (M.M.W) and U01 AI101990-01 (JHB).
We thank Dr. Thomas Hawn for providing the custom designed Taqman assay for genotyping of rs5743618.
Abbreviations
- TLR
Toll-like Receptor
- Pam3CSK4
N-palmitoyl-S-dipalmitoylglyceryl Cys-Ser-(Lys)4
- Treg
regulatory T cell
- Teff
effector T cell
- PGN
Peptidoglycan
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Kawai T, Mühlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 4.Guan Y, Ranoa DRE, Jiang S, Mutha SK, Li X, Baudry J, Tapping RI. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J Immunol. 2010;184:5094–5103. doi: 10.4049/jimmunol.0901888. [DOI] [PubMed] [Google Scholar]
- 5.Mayerle J, den Hoed CM, Schurmann C, Stolk L, Homuth G, Peters MJ, Capelle LG, Zimmermann K, Rivadeneira F, Gruska S, Völzke H, de Vries AC, Völker U, Teumer A, van Meurs JBJ, Steinmetz I, Nauck M, Ernst F, Weiss F-U, Hofman A, Zenker M, Kroemer HK, Prokisch H, Uitterlinden AG, Lerch MM, Kuipers E. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 2013;309:1912–1920. doi: 10.1001/jama.2013.4350. [DOI] [PubMed] [Google Scholar]
- 6.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U, Davey-Smith G, Timpson NJ, Tung JY. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bønnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, Linneberg A, Curtin JA, Warrington NM, Standl M, Kerkhof M, Jonsdottir I, Bukvic BK, Kaakinen M, Sleimann P, Thorleifsson G, Thorsteinsdottir U, Schramm K, Baltic S, Kreiner-Møller E, Simpson A, St Pourcain B, Coin L, Hui J, Walters EH, Tiesler CMT, Duffy DL, Jones G, Ring SM, McArdle WL, Price L, Robertson CF, Pekkanen J, Tang CS, Thiering E, Montgomery GW, Hartikainen A-L, Dharmage SC, Husemoen LL, Herder C, Kemp JP, Elliot P, James A, Waldenberger M, Abramson MJ, Fairfax BP, Knight JC, Gupta R, Thompson PJ, Holt P, Sly P, Hirschhorn JN, Blekic M, Weidinger S, Hakonarsson H, Stefansson K, Heinrich J, Postma DS, Custovic A, Pennell CE, Jarvelin M-R, Koppelman GH, Timpson N, Ferreira MA, Bisgaard H, Henderson AJ. Australian Asthma Genetics Consortium (AAGC), and EArly Genetics and Lifecourse Epidemiology (EAGLE) Consortium. 2013. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 45:902–906. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heffelfinger C, Pakstis AJ, Speed WC, Clark AP, Haigh E, Fang R, Furtado MR, Kidd KK, Snyder MP. Haplotype structure and positive selection at TLR1. Eur J Hum Genet. 2014;22:551–557. doi: 10.1038/ejhg.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, Jarvik GP, Hajjar AM, Nickerson DA, Rieder M, Sevransky J, Maloney JP, Moss M, Martin G, Shanholtz C, Garcia JGN, Gao L, Brower R, Barnes KC, Walley KR, Russell JA, Martin TR. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E, Hamann L, Schumann RR, Tapping RI. Cutting edge: A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol. 2007;178:7520–7524. doi: 10.4049/jimmunol.178.12.7520. [DOI] [PubMed] [Google Scholar]
- 11.Hart BE, Tapping RI. Cell surface trafficking of TLR1 is differentially regulated by the chaperones PRAT4A and PRAT4B. J Biol Chem. 2012;287:16550–16562. doi: 10.1074/jbc.M112.342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikacenic C, Reiner AP, Holden TD, Nickerson DA, Wurfel MM. Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun. 2013;41:52–57. doi: 10.1038/gene.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawn TR, Misch EA, Dunstan SJ, Thwaites GE, Lan NTN, Quy HT, Chau TTH, Rodrigues S, Nachman A, Janer M, Hien TT, Farrar JJ, Aderem A. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol. 2007;37:2280–2289. doi: 10.1002/eji.200737034. [DOI] [PubMed] [Google Scholar]
- 14.Oberg H-H, Ly TTH, Ussat S, Meyer T, Kabelitz D, Wesch D. Differential but direct abolishment of human regulatory T cell suppressive capacity by various TLR2 ligands. J Immunol. 2010;184:4733–4740. doi: 10.4049/jimmunol.0804279. [DOI] [PubMed] [Google Scholar]
- 15.Nyirenda MH, Sanvito L, Darlington PJ, O’Brien K, Zhang G-X, Constantinescu CS, Bar-Or A, Gran B. TLR2 stimulation drives human naive and effector regulatory T cells into a Th17-like phenotype with reduced suppressive function. J Immunol. 2011;187:2278–2290. doi: 10.4049/jimmunol.1003715. [DOI] [PubMed] [Google Scholar]
- 16.Schneider A, Buckner JH. Assessment of suppressive capacity by human regulatory T cells using a reproducible, bi-directional CFSE-based in vitro assay. Methods Mol Biol. 2011;707:233–241. doi: 10.1007/978-1-61737-979-6_15. [DOI] [PubMed] [Google Scholar]
- 17.Schneider A, Rieck M, Sanda S, Pihoker C, Greenbaum C, Buckner JH. The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J Immunol. 2008;181:7350–7355. doi: 10.4049/jimmunol.181.10.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 19.Goodman WA, Levine AD, Massari JV, Sugiyama H, McCormick TS, Cooper KD. IL-6 signaling in psoriasis prevents immune suppression by regulatory T cells. J Immunol. 2009;183:3170–3176. doi: 10.4049/jimmunol.0803721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider A, Long SA, Cerosaletti K, Ni CT, Samuels P, Kita M, Buckner JH. In Active Relapsing-Remitting Multiple Sclerosis, Effector T Cell Resistance to Adaptive Tregs Involves IL-6–Mediated Signaling. Sci Transl Med. 2013;5:170ra15–170ra15. doi: 10.1126/scitranslmed.3004970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.