Summary
ATP-dependent chromatin remodelers regulate chromatin structure during multiple stages of transcription. We report that RSC, an essential chromatin remodeler, is recruited to the open reading frames (ORFs) of actively transcribed genes genome-wide, suggesting a role for RSC in regulating transcription elongation. Consistent with such a role, Pol II occupancy in the ORFs of weakly transcribed genes is drastically reduced upon depletion of the RSC catalytic subunit Sth1. RSC inactivation also reduced histone H3 occupancy across transcribed regions. Remarkably, the strongest effects on Pol II and H3 occupancy were confined to the genes displaying the greatest RSC ORF enrichment. Additionally, RSC recruitment to the ORF requires the activities of the SAGA and NuA4 HAT complexes and is aided by the activities of the Pol II CTD Ser2 kinases Bur1 and Ctk1. Overall, our findings strongly implicate ORF-associated RSC in governing Pol II function and in maintaining chromatin structure over transcribed regions.
Introduction
The ATP-dependent chromatin remodeling complex Remodels Structure of Chromatin (RSC) is an essential, abundant, fifteen-subunit complex with homology to SWI/SNF (Cairns et al., 1996; Cairns et al., 1999). RSC regulates the transcription of a wide array of Pol II and Pol III transcribed genes, including genes involved in metabolism, stress response, cell wall synthesis and mitochondrial function, as well as cell cycle progression (Angus-Hill et al., 2001; Campsteijn et al., 2007; Damelin et al., 2002; Govind et al., 2005; Mas et al., 2009; Ng et al., 2002; Van de Vosse et al., 2013). In addition to transcription, RSC is important for chromosome segregation (Hsu et al., 2003), replication, and the response to DNA damage (Charles et al., 2011; Liang et al., 2007).
RSC possesses multiple domains that may be involved in recruiting the complex to its target genes. For example, the DNA binding domains of Rsc3 and Rsc30 recruit RSC to CGCG motifs in promoter regions (Badis et al., 2008; Floer et al., 2010). Rsc1 and Rsc2 possess A-T hook motifs (Cairns et al., 1999) that likely stabilize RSC-DNA interactions, and the SWIRM DNA binding domain of Rsc8 potentially mediates RSC-nucleosome interactions (Da et al., 2006). The bromodomains of Sth1, Rsc1, Rsc2 and Rsc4 are also likely candidates for facilitating the recruitment of RSC to chromatin (Cairns et al., 1999; Kasten et al., 2004). In addition to the recognition of DNA binding motifs or histone modifications, the interaction of RSC with activators such as Gcn4 (Swanson et al., 2003) and Hog1 (Mas et al., 2009), as well as the interaction of RSC with all three RNA polymerases (Soutourina et al., 2006) potentially recruits RSC to its target sites.
While the role of RSC in transcription of Pol II and Pol III genes has been well established (Angus-Hill et al., 2001; Damelin et al., 2002; Ng et al., 2002; Parnell et al., 2008), whether RSC specifically modulates a particular stage of the process remains unclear. The presence of RSC in intergenic regions has been linked with low nucleosome occupancy and is required for normal mRNA levels (Badis et al., 2008; Hartley and Madhani, 2009; Parnell et al., 2008), suggesting that it promotes initiation. Additionally, upon induction of carbohydrate metabolism genes, RSC binds to the promoters prior to TFIIB, suggesting that RSC facilitates initiation at these genes (Ng et al., 2002). Mutations in the RSC complex resulted in reduced recruitment of Pol II and TBP at the promoter of the Gcn4 target gene ARG1 (Govind et al., 2005), and Rsc3 was shown to facilitate activator binding at the GAL1/GAL10 locus (Floer et al., 2010), suggesting that the RSC complex regulates initiation at these genes as well.
Several recent in vitro and in vivo studies suggest that RSC facilitates transcription elongation in addition to initiation. RSC promotes transcription through a nucleosomal template in vitro (Carey et al., 2006) and the MAP kinase, Hog1, was shown to recruit RSC to the ORFs of osmo-stress-responsive genes to facilitate transcription (Mas et al., 2009). Furthermore, reduced RSC occupancy in the coding sequence of GAL1 in a gcn5Δ/esa1 mutant was coupled with a reduced rate of elongation and histone eviction (Ginsburg et al., 2009), suggesting that RSC function in the coding sequence facilitates elongation at that gene.
In this study, we have examined the genome-wide localization of RSC under three different growth conditions, including amino acid starvation stress. We report that RSC is enriched at two classes of genes, one characterized by intergenic binding and another in which RSC is predominantly localized to the transcribed regions. RSC recruitment is mediated by the HATs Gcn5 and Esa1, and by the Ser2 Pol II CTD kinases Bur1 and Ctk1, predominantly in the ORFs of weakly transcribed genes where RSC modulates both Pol II and histone occupancy. Collectively, our data strongly indicate a role for RSC in regulating post-initiation steps of transcription.
Results
RSC is cotranscriptionally recruited to coding sequences under amino acid starvation stress
While various studies have concluded that RSC binds predominantly to intergenic regions (Damelin et al., 2002; Ng et al., 2002), in vitro studies indicate a role for RSC in promoting transcription elongation (Carey et al., 2006; Kuryan et al., 2012). We observed recruitment of multiple RSC subunits, including the catalytic subunit Sth1, primarily to the open reading frame (ORF) of a Gcn4 target gene, ARG1 (Figures 1A and S1). To determine whether these results are indicative of a more global phenomenon, we utilized an inducible system in which Gcn4-target genes are upregulated under amino acid starvation stress (Natarajan et al., 2001).
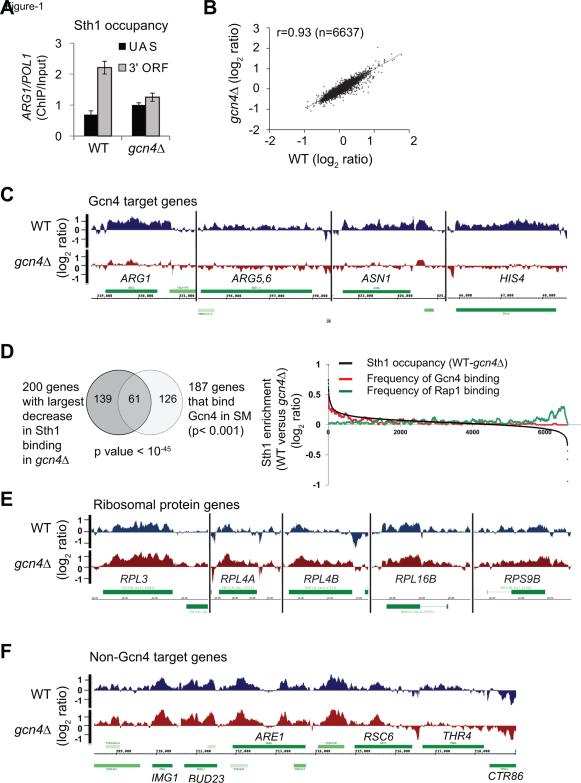
Figure 1. RSC is recruited to coding sequences during amino acid starvation.
A) Myc-tagged Sth1 (Sth1) occupancy measured by chromatin immunoprecipitation (ChIP) at the UAS and 3’ ORF of ARG1 in WT and gcn4Δ cells after inducing with SM (sulfometuron methyl). Error bars represent SEM (n=3). B) Scatter plot of Sth1 enrichment ([ChIP/Input] log2 ratios) in the coding regions, under inducing conditions (SM) in WT and gcn4. C) Snapshots of high resolution ChIP-chip indicating that Sth1 binds to the coding regions of Gcn4-target genes in WT (top) but not gcn4Δ cells (bottom). D) Venn diagram (left) and frequency plot (right) showing enrichment of Gcn4 target genes (red) among the genes showing the largest reduction in Sth1 binding (over ORFs) in gcn4Δ, and Rap1-target genes (green) enriched among the genes showing increased Sth1 binding (over ORFs) in gcn4Δ. E-F) Snapshots showing Sth1 enrichment in WT and gcn4Δ at ribosomal protein genes (E) and at non-Gcn4 target genes along with the Gcn4 target gene THR4 (F).
See also Figure S1.
WT and gcn4Δ cells were treated with sulfometuron methyl (SM; inhibits isoleucine/valine biosynthesis) to induce transcription of Gcn4 target genes, and Sth1 binding was determined by high resolution ChIP-chip (Affymetrix). Since these genes are minimally transcribed in gcn4Δ cells, we could measure transcription-dependent recruitment of RSC to Gcn4 target genes. The average Sth1 occupancy in coding regions genome-wide was quite similar (Pearson correlation, r=0.93) in WT and gcn4Δ cells (Figure 1B) suggesting that deleting GCN4 per se did not substantially alter RSC occupancy at non-Gcn4 targets genome-wide. In agreement with our gene-specific ChIP results (Figures 1A and S1), Sth1 bound across the coding regions of many Gcn4 target genes, including ARG1 and HIS4 in WT but not gcn4Δ cells (Figure 1C). Consistently, genes exhibiting the largest reduction in Sth1 ORF occupancy in gcn4Δ cells were enriched for Gcn4 targets (Harbison et al., 2004) (Figure 1D), indicating that RSC localizes to the transcribed regions of Gcn4-regulated genes during active transcription.
We also observed increased Sth1 occupancy in the coding regions of some genes in gcn4Δ versus WT cells. These genes were enriched for Rap1 targets (p value < 6.4 × 10−22 for overlap) (Harbison et al., 2004) (Figure 1D, right), consistent with the observation that Gcn4 mediates transcriptional repression of ribosomal protein genes, which are Rap1 targets, under amino acid starvation conditions (Joo et al., 2011). Thus, loss of Gcn4 causes up-regulation of these Rap1 targeted genes, with concomitant increase in Sth1 binding. RSC was previously shown to localize to the promoters of the ribosomal protein genes (Ng et al., 2002), whereas we find that RSC binds to the coding regions of these genes as well (Figure 1E). Sth1 binding was additionally observed in the coding regions of many non-Gcn4 target genes, independent of Gcn4, indicating that RSC binds to the ORFs of multiple classes of genes (Figure 1F). Collectively, our results showing diminished Sth1 binding in gcn4Δ cells at Gcn4 target genes and increased binding at Rap1 target genes that exhibit Gcn4-dependent repression strongly suggest that RSC binds to the coding regions of actively transcribing genes, at least under amino acid starvation stress.
RSC is recruited to the coding sequences of actively transcribing genes
To determine if RSC localization to coding regions is a general phenomenon and is not limited to amino acid starvation conditions, we measured Sth1 occupancy in synthetic complete (SC) media with and without SM induction by ChIP-chip using Agilent 4×44K arrays. A previous study reported differences in Mediator binding in SC versus YPD, indicating condition-specific association of general regulatory factors with their targets (Fan and Struhl, 2009). We therefore measured Sth1 occupancy in YPD as well as SC, and Rpb3 occupancy in all three growth conditions to validate the link between RSC occupancy and active transcription.
Heat-maps depicting genome-wide Sth1 binding reveal robust RSC enrichment in coding regions under all three growth conditions (Figure 2A). Consistent with a recent study showing RSC binding to the first three genic nucleosomes (Yen et al., 2012), RSC binding is slightly biased toward the 5’ ORF. Sth1 enrichment in the coding sequences appears to be greater than that of the promoter, at least at the genes showing the highest Pol II enrichment in all three conditions (Figure 2A), indicating that RSC localizes to the coding sequences of highly transcribed genes. While Rpb3 occupancy increased at Gcn4-regulated genes in SM-treated cells (Figure S2A), it was noticeably reduced in the coding regions of many genes (Figure 2B, compare SC and SM Rpb3). RSC enrichment was generally lower across the genome in SM, consistent with the observed reduction in Rpb3 occupancy (Figure 2B). These last findings further support a link between RSC ORF occupancy and active transcription. Consistent with this finding, Rpb3 ORF occupancy correlated better with Sth1 in ORFs than in promoters (Pearson correlation r=0.64 vs r=0.49) across all three growth conditions (Figures 2C and S2B). Similarly, transcription frequency (taken from (Holstege et al., 1998)) was more strongly correlated with Sth1 ORF occupancy than promoter occupancy (Figure 2D). Altogether, the large number of genes enriched for RSC in the coding region (Figure 2A), as well as the strong correlation between Sth1 and Rpb3 ORF occupancy (Figure 2C), indicate that RSC is globally recruited to the coding regions of actively transcribed genes.
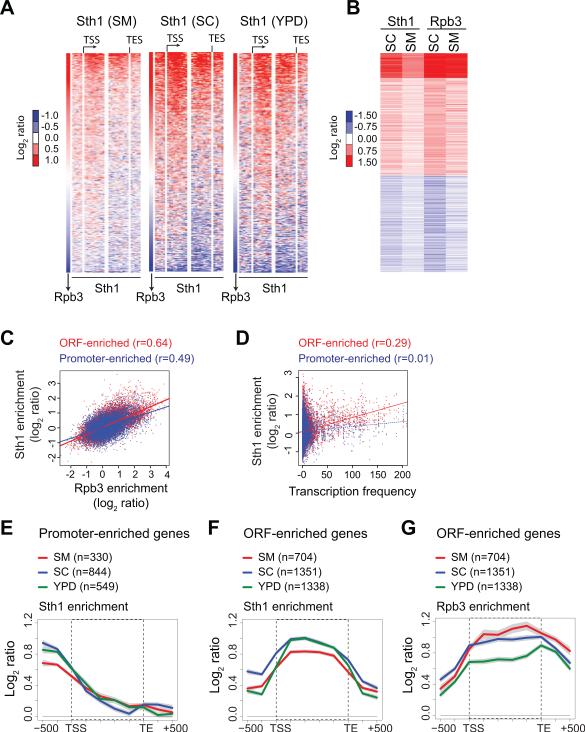
Figure 2. RSC is recruited to transcribed regions under multiple growth conditions.
A) Sth1 and Rpb3 binding was determined by ChIP-chip (Agilent 4×44 arrays) in WT cells grown in YPD, synthetic complete (SC) or SC+SM medium. Heat maps depicting Sth1 binding across the genes (n=5474) sorted from the highest to the lowest average ORF Rpb3 values in each growth condition; SM (left), SC (middle) and YPD (right). The genes were computationally split in the center and the first half was aligned to the transcription start site (TSS) while the other half was aligned to the transcription end site (TES). B) Heat-maps showing Sth1 and Rpb3 binding (averaged over ORF) in SC and SM (n=5474). C-D) Scatter plots showing the correlation between Sth1 binding of the ORF-enriched and promoter-enriched classes versus the Rpb3 occupancies (averaged over ORF) (C) and versus transcription frequencies (from Holstege et al., 1998) (D). The Pearson correlations are shown. E-G) Gene averaged profiles of Sth1 binding at the promoter-enriched genes (E), at the ORF-enriched genes (F) and Rpb3 binding profile for the ORF-enriched genes (G). The gray shading represents SEM.
See also Figure S2.
To further examine Sth1 binding profiles, we selected genes displaying average Sth1 binding greater than log2 (ChIP/Input) of 0.5 in the promoter (promoter-enriched) or in the coding region (ORF-enriched). The analyses described below were performed with these RSC-enriched genes unless indicated otherwise. We identified 330 genes in SM, 844 genes in SC and 545 genes in YPD that were promoter-enriched, while 704 genes in SM, 1351 genes in SC and 1338 genes in YPD exhibited Sth1 enrichment in coding regions. The gene-averaged profile of Sth1 binding in the promoter-enriched class showed maximum binding in the region upstream of the transcription start site (TSS), as expected, suggesting that RSC binds primarily to the promoters of these genes (Figure 2E). In contrast, the ORF-enriched class showed greater Sth1 binding across the coding region, peaking slightly downstream of the TSS and declining just prior to the transcription end site (TES) (Figure 2F). The Rpb3 binding profile closely mirrored the Sth1 binding profile (Figure 2G) suggesting that RSC occupies the coding regions of actively transcribing genes. Only a small fraction of RSC-enriched genes (9-12%, Figure S2C) met the threshold for Sth1 enrichment at both the promoter and the ORF, implying that RSC generally binds robustly to either promoter or coding regions genome-wide.
Interestingly the highly expressed ribosomal protein genes were more prevalent among the ORF-enriched class (82%, p=4.6 × 10−46) than the promoter-enriched class (34%, p=0.003) in SC, suggesting that highly transcribed genes recruit RSC to their coding sequences. Consistent with this observation, the coding sequences of Gcn4-target genes, which are upregulated under amino acid starvation stress, were highly enriched for RSC in SM (Figure S2D). We also, however, observed a number of highly transcribing genes (Rpb3 > 0.5 log2 ratio) in all three growth conditions that were devoid of Sth1 (Figure S2E), implying that RSC is not recruited to the ORF of all highly transcribed genes. Overall, the substantial Sth1 ORF enrichment observed at a large number of genes in all three growth conditions, coupled with the strong correlation between Sth1 and Rpb3 strongly indicates that RSC is recruited to the coding sequences of actively transcribing genes.
Histone acetyltransferases promote RSC recruitment to weakly transcribed genes
Having determined that RSC binds to transcribed coding regions, we asked how RSC might be recruited to these regions. Four of the RSC subunits, including Sth1, account for almost half of the bromodomains found in S. cerevisiae (Kasten et al., 2004), and histone acetylation enhances RSC binding to nucleosomes in vitro (Carey et al., 2006; Chatterjee et al., 2011). These studies and others suggest a potential role of histone acetylation in RSC recruitment. Whether histone acetylation promotes RSC binding to chromatin in general or specifically to coding regions in vivo has not been addressed.
Gcn5-containing SAGA and Esa1-containing NuA4 are the two major histone H3 and H4 histone acetyltransferase (HAT) complexes in yeast. Both SAGA and NuA4 promote RSC-stimulated transcription in vitro (Carey et al., 2006) and the HAT double mutant gcn5Δ/esa1ts reduces RSC occupancy at the GAL1 ORF (Ginsburg et al., 2009). We thus determined Sth1 and Rpb3 occupancy in gcn5Δ/esa1ts in both SC and SM to examine the role of histone acetylation in recruiting RSC genome-wide. Heat-maps representing Sth1 binding at the ORF-enriched class (1351 genes in SC and 704 genes in SM) displayed a general reduction in RSC ORF enrichment in gcn5Δ/esa1ts (Figure 3A). Gene-averaged profiles revealed that the decrease in Sth1 ORF binding in SC was accompanied by reduced Rpb3 binding (Figure 3B), suggesting that the reduction in RSC binding could stem primarily from impaired transcription in the HAT mutant. However, the RSC binding defect in SM, although less pronounced than in SC, is not attributable to reduced Rpb3 binding (Figure 3B). This finding indicates that Rpb3 and Sth1 are recruited to some genes by distinct pathways (varying in dependence on Gcn5/Esa1) in yeast grown in SM. Why this is not the case in SC is not clear, but may reflect a distinct cohort of transcriptional activators underlying the different transcriptional programs in these two conditions.
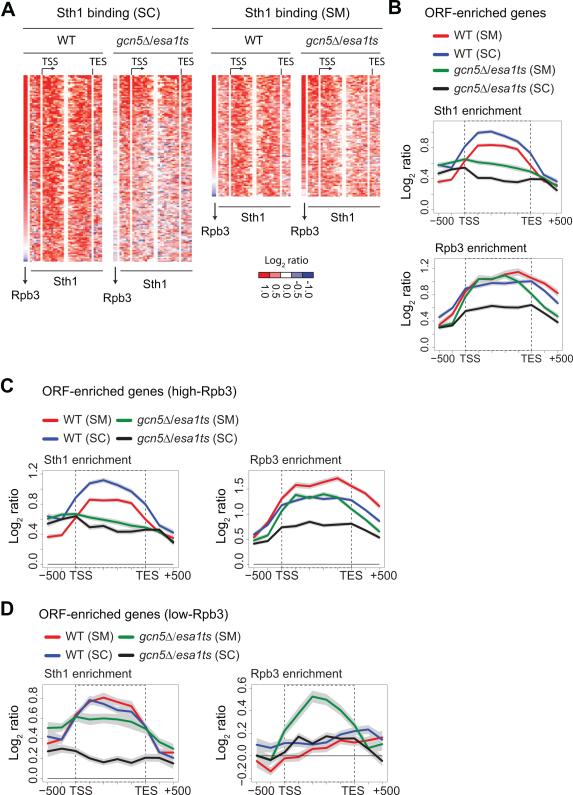
Figure 3. Histone acetyltransferases Gcn5 and Esa1 promote RSC recruitment to the coding region.
A) Heat-maps depicting Sth1 distribution across the ORF-enriched genes (Sth1 average ORF occupancy greater than log2 0.5) ranked on the basis of Rpb3 enrichment in WT from the highest to the lowest in WT and gcn5Δ/esa1ts in SC (left) and SM (right). B) Sth1 (top) and Rpb3 (bottom) binding profiles at the metagene comprised of the ORF-enriched genes in SC and SM for WT and gcn5Δ/esa1ts are shown. C-D) Metagene profiles of Sth1 and Rpb3 binding are shown at the ORF-enriched genes with Rpb3 occupancies greater than log2 0.5 (high-Rpb3) (C) and less than log2 0.5 (low-Rpb3) (D) for WT and the gcn5Δ/esa1 mutant in SC and SM. The gray shading represents SEM.
See also Figure S3.
Interestingly, in contrast to the gene average profile analysis, k-means clustering (k=4) of the changes in Sth1 and Rpb3 occupancies (HAT mutant versus WT) in SC revealed reduced Sth1 occupancy at genes weakly occupied by Pol II without corresponding defects in Rpb3 binding (Figure S3A; cluster 4). This result suggests that HATs are important for recruiting RSC to the coding regions of weakly transcribed genes. To examine this observation further, we divided the ORF-enriched genes into two classes based on average Rpb3 ORF occupancy (log2 (ChIP/Input) >0.5, high-Rpb3; log2 (ChIP/Input) <0.5, low-Rpb3). Metagene analysis at the high-Rpb3 genes revealed similar reductions in Sth1 and Rpb3 occupancy in both SC and SM (Figure 3C). In contrast, at the low-Rpb3 genes, the gcn5Δ/esa1ts mutant elicited a marked reduction in Sth1 occupancy (Figure 3D, left), whereas Rpb3 occupancy was largely unaffected or even increased (Figure 3D, right).
Given that promoter regions are generally enriched for acetylated histones as compared to their corresponding ORFs (Pokholok et al., 2005), we examined whether RSC binding was reduced at the promoter-enriched genes in gcn5Δ/esa1. Remarkably, Sth1 binding in the region upstream of the TSS of these genes was largely unaffected by the HAT mutations in SM, and was only slightly reduced in SC (Figure S3B). The reduction in SC was accompanied by a similar reduction in Rpb3. Thus, a direct role of HATs in recruiting RSC is evident only for weakly transcribed ORFs. Acetylated histones might also enhance RSC recruitment at highly transcribed genes, but the importance of HATs for high-level of Pol II occupancy obscures this possibility.
Serine 2 kinases promote RSC binding to the coding regions of weakly transcribed genes
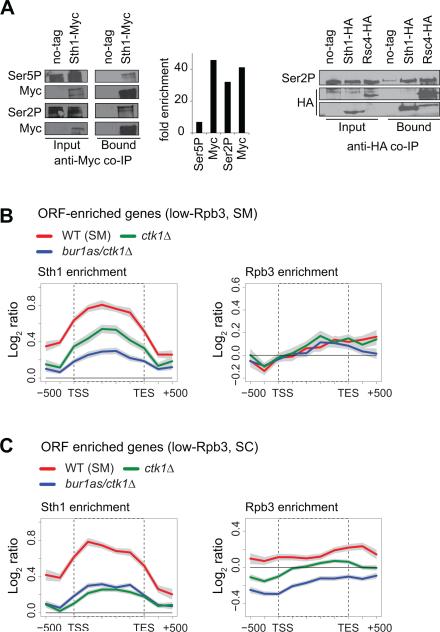
The phosphorylated Pol II C-terminal domain (CTD) has been shown to recruit several factors involved in Pol II elongation (Jeronimo et al., 2013). The Pol II CTD is phosphorylated at Ser5 (Ser5P) by Kin28 very early during Pol II elongation, and later at Ser2 (Ser2P) by Ctk1. Ser2 is also phosphorylated by Bur1 at the 5’ region of transcribed genes (Qiu et al., 2009). As such, both Ser5P and Ser2P occur during early elongation, and either of these modifications could potentially facilitate RSC recruitment to coding sequences. Since RSC interacts with all three RNA polymerases (Soutourina et al., 2006), we asked if RSC interacts with phosphorylated Pol II. Our results show that multiple subunits of RSC coimmunoprecipitate Ser5P and Ser2P Pol II (Figure 4A), suggesting that phosphorylated Pol II could promote RSC recruitment to the coding sequences.
Figure 4. Ser2 kinases promote RSC recruitment to the coding regions of low transcribed genes.
A) Myc-tagged Sth1 (left) or HA-tagged Sth1 and Rsc4 (right) were immunoprecipitated using anti-Myc or anti-HA antibodies, and the immunoprecipitates were detected by western blot for the presence of Pol II phosphorylated at Ser5 (Ser5P) or Ser2 (Ser2P). Untagged WT was used as a control. Ser5P, Ser2P and Myc signals were quantified after correcting for inputs are presented as fold enrichment. B-C) Gene average profile of Sth1 and Rpb3 at the ORF-enriched genes with low Rpb3 occupancy (log2 <0.5) in SM (B) and in SC (C) for the WT and the Serine2 kinase mutants are shown. The Bur1 kinase activity was inhibited by treating bur1as/ctk1Δ cells with the ATP-analog 3MB-PP1. The bur1as/ctk1Δ strain untreated with 3MB-PP1 is labeled ctk1Δ. The gray shading represents SEM.
See also Figure S4.
To test this hypothesis, we analyzed Sth1 recruitment in Pol II CTD kinase mutants. To determine whether Ser5P was important for recruiting RSC to coding regions, we measured genome-wide Sth1 occupancy in kin28as/bur2Δ (as; analog sensitive), a mutant which elicits greater reductions in Ser5P than the single kin28as mutant (Bataille et al., 2012). As expected, treating kin28as/bur2Δ with the ATP-analog NA-PP1 substantially reduced Ser5P levels at ARG1 (Figure S4A). Impairing Ser5 phosphorylation in SC led to a uniform reduction in Sth1 occupancy at ORF-enriched genes, but elicited a comparable reduction in Rpb3 binding (Figures S4C and S4D). Thus, the reduction in Ser5 phosphorylation could affect RSC recruitment indirectly by reducing transcription, making it unclear whether Ser5P has a direct role in RSC recruitment. Examining Sth1 and Rpb3 occupancy in Gcn4 inducing conditions revealed essentially no effect of reducing Ser5P on RSC occupancy (Figures S4E and S4F).
To examine the role of Ser2 phosphorylation in recruiting RSC, we determined Sth1 and Rpb3 occupancy by ChIP-chip in the Ser2 kinase double mutant bur1as/ctk1Δ after treating the cells with the ATP-analog 3MB-PP1 to inactivate Bur1 kinase. This mutant produced similar reductions in Sth1 binding at the ARG1 3’ ORF with or without ATP-analog treatment (Figure S4B), suggesting that Ctk1-mediated Ser2P is important for RSC recruitment at this gene. We thus analyzed Sth1 binding genome-wide in the bur1as/ctk1Δ mutant without 3MB-treatment (hereafter referred to as ctk1Δ).
The Ser2 kinase mutant (analog treated or untreated) reduced Sth1 occupancy across the coding regions of the ORF-enriched class of genes (Figure S4C), but diminished Rpb3 occupancy as well (Figure S4D), suggesting that the reduction in Sth1 was likely due to a defect in transcription in the absence of Ser2P. Under amino acid starvation stress, while both bur1as/ctk1Δ and ctk1Δ produced similar reductions in Rpb3 occupancy at the ORF-enriched genes (Figure S4F), Sth1 binding was reduced further in the double mutant (Figure S4E), suggesting that both kinases promote RSC recruitment to the transcribed regions. To distinguish the Sth1 binding defect from the effect of Ser2 kinase mutants on transcription, we analyzed Sth1 and Rpb3 binding profiles by average Rpb3 ORF occupancy (log2 ratio >0.5 high-Rpb3; <0.5 low-Rpb3). Under amino acid starvation stress, Sth1 binding was distinctly reduced at the low-Rpb3 genes in the Ser2 kinase mutants without any reduction in Rpb3 occupancy (Figure 4B). Similarly, Sth1 binding was diminished under normal growth conditions (Figure 4C), although this reduction could be partially attributed to Pol II binding defects particularly in the bur1as/ctk1Δ mutant. The defect in Sth1 binding at the high-Pol II occupied genes was indistinguishable from the Rpb3 binding defect (data not shown), suggesting that either Ser2 kinases are not required for RSC recruitment to these genes, or that their effect is masked by the reliance of these genes on Ser2 kinase activity for transcription. Together, our results implicate Ser2 kinases in promoting RSC binding to the coding sequences of weakly transcribed genes.
ORF-bound RSC is required for the transcription of low-Pol II occupied genes
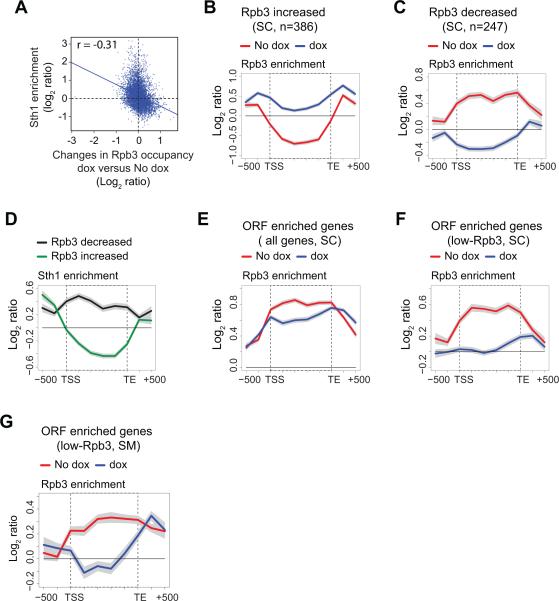
Considering that RSC occupies coding regions, and that Sth1 and Pol II occupancy are highly correlated (Figure 2), we asked whether RSC facilitates transcription. To do so, we depleted RSC catalytic subunit Sth1 and measured Rpb3 occupancy genome-wide in both SC and SM. We depleted Sth1 by using an STH1-TET strain in which the endogenous promoter was replaced by a tetracycline-repressible promoter. Sth1 protein levels were greatly reduced in STH1-TET cells treated with doxycycline in both SM and SC (Figure S5A). Sth1 ORF enrichment in WT cells was mildly anti-correlated with the changes in average Rpb3 ORF occupancy upon Sth1-depletion in both SC (Pearson correlation r = −0.31, p-value < 2.4 × 10−122) (Figure 5A) and SM (data not shown). These results suggest that ORF-associated RSC promotes transcription by regulating Pol II occupancy in coding sequences of at least some genes.
Figure 5. Sth1-depletion reduces Pol II occupancies at RSC occupied genes.
Rpb3 occupancy was determined by ChIP-chip in STH1-TET cells grown in the presence of doxycycline (dox) to deplete Sth1, or in the absence (No dox). A) Scatter plot showing the correlation between the changes in Rpb3 binding upon Sth1 depletion versus Sth1 enrichment in coding regions in the WT control cells grown in SC genome-wide (n=5464). B-C) Rpb3 binding profiles at the genes exhibiting an increase in Rpb3 occupancy (> 0.5 log2 ratio, n=386) or decrease (> −0.5 log2 ratio, n=247) upon Sth1 depletion. D) Sth1 binding profiles at the genes analyzed in panels B and C. E-F) Metagene profile of Rpb3 occupancy at the genes enriched for RSC in the coding regions (ORF-enriched genes; Sth1 binding >0.5 log2 ratio, n=1351) and a subset of ORF-enriched, low-Rpb3 occupied genes (Rpb3 <0.5 log2 ratio, n=356). G) Rpb3 occupancy profile at the low-Rpb3 occupied genes (n=268) in dox and No dox cells induced by SM. The gray shading represents SEM.
See also Figure S5.
To further investigate this phenomenon, we analyzed Rpb3 occupancy at genes that exhibited a change in average Rpb3 ORF occupancy (> log2 0.5; dox vs No dox) upon depleting Sth1. Rpb3 occupancy increased in the coding regions of 386 genes and decreased at 247 genes in SC (Figures 5B and 5C). Since these genes were selected solely on the basis of the change in Rpb3 occupancy, we asked whether these genes were actually enriched for RSC. The gene-averaged Sth1 binding profiles revealed that genes with decreased Pol II were indeed enriched for Sth1 in the coding regions (Figure 5D). In contrast, the genes exhibiting increased Pol II on Sth1 depletion were largely devoid of Sth1 in the ORF in WT cells. Similarly, genes showing reduced Pol II occupancy upon Sth1 depletion in SM were enriched for RSC in the coding regions, whereas the genes showing increased Rpb3 were not (Figures S5B-D). Altogether, these results strongly suggest that RSC recruited to coding regions promotes transcription.
Having shown that ORF-bound RSC affects transcription of a subset of genes (Figures 5C and S5C), we analyzed the effect of Sth1-depletion on genes specifically enriched for RSC in the ORF. In SC, Sth1-depletion reduced Pol II occupancy in the coding region downstream of the TSS of the ORF-enriched genes (Figure 5E), indicating a role for RSC in Pol II elongation-coupled events. To examine if RSC recruitment to coding regions is equally important for genes transcribed at high and low levels, we examined the impact of Sth1-depletion on Rpb3 occupancy at high and low Pol II occupied genes (log2 > 0.5 and <0.5 Rpb3). Rpb3 was only mildly reduced in the coding regions of the high-Rpb3 genes (Figure S5E), suggesting that RSC is dispensable for the transcription of highly expressed genes. Interestingly, Rpb3 occupancy was severely reduced in the coding regions of low-Pol II occupied genes in the absence of Sth1 (Figure 5F, compare dox and No dox), indicating that the transcription of weakly transcribed genes is strongly dependent on the recruitment of RSC to the coding regions. Consistent with this observation, Rpb3 binding was greatly diminished only from the coding regions of the ORF-enriched low-Rpb3 occupied genes upon Sth1-depletion in SM (Figure 5G). Similar to SC, Rpb3 occupancy at the high Pol II-occupied genes was largely unaffected by Sth1-depletion in SM (Figure S5F). Overall, our results indicate that RSC recruitment to coding sequences is critical for efficient transcription of weakly transcribed genes.
RSC is recruited to coding regions to maintain co-transcriptional histone occupancy
Having seen that RSC recruitment to transcribed regions promotes transcription of low-Pol II occupied genes, we considered whether RSC might promote histone eviction to allow efficient transcription of these genes. ChIP-chip of histone H3 occupancy in Sth1-depleted and non-depleted cells grown in SC revealed that Sth1 enrichment in coding regions was negatively correlated with changes in H3 occupancy in the ORF upon Sth1-depletion (r = −0.49, p-value <2.3 × 10−76) (Figure 6A). This result implies that histone occupancy decreases at RSC-occupied genes in the absence of RSC activity. Consistent with this finding, H3 occupancy at ORF-enriched genes (Sth1 enrichment > 0.5 log2 ratio) was reduced primarily in coding regions at both high and low Pol II-occupied genes on Sth1 depletion (Figures 6B and 6C). To determine whether the reduction in H3 binding was linked to the presence of Sth1 in coding regions, we analyzed changes in H3 binding upon Sth1-depletion according to the WT Sth1 enrichment in deciles. This analysis revealed that the maximum histone depletion in the coding region was associated with the higher deciles of Sth1 occupancy whereas the lower deciles actually showed elevated histone occupancy in the ORF (Figure 6D). This data suggests that RSC localizes to coding regions to preserve ORF histone occupancy by stabilizing nucleosomes or by facilitating cotranscriptional histone deposition rather than disassembly.
Figure 6. ORF-bound RSC maintains cotranscriptional histone occupancies.
A) Scatter plot showing the negative correlation between Sth1 enrichment in the coding regions in WT control cells grown in SC versus the change in histone H3 binding in Sth1-depleted cells. B-C) Gene averaged H3 occupancy profile at the ORF-enriched genes with high Rpb3 >0.5 log2 (B) and low Rpb3 (<0.5 log2) (C) in dox and No dox treated cells. D) The ORF occupancies of Sth1 were grouped into deciles, and the changes in H3 binding under Sth1-depleting conditions are shown in the boxplot. A dashed line (blue) represents no change in H3 occupancy and the notches represent the 95% confidence interval for each median. E) H3 binding profile at the promoter enriched genes. The gray shading represents SEM. F) Venn diagram showing the overlap of Rsc3 binding site containing promoters (identified in Badis et al., 2008) with the genes showing enrichment of Sth1 (> 0.5 log2 ratio) at the promoters.
See also Figure S6.
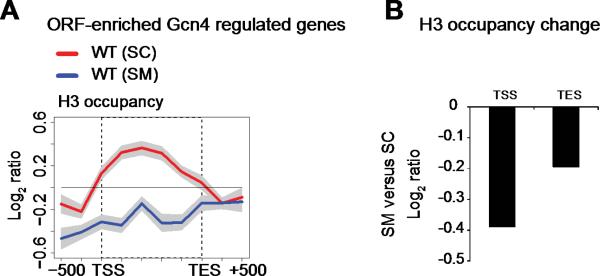
Our finding that Rpb3 binding was substantially decreased at the ORFs of weakly transcribed genes (Figure 5F) indicates a reduction in H3 occupancy is associated with reduced transcription of these genes. This is surprising because a reduction in nucleosome occupancy would be expected to enhance transcription. Hence, the reduced Pol II occupancy at weakly transcribed ORF-enriched genes evoked by RSC inactivation does not arise simply from a defect in cotranscriptional nucleosome disassembly. Our finding suggests a dual role for RSC in regulating chromatin structure. RSC may enhance transcription by remodeling at specific sites or stages during transcriptional activation, while regulating histone reassembly during ongoing transcription. Indeed, histone density is reduced at the ORF-enriched Gcn4-regulated genes upon activation of Gcn4 (SM) (Figure S6), likely reflecting activities of various chromatin remodeling activities, possibly including RSC (Yen et al., 2012). Similar to our findings, reduced occupancy by Pol II and histones has been reported upon the loss of Spt6 histone chaperone function (Ivanovska et al., 2011).
H3 binding was surprisingly decreased at promoter-enriched genes upon Sth1-depletion (Figure 6E) in contrast to a previous study in which increased nucleosome occupancy was observed in a rsc3ts mutant at a subset of promoters containing Rsc3 binding sites (Badis et al., 2008). Only a fraction of promoters containing Rsc3 binding sites, however, were present among our list of Sth1 promoter-enriched genes (Figure 6F). Additionally, the observed increase in nucleosomal occupancy in the rsc3 mutant was associated with a transcriptional defect, implying that impaired transcription might have contributed to the increase. Overall, we find that Sth1 depletion results in a general decrease in H3 occupancy, indicating a role for RSC in maintaining nucleosome occupancy in both promoter and coding regions.
Discussion
In this study, we show that RSC is recruited to the coding regions of actively transcribing genes under various growth conditions. RSC recruitment to the ORF is aided by Gcn5 and Esa1 containing HAT complexes, consistent with the preponderance of bromodomains in the RSC complex. Phosphorylation of the Pol II CTD at Ser2 additionally contributes to the association of RSC with coding regions. We further show that the transcription of weakly transcribed genes is highly dependent on the presence of RSC in their coding sequences. Finally, our results underscore the role of RSC in maintaining histone occupancy in transcribed regions genome-wide.
RSC binds to the coding sequences of actively transcribing genes
We find that RSC binding to coding regions is linked to active transcription. RSC binds to the ORFs of Gcn4 target genes in WT but not gcn4Δ cells upon induction (Figure 1C). RSC ORF occupancy at Rap1 target genes, which are repressed by Gcn4 under amino acid starvation, increased in the absence of Gcn4 (Figures 1D and 1E), linking RSC ORF-occupancy to derepression. Additionally, RSC binding was well correlated with Pol II occupancy genome-wide under both amino acid starvation stress and normal growth conditions (Figure 2). Lastly, a general downregulation of transcription in SM-treated cells corresponded with reduced Sth1 ORF binding. Our findings that RSC localizes to the coding regions of a large proportion of genes strongly implicate RSC in regulating post-initiation steps of transcription.
In addition to the ORFs, RSC binds predominantly to promoter regions at a smaller proportion of genes (Figures 2E), consistent with earlier studies (Damelin et al., 2002; Ng et al., 2002; Parnell et al., 2008). RSC targets in these studies, however, were determined by intergenic (promoter) arrays that contained only 64 ORFs. In our study, the size of the ORF-enriched class exceeds that of the promoter-enriched class across three different growth conditions, implicating RSC in modulating transcription or chromatin structure post-initiation, which is consistent with an in vitro study demonstrating that RSC promotes Pol II elongation through a nucleosomal template (Carey et al., 2006).
HATs and Serine 2 kinases promote RSC binding to coding regions
Multiple studies have suggested a role for histone acetylation in recruiting RSC to chromatin based on the interaction of RSC with acetylated histones in vitro (Carey et al., 2006; Chatterjee et al., 2011; Kasten et al., 2004). Whether acetylated histones are indeed required for promoting RSC recruitment in vivo, however, is unclear. One study has shown impaired RSC recruitment in HAT mutants, but only to one gene (GAL1) (Ginsburg et al., 2009). Our genome-wide analysis of RSC binding in gcn5Δ/esa1 cells, which reduces both H3 and H4 acetylation, indicates that histone acetylation is globally important for efficient RSC recruitment (Figure 3). Strikingly, the gcn5Δ/esa1 mutant reduced Sth1 binding to the coding sequences of weakly transcribed genes without reducing Pol II occupancy (Figure 3D), whereas Sth1 and Pol II occupancy was equally impaired in the ORFs of strongly transcribed genes. These findings suggest either that HATs are not important for RSC binding at strongly transcribed genes, or that the effect of depleting HAT activity is not visible at these genes due to their reliance on histone acetylation for transcription. Alternatively, ORF nucleosomes at strongly transcribed genes are potentially subject to rapid deacetylation, such as ARG1, where multiple HDACs, including Hos2 and Hda1, are recruited under activating conditions (Govind et al., 2010). Interestingly, hos2Δ and hda1Δ partially suppress the phenotype associated with Rsc4 bromodomain mutants (Kasten et al., 2004). A pathway that is not related to histone acetylation may thus be more important for recruiting RSC to strongly transcribed genes, whereas the persistence of acetylated histones in the ORFs of weakly transcribed genes likely renders acetylation-dependent recruitment a more predominant mechanism there.
Although promoter regions are generally enriched for acetylated nucleosomes (Pokholok et al., 2005), the gcn5Δ/esa1 mutant unexpectedly did not impair RSC binding at Sth1 promoter-enriched genes (Figure S3B) or in the region upstream of the TSS of ORF-occupied genes (Figures 3C and 3D). RSC recruitment to the promoter by sequence specific DNA binding proteins, such as Reb1 and Abf1 (Hartley and Madhani, 2009), or by its Rsc3/Rsc30 subunits (Badis et al., 2008) could explain the failure of the gcn5Δ/esa1ts mutation to abrogate RSC binding to promoters. Overall, our data indicate that histone acetylation enhances RSC recruitment to the transcribed regions of many genes.
The strong correlation observed between Rpb3 and RSC occupancy in coding regions (Figure 2A and 2F-G), as well as the interaction of RSC with Pol II (Soutourina et al., 2006) suggests that elongating Pol II promotes RSC recruitment to coding regions. Both Ser2 and Ser5 phosphorylated Pol II coimmunoprecipitate with RSC (Figure 4A) consistent with the presence of these modifications in transcribed regions (Jeronimo et al., 2013). Analysis of RSC binding in Pol II CTD kinase mutants revealed the importance of Ctk1 in promoting RSC recruitment to the coding sequences of weakly transcribed genes (Figures 4B and 4C). We were unable to detect any preferential binding of TAP-purified Sth1 or Rsc2 with phosphorylated CTD peptides (data not shown). It is, therefore, unlikely that Ser2 phosphorylation recruits RSC directly. We thus speculate that Ser2 phosphorylation indirectly stimulates RSC binding to coding sequences. The phosphorylated CTD is a well-defined scaffold for recruiting various elongation factors to coding sequences (reviewed in (Jeronimo et al., 2013)) that potentially recruits an additional factor required for bringing RSC to ORFs. Both Spt6 and the Paf1 complex, which rely on Ser2 phosphorylation for recruitment, could potentially mediate RSC association with coding regions (Burugula et al., 2014; Qiu et al., 2012).
Our finding that RSC binding is greatly reduced only at weakly transcribed genes in both HAT and Ser2 kinase mutants suggest that additional factors can contribute to RSC recruitment at highly expressed genes that can compensate for the reduction in histone acetylation or Ser2 phosphorylation. As such, either impairing histone acetylation or Ser2 phosphorylation severely reduces RSC association with the coding regions of weakly transcribed genes.
ORF-bound RSC regulates Pol II and histone occupancy
Earlier genome-wide studies suggested a role for RSC in transcription activation as well as in the repression of many genes (Damelin et al., 2002; Ng et al., 2002). Consistent with these studies, Sth1 depletion increased as well as reduced Pol II occupancies at many genes (Figures 5B-5C and S5B-C). While the genes exhibiting increased Pol II were largely devoid of RSC in their coding regions (Figures 5D and S5D), the Sth1 binding profile at these genes resembled that of the promoter-enriched genes, suggesting that RSC might repress transcription initiation at these genes.
Remarkably, only the genes displaying reduced Pol II occupancy upon Sth1-depletion were enriched for RSC in the coding regions (Figures 5D and S5D), suggesting that ORF-bound RSC promotes transcription of this set of genes. Consistent with this finding, Pol II was drastically reduced upon Sth1 depletion in the coding regions of weakly transcribed genes specifically enriched for RSC in the ORF (Figures 5F and 5G). We propose that the chromatin remodeling activity of RSC, which may include histone eviction, maximizes the transcription rate. In the absence of RSC activity, the transcription of weakly expressed genes is more severely attenuated than that of strongly transcribed genes. The finding that Sth1 binding is anti-correlated with changes in Rpb3 upon Sth1 depletion (Figure 5A), coupled with the observation that Rpb3 preferentially declines near the TSS (Figure 5E), suggests that RSC is recruited to the ORF to regulate elongation or other post-initiation activities. Our results support studies showing stimulation of Pol II elongation through acetylated nucleosomal templates by RSC in vitro (Carey et al., 2006), and reduced Pol II ORF occupancy in RSC mutants at specific genes in vivo (Ginsburg et al., 2009; Mas et al., 2009).
Previous studies indicated that impairing RSC function leads to smaller nucleosome free regions (NFRs) (Badis et al., 2008; Hartley and Madhani, 2009), suggesting that RSC is involved in removing nucleosomes from these regions. We observe a reduction in H3 occupancy in coding regions as well as at the promoters of RSC occupied genes upon Sth1 depletion (Figures 6), suggesting that RSC is largely responsible for maintaining H3 occupancy. Our results are not inconsistent with prior studies, as the promoters displaying constricted NFRs were not definitively shown to be bound by RSC (Badis et al., 2008). In fact, consistent with our results, reduced nucleosome occupancy was observed at promoters lacking Rsc3 binding sites in a rsc3-1 mutant (Badis et al., 2008). Additionally, the elevated nucleosome occupancy observed in rsc3-1 in that study was associated with a transcription defect, rendering it difficult to ascertain whether the increase arose from the loss of RSC or from a general defect in transcription. It is possible that RSC complexes recruited to promoters through the DNA binding activity of Rsc3 perform a different function than those recruited to coding regions by HATs, Ser2 kinases or other factors. In support of this hypothesis, the RSCa complex lacks the Rsc3/Rsc30 dimer (Angus-Hill et al., 2001). Our finding that H3 occupancy generally decreases upon Sth1 depletion is additionally supported by a recent study showing depletion of nucleosomes in BRG1 and SNF5 deletion mutants lacking functional SWI/SNF complex (Tolstorukov et al., 2013)
We find that the overall reduction of H3 from coding regions upon RSC inactivation is strongly dependent on robust RSC enrichment under normal conditions (Figure 6D), suggesting that RSC is not only involved in remodeling or evicting histones but is also important for maintaining nucleosome density across coding regions during transcription. We propose that RSC coordinates with histone chaperones to efficiently reestablish chromatin structure perturbed by transcription. In support of this hypothesis, the histone chaperone NAP1 was shown to function coordinately with the RSC complex to maintain nucleosome occupancy in vitro (Kuryan et al., 2012). Additionally, Sth1 genetically interacts with the histone chaperone Spt6 (Du et al., 1998), and inactivation of either RSC or Spt6 reduces both Pol II and histone occupancy in coding regions (Figure 5 and 6) (Ivanovska et al., 2011). Alternatively, RSC inactivation could promote excessive histone eviction. Structural studies have shown that RSC can accommodate an entire nucleosome in its central cavity (Chaban et al., 2008), potentially insulating the nucleosome from the activity of other chromatin remodelers. In support of this hypothesis, multiple chromatin remodeling complexes were recently shown to act together at a single gene (Yen et al., 2012). It is possible that the loss of RSC function exposes normally protected nucleosomes to abnormal eviction by other chromatin remodelers.
Collectively our results strongly implicate RSC in regulating transcription post-initiation. The robust enrichment of RSC in coding regions, mediated in part by HATs and Ser2 kinases, the strong link between Sth1 and Rpb3 binding, and the reduction in Pol II and histone occupancy in the coding regions of weakly transcribed genes in Sth1-depleted cells, all support a role for RSC in promoting transcription and in maintaining nucleosome occupancy in coding regions.
Experimental Procedures
Yeast strains and growth conditions
Yeast strains utilized in this study are listed in Table S1. Cells were grown in SC and YPD to an absorbance A600nm of 0.7. For Gcn4 induction, cells grown in SC were treated with 0.65 μM sulfometuron methyl (SM) for 30 min. To deplete Sth1, STH1-TET cells were treated with 50 μg/ml doxycycline for ~6 hr. bur1as/ctk1Δ cells were treated with 12 μM 3MB-PP1 for 80 minutes and kin28asbur2Δ cells with 6 μM NAPP1 for 35 min to inactivate kinase activities of Bur1 and Kin28, respectively. gcn5Δ/esa1ts cells were grown at 25°C to an A600nm of 0.5 and moved to 37°C for 1.5-2 hr. to inactivate Esa1 activity.
ChIP and ChIP-chip
Chromatin immunoprecipitation experiments were performed as described previously (Govind et al., 2012) using the following antibodies. Anti-myc (Roche), anti-Rpb3 (Neoclone), anti-Ser5P (H14; Covance), anti-Ser2P (Bethyl), anti-H3 (Abcam). Primers used for ChIP are listed in Table S2. Affymetrix tiling arrays (P/N520055) and Agilent 4×44K arrays (G4493A) were used for ChIP-chip experiments. ChIP and input DNA were amplified using the GenomePlex WGA-2 kit (Sigma). The samples (for Affymetrix arrays) were further fragmented to 30-70 bp before labeling and then hybridized to the arrays as per the manufacturer instructions and processed as described previously (Lee et al., 2007; Venkatesh et al., 2012).
Supplementary Material
Highlights.
RSC is recruited to the coding sequences of actively transcribing genes
HATs and serine 2 kinases promote RSC recruitment to coding regions
ORF-bound RSC regulates RNA polymerase II and histone occupancies
Acknowledgments
We thank Alan Hinnebusch for bur1as/ctk1Δ and Francois Robert for kin28as/bur2Δ. We also thank Swaminathan Venkatesh for sharing R and PERL scripts for the analysis of the genomic data. We acknowledge assistance from the Wadsworth Center Bioinformatics and Microarray Core Facilities. This work is supported by a grant to CKG from The National Institutes of Health (NIH, GM095514) and by a grant from The National Science Foundation to RHM (MCB0949722).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The Gene Expression Omnibus accession number for the data reported in this paper is GSEXXX.
References
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- Burugula BB, Jeronimo C, Pathak R, Jones JW, Robert F, Govind CK. HDACs and Phosphorylated Pol II CTD recruit Spt6 for cotranscriptional histone reassembly. Mol Cell Biol. 2014 doi: 10.1128/MCB.00695-14. doi:10.1128/MCB.00695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two Functionally Distinct Forms of the RSC Nucleosome-Remodeling Complex, Containing Essential AT Hook, BAH, and Bromodomains. MolCell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Campsteijn C, Wijnands-Collin AM, Logie C. Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS Genet. 2007;3:e92. doi: 10.1371/journal.pgen.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban Y, Ezeokonkwo C, Chung W-H, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles GM, Chen C, Shih SC, Collins SR, Beltrao P, Zhang X, Sharma T, Tan S, Burlingame AL, Krogan NJ, et al. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proc Natl Acad Sci U S A. 2011;108:10620–10625. doi: 10.1073/pnas.1019735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011;39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, WIlson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA. The Genome-Wide Localization of Rsc9, a Component of the RSC Chromatin-Remodeling Complex, Changes in Response to Stress. Molecular Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 Lysine Acetyltransferase Esa1 Is Targeted to Coding Regions and Stimulates Transcription Elongation with Gcn5. Mol Cell Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Ginsburg D, Hinnebusch AG. Measuring Dynamic Changes in Histone Modifications and Nucleosome Density during Activated Transcription in Budding Yeast. Methods Mol Biol. 2012;833:15–27. doi: 10.1007/978-1-61779-477-3_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that Specify Promoter Nucleosome Location and Identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the Regulatory Circuitry of a Eukaryotic Genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hsu J.-m., Huang J, Meluh PB, Laurent BC. The Yeast RSC Chromatin-Remodeling Complex Is Required for Kinetochore Function in Chromosome Segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Jacques PE, Rando OJ, Robert F, Winston F. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol Cell Biol. 2011;31:531–541. doi: 10.1128/MCB.01068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Bataille AR, Robert F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev. 2013;113:8491–8522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- Joo YJ, Kim JH, Kang UB, Yu MH, Kim J. Gcn4p-mediated transcriptional repression of ribosomal protein genes under amino-acid starvation. Embo j. 2011;30:859–872. doi: 10.1038/emboj.2010.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. Embo J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryan BG, Kim J, Tran NN, Lombardo SR, Venkatesh S, Workman JL, Carey M. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci U S A. 2012;109:1931–1936. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC Functions as an Early Double-Strand-Break Sensor in the Cell's Response to DNA Damage. Current Biology. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G, de Nadal E, Dechant R, de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. Embo J. 2009;28:326–336. doi: 10.1038/emboj.2008.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. Embo J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Gaur NA, Hinnebusch AG. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 2012;31:3494–3505. doi: 10.1038/emboj.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 Enhances BUR1/BUR2 Recruitment and Ser2 CTD Phosphorylation Near Promoters. Molecular Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim S-J, Natarajan K, Yoon S, Hinnebusch AG. A Multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. MolCellBiol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proceedings of the National Academy of Sciences. 2013;110:10165–10170. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 2013;152:969–983. doi: 10.1016/j.cell.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.