Abstract
Advanced gastric cancer (GC) is one of the most aggressive gastrointestinal malignancies, ADAM (A Disintegrin and Metalloproteinase)-9 is a cell-surface membrane glycoprotein with oncogenic properties that is overexpressed in several cancers. Herein, we investigated the biological mechanism of ADAM9 in the progression, proliferation and invasion of GC. First, we detected ADAM’s expression, processing and protease activity in GC cells. Protease activity was moderately correlated with ADAM9 protein expression, but was better related to a processed smaller molecular weight (84 kDa) form of ADAM9. Knockdown of ADAM9 or specifically targeted monoclonal antibody (RAV-18) suppressed cancer cell proliferation and invasion in high ADAM9 expressing cells, not in low expressing cells. RAV-18 showed in vivo antitumor activity in a GC xenograft model. Hypoxia (1% oxygen) induced ADAM9 expression and functional activity in low expressing GC cells that was inhibited by siRNA knockdown or RAV-18 antibody to levels in normoxic cells. Overall, our studies show that ADAM9 plays an important role in GC proliferation and invasion, and that while expressed in some GC cells at high levels that are responsive to functional inhibition and antitumor activity of a catalytic site directed antibody, other GC cells have low levels of expression and only when exposed to hypoxia do ADAM9 levels increase and the cells become responsive to ADAM9 antibody inhibition. Therefore, our findings suggest that ADAM9 could be an effective therapeutic target for advanced GC.
Keywords: ADAM9, invasion, gastric cancer, RAV-18
INTRODUCTION
Gastric cancer (GC) is the second leading cause of cancer-related mortality worldwide with a median survival of only around a year after diagnosis of advanced disease. This dismal prognosis mainly results from early invasion and high metastatic activity with an unknown molecular basis. Cancer metastasis is a complex, multi-step process involving the interplay between cancer cells and surrounding components of extracellular matrix (ECM), and ‘invasion’ is the first step by which cancer cells break away from the primary tumor and infiltrate surrounding stoma(1). Proteolysis has emerged as a key post-translational regulator of invasion in GC(2, 3). However, the mechanism and major players in invasion have not yet been fully elucidated.
A disintegrin and metalloproteases (ADAMs) are modular type I transmembrane proteins that contain a metalloprotease and a disintegrin-like domain, and recent studies have demonstrated a relationship between increased ADAMs and cancer progression(4). Twenty-one members of ADAM family have been identified in human, 13 of which have functional proteases. ADAM9 is shown to possess potent biological activities and is highly expressed in cancers including prostate, pancreas, breast, and colon, associated with cancer progression and poor clinical out-come(5–9). In prostate cancer, microenvironmental stress on tumor cells leads to shedding of proheparin-binding epidermal growth factor (EGF), which is processed by ADAM9. The metalloprotease domain of ADAM9 also cleaves the insulin β-chain, tumor necrosis factor (TNF)-α, transforming growth factor (TGF)-α, gelatin, β-casein, and so on, and induces the shedding of EGF, fibroblast-growth factor receptor 2IIIB, and heparin-binding EGF-like growth factor(10, 11). Therefore, the abilities of ADAM9 to degrade specific ECM substrates, release active growth factors, interact with key regulatory factors, and its expression at the invasion edges of several cancer metastases suggests that ADAM9 regulates several cancer-related processes – including cell growth and invasion.
A previous report has shown that the transcription of ADAM9 is up-regulated in GC(12), however, its biological function and potential as a therapeutic target are largely unknown. In this study, we investigated the biological role of ADAM9 in GC invasion, and explored a therapeutic approach of anti-metastasis treatment for high ADAM9-expressing cancers using an ADAM9-specific inhibitory antibody.
MATERIALS and METHODS
Reagents and antibodies
Matrigel was supplied by BD science (Franklin Lakes, NJ). Recombinant ADAM9 protein was from R&D Systems Inc. (Minneapolis, MN). ADAM9 antibodies were from Cell Signaling (Danver, MA) for immunoblot and GeneTex (Irvine, CA) for immunohistochemistry. Antibodies of HIF-1α and total/phospho-forms of EGFR and ERK were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against GAPDH and α-tubulin were purchased form Abcam (Cambridge, MA) and Sigma-Aldrich (St. Louis, MO). Total and phospho-Akt antibodies were purchased from Cell Signaling (Danver, MA). RAV-18, an anti-ADAM9-specific blocking antibody, was kindly provided by Macrogenics Inc. (South San Francisco, CA).
Cell culture
The human GC cell lines AGS, NCI-N87, Hs746T and KATO-III and the cervical cancer cell line HeLa, positive control of ADAM9 were purchased from American Type Culture Collection (ATCC, Rockville, Maryland, USA) in 2012. SNU-1, -5 and -16 in 2006, SNU-216, -638 and -668 in 2007, SNU-484 in 2008, MKN-1, -28, -45 and -74 were supplied from Korean Cell Line Bank (KCLB, Seoul, Korea). Above cell lines were authenticated by standard short tandem repeat DNA typing (STR) methodology before purchased at ATCC and KCLB. YCC-1, -2 and -3 were established in 1989, YCC-6 and -7 were in 1994 and YCC-9, and -10, -11 and -16 were established in 2000 by the Cancer Metastasis Research Center at Yonsei University College of Medicine (Seoul, Korea) from the as cites or peripheral blood (YCC-16) of advanced GC patients. YCC cell lines were authenticated by STR in July 20th 2012 that had been outsourced to KCLB. All cells were maintained in RPMI-1640 (Nissui, Tokyo, Japan) and supplemented with 10% FBS (Omega Scientific, Inc., Tarzana, CA), 100 units/mL ampicillin, 100 ug/mL streptomycin, and 2 mM glutamine (Gibco, Grand island, NY) in 5% CO2 humidified atmosphere at 37°C. All cell lines were expanded and cryopreserved in liquid nitrogen in our laboratory.
RNA extraction and RT-PCR
Total RNA was extracted using Trizol reagent (Qiazen Science, Hilden, Germany) according to the manufacturer’s protocol. Briefly, 500 µl of Trizol were added to 1×107 cells, and after adding 200 µl chloroform, cell homogenates were centrifuged at 13,200 rpm for 25 min at 4°C, and the aqueous phases were collected. Total RNA was precipitated with the same volume of isopropanol for 60 min at −20°C and centrifuged for 30 min. The pellets were washed once with 70% ethanol and suspended in RNase-free water. The quantity of total RNA was measured using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE).
To synthesize the cDNA, 4 µg of total RNA from each cell line was mixed with the oligo-dT primer and incubated at 65°C for 10 min. After adding the SuperScript II, 5X first strand buffer, 100 mM DTT, and 10 mM dNTP mix to the RNA/oligo-dT mixture, a reverse transcription process was performed at 42°C for 90 min. The remaining RNA was hydrolyzed by incubation at 65°C for 15 min in 0.1 N NaOH. The reaction was then neutralized by the addition of an equal volume of 0.1 N HCl.
Amplification reactions were performed in an Eppendorf Mastercycler Grandient (Eppendorf, Hamburg, Germany). PCR was performed using 1 µg cDNA, 2.5 mM dNTPs, 1.5 mM 10X PCR buffer with MgCl2, and 5U Taq polymerase (Invitrogen, Carlsbad, CA) to total 50 µL of reaction volume. The primer sequences used were as follows: ADAM9 (Forward: 5’-AGT GGC GGG AAA AGT TTC TT-3', Reverse: 5’-CCA GCG TCC ACC AAC TTA TT-3'). The conditions for PCR amplification were as follows: 94°C for 2 min followed by 94°C for 40 sec, 60°C for 50 sec and 72°C for 1 min, then 72°C for 2 min, and repeated for 30 cycles. PCR products were separated on ethidium bromide gels containing 1.2% agarose.
Exposure to hypoxic conditions
When cells reached 80% confluence in a 25 cm2 flask, the flask was placed in a hypoxic chamber (Personal O2/CO2 Incubator, Aspect, Fukuoka, Japan). Cells were exposed to 1% O2, 5% CO2, and 37°C for 24 h. After that, cells were collected and transferred to a Matrigel-coated 24-well plate for invasion assay. Invaded cells were stained using 0.5% crystal violet and counted after 24 h. Protease activity assay, immunobloting and proliferation assay were conducted using cells exposed to hypoxic conditions in the same way.
Immunoblotting
Subconfluent cells were harvested and suspended in a whole cell lysis buffer [50 mM Tris (pH 8.0), 10% glycerol, 2 mM EDTA, 5 mM NaF, 1% NP-40, 150 mM NaCl, 1 mM sodium orthovanadate, 1 mM NaF, 10 µg/mL aprotinin, 10 µg/mL leupeptin, 1 mM phenyl-methyl-sulfonyl fluoride]. The lysates were subjected to SDS-PAGE and blotted onto the PVDF membrane. The membrane was then incubated with the appropriately diluted primary antibody overnight at 4°C, followed by a horseradish peroxidase (HRP)-conjugated secondary antibody for 60 min at room temperature. The membrane was visualized with an ECL Western blotting reagent kit (Amersham Bioscience, Buckinghamshire, UK).
Measurement of ADAM9 protease activity
Protease activity of ADAM9 was measured by a modification of the method of Cisse et al.(13). Briefly, 5×104 cells were seeded in each well of a 96-well plate (BD Falcon, Franklin Lakes, NJ) and cultured for one day. The cells were rinsed with PBS twice, and 100 uL PBS containing 1 µM ADAM9 substrate (R&D systems. Inc., Minneapolis, MN) was added and incubated at 37°C for 8 h. Supernatant was transferred to a black 96-well plate and the fluorescence activity was measured at 320/405 nm using a Spectro-photometer (Perkin Elmer, Waltham, MA). To normalize the fluorescent activity to cell number, the cells remained in the plate and were treated with a MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma, Saint Louis, MO) solution. After incubation for 4 h, the MTT solution was discarded and the MTT-formazan crystals were dissolved in 150 µL DMSO. The absorbance was measured at 570 nm using a multi-well ELISA automatic Spectrometer (Behring ELISA Processor II, Marburg, Germany). The protease activity was calculated using the following equation: Protease activity = (fluorescence at 320/405 nm)/ absorbance of MTT.
ADAM and HIF-1α knockdown experiment
Small interfering RNA (siRNA) against ADAM9 was purchased from Origene (Rockville, MD, USA) and was also independently designed using the Genescript siRNA Finder (https://www.genscript.com/ssl-bin/app/rnai). HIF-1α siRNA was designed by Genescript siRNA Finder. The ADAM9 and HIF-1α siRNA target sequence are as follows: ADAM9; 5’-GACTGTCGGTTCCTTCCAG-3’ and HIF-1α; 5‘-GTCTGCAACATGGAAGGTA-3’. ADAM-10 and -17 siRNA were supplied from Bioneer Corporation (Daejeon, Korea). Cells were transfected with Lipofectamine MAX (Invitrogen) mixed with 50 nM siRNA. After 48 h, cells were harvested for further analysis.
Matrigel™ trans-well assay
The lower side of an insert (diameter, 6.5 mm; pore size, 8 µm) (Becton Dickinson, Franklin Lakes, NJ) was coated with 8 µg/µL Matrigel. Filters were placed on a 24-well containing media supplemented with 0.1% bovine serum albumin (BSA). Cells were harvested with a cell dissociation solution (Sigma, Saint Louis, MO) and suspended in a medium with 3% BSA. Cells (1×105) were then placed in the upper part of a trans-well chamber and allowed to migrate for 24 h.
For ADAM9 inhibitor treatment, RAV-18 was added to the upper compartment in serial doses. After 24 h, non-invaded cells were swabbed off, and invaded cells on the lower side of the membrane were fixed in 4% paraformaldehyde in PBS and stained with 0.5% crystal violet at room temperature for 10 min. Invasion activity was quantified by counting the cells on three inserts. The data are expressed as the average number of cells per insert.
Adhesion assay
Harvested cells were suspended in media containing 0.1% BSA, and 1×105 cells were seeded into 8 µg/µL Matrigel-precoated 96-well plates that were then incubated for 2 h with or without RAV-18. Media was removed and the attached cells were stained with 0.5% crystal violet at room temperature for 10 min. Stained cells were dissolved in 0.1 M sodium citrate and measured by using ELISA reader at 570 nm.
Survival assay
Cells were seeded into a 96-well plate and incubated at 37°C for 24 h. RAV-18 was serially diluted with media and added to each well. Following 72 h of incubation, 50 µL (2 mg/mL) of MTT solution was added and incubated for an additional 4 h. After centrifugation at 400 ×g for 10 min, the media and MTT were removed from the wells and the remaining MTT-formazan crystals were dissolved by addition of 150 µL DMSO. Following 10 min of shaking incubation, the absorbance at 570 nm was measured with multi-well ELISA automatic spectrometer. Results were expressed as percent cell survival, which was calculated using the following formula: % survival = [(mean absorbance of test wells – standard absorbance) / (mean absorbance of control wells – standard absorbance)] × 100. Control wells were treated with the medium alone (without the drug).
Proliferation assay
Cells (5×104) were seeded in a 24-well plate with or without treatment of RAV-18. Cells were counted every day for seven days. Cellular doubling time (DT) was calculated by the following equation: DT = (Time after cell growth – Time before cell growth) / log2 (Number of cells after growth / Number of cells before growth).
In vivo xenograft model
BALB/c nu/nu mice (female, 7 weeks old, SLC Inc., Shizuoka, Japan) were housed under specific-pathogen-free conditions. Experiments were performed according to the standard guidelines for animal experiments of Yonsei University College of Medicine (Seoul, Korea). The effect of RAV-18 on the xenograft model was examined as follows: 1×107 MKN-28 cells were inoculated subcutaneously (SC) in the flank of the mouse or injected in the peritoneum (IP). The mice were divided into four groups: a control group of SC (PBS i.p., n=7), a RAV-18-treated group of SC (50 mg/kg i.p., 5 times for 2 weeks, n=7), a control group of IP (PBS i.p., n=7), and a RAV-18-treated group of IP (50 mg/kg i.p., 5 times for 2 weeks, n=7). The treatment was started on day 21 after cell inoculation and mice were sacrificed after eight weeks. Tumor volume and body weight were measured twice weekly. The tumor volume was calculated using the formula: volume = length × width × width × 0.5. At the end of the experiment, tumors and peritoneal nodules were collected. The weights of collected samples were measured and the peritoneal nodules were counted.
Immunohistochemistry (IHC)
Tumor specimens were fixed in 10% formaldehyde and embedded in paraffin. All samples were cut into 5-µm-thick sections for IHC. The sections were stained with H&E and immunostained with anti-ADAM9 (1:100), anti-pEGFR (1:200) and anti-pERK (1:100) antibodies at RT for 90 min. The sections were reacted with an EnVision reagent (Dako Co, Japan) for visualization. The results of immunostaining were categorized as follows: staining in less than 10% of the tumor cells was scored as 0; staining in more than 10% of the tumor cells as scored as 1+; weak to moderate staining in more than 10% of the tumor cells was scored as 2+; strong staining in more than 10% of the tumor cells was scored as 3+.
Statistical analysis
Quantitative data were represented as the mean ± standard deviation (SD) of at least three independent experiments. Statistical comparison between groups was done using Student's t-test. Differences were regarded as statistically significant when the p-value < 0.05.
RESULTS
Screening of ADAM9 expression and protease activity in the GC cell panel
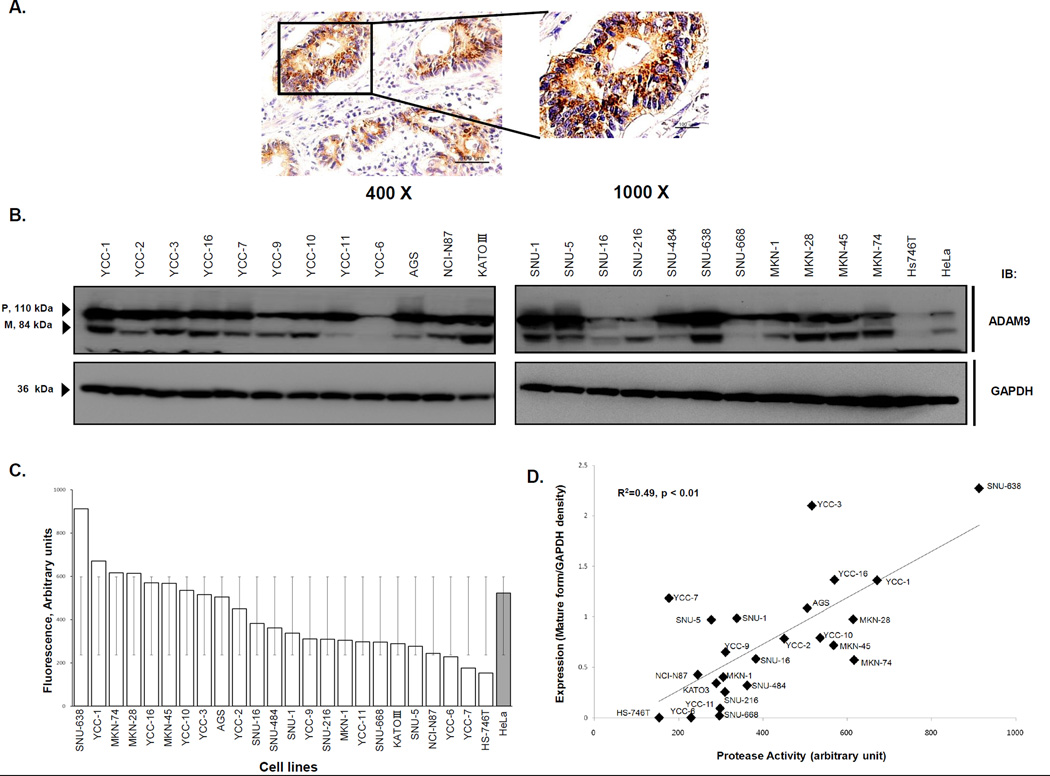
We first carried out immunohistochemistry on paraffin-embedded tumor sections from ten GC patients with tumor infiltrating beyond subserosa (≥ T3). Four of 10 (40%) cancers expressed ADAM9 whereas no expression was found in adjacent non-cancerous tissue. ADAM9 expression was highest in cells along infiltrating margins bordering non-cancerous epithelium, and was located at the membrane and in the cytoplasm (Fig 1A).
Figure 1. ADAM9 protease activity and expression in GC.
(A) ADAM9 expression is shown the cell membrane and cytoplasm in GC tissues. (B) Protein levels of ADAM9 varied in GC cell lines (P: pro-form and M: mature-form). HeLa cell lysate were used in positive control. Additionally, (C) ADAM9 protease activities varied in GC cell lines. (D) ADAM9 protease activities were correlated with protein expressions of ADAM9 mature form in GC cell lines (R2 = 0.49, p < 0.01). High and low activities were distinguished as being outside one standard deviation of the mean of all cell lines’ protease activity.
Then, we screened for ADAM9 expression in 24 GC cell lines. The mRNA levels varied (Fig. S1) and did not correlate with ADAM9-protein expression in the cell lines (R2=0.106, p = 0.203). In immunoblots, ADAM9 was detected as two bands: a pro-form (110 kDa) and mature form (84 kDa) (Fig. 1B)(14). For the further experiments, we selected four cells with high ADAM9 protein expression and protease activity (SNU-638, YCC-1, MKN-74, and MKN-28) and three cells with low ADAM9 protein expression and protease activity (YCC-6, YCC-7, and Hs746T) as being outside of 1-SD of the mean value of protease activity tested (Fig.1C). Protease activity and protein expression of the mature form were correlated each other (Fig. 1D; R2=0.493, p < 0.01).
Effect of ADAM9 suppression in GC cells
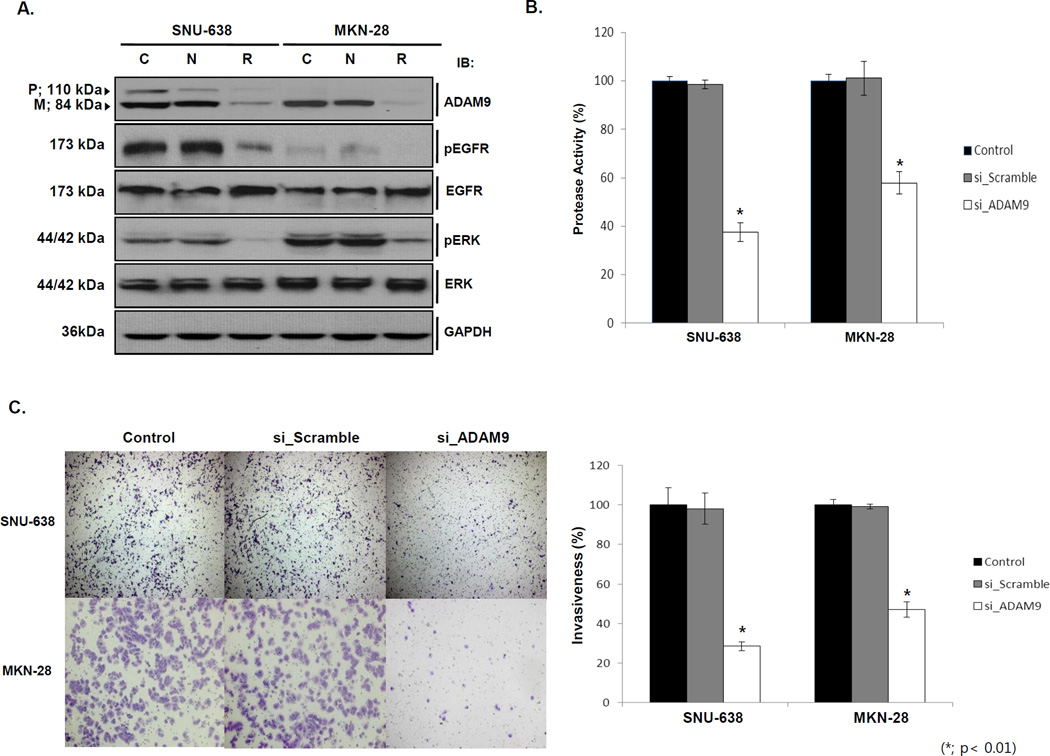
First, we screened four siRNA against ADAM9 and selected the most efficient one for further experiment (Fig S2). In cells with high ADAM9 expression, protein levels were reduced, and its protease activity was also decreased significantly by 61.8±3.9% in SNU-638 and 42.7±4.5% in MKN-28(Fig. 2A and B; p < 0.01). The knockdown of ADAM9 led to decline in phospho-EGFR and phospho-ERK (Fig. 2A), Then, we examined the invasion after siADAM9 transfection. Cells transfected with siADAM9 showed a reduced invasion activity through Matrigel compared to control cells by 71.4±2.2% in SNU-638 and by 53.0±3.9% in MKN-28 (Fig. 2C; p < 0.01), which suggests that the decreased invasion may be a consequence of inhibited EGFR/ERK signaling.
Figure 2. Effects of ADAM9 expression modulation by transient ADAM9 inhibition to protease activity and cell invasiveness.
(A) When siADAM9 was transfected to high ADAM9 cell lines, SNU-638 and MKN-28, ADAM9 expression was decreased. Phospho-EGFR and Phospho-ERK expression also decreased when siADAM9 was transfected to high ADAM9 cell lines. (C: control, N: scrambled siRNA, and R: siADAM9) (B) Protease activity was significantly decreased in siADAM9 transfected cells (62.9±3.9%; SNU-638 and 43.7±.5%; MKN-28 at siADAM9 transfection, p < 0.001). (C) When high ADAM9 cells were transfected with siRNA to ADAM9, invasiveness decreased (71±2.2%; SNU-638 and 53±3.9%; MKN-28 at siADAM9 transfection, p < 0.01).
Effect on the proliferation and invasion by inhibiting protease activity of ADAM9
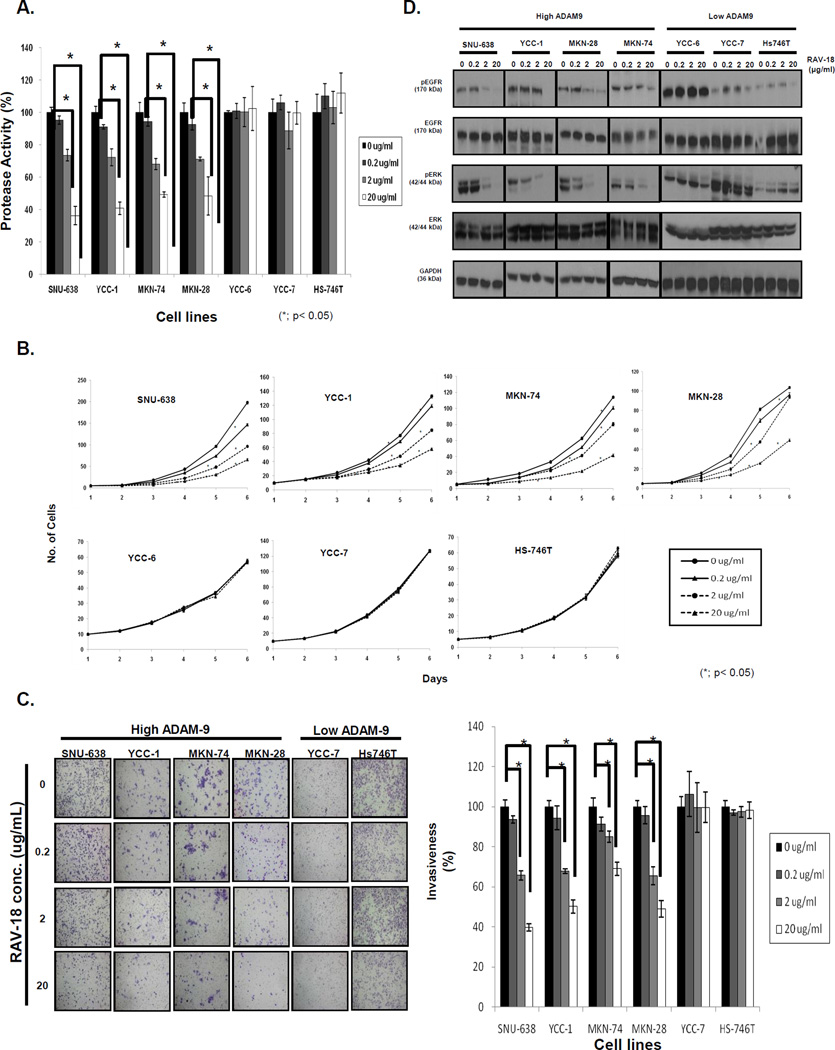
ADAM9 is a proteolytically active enzyme and the protease activity is currently the well-defined function of ADAMs, with most of the putative substrates being trans-membrane proteins. Therefore, we focused on the proteolytic domain of ADAM9 as a biomarker and therapeutic target using RAV18, an ADAM9-specific blocking antibody, for our studies. GC cells with high-expression of ADAM9 treated with RAV-18 showed a dose-dependent decrease in ADAM9 protease activity (Fig. 3A; 68.7±0.8% in SNU-638, 62.1±2.6% in MKN-28 and 50.3±2.1% in MKN-74 at 20 µg/mL RAV-18, p < 0.05), however there was no decrease in ADAM9 protease activity in cells with low expression of ADAM9.
Figure 3. Changes in ADAM9 protease activities, invasion, and proliferation with treatment of RAV-18.
(A) When cells with high ADAM9 protease activity were treated with RAV-18, a dose-dependent decrease in protease activity was seen (from SNU-638 to MKN-28). However, cell lines with low ADAM9 protease activity showed no change in protease activities (from YCC-6 to Hs746T). (B) Cell lines with high ADAM9 activity treated with RAV-18 showed dose-dependent inhibition of proliferation. However, the proliferation of cell lines with low ADAM9 activity was not inhibited. (C) RAV-18 also dose-dependently reduced invasiveness in cell lines with high ADAM9 activity, but did not influence invasiveness of low activity cells (*; p < 0.05). (D) When cell lines were treated with RAV-18, pEGFR and pERK expression was dose-dependently decreased in cell lines expressing high ADAM9; low-expressing cell lines did not demonstrate a change.
Next, we investigated the effect of RAV-18 on cell proliferation. For SNU-638 cells, whose baseline doubling time (DT) was 22.6±0.1 h, after treatment with 20 µg/mL RAV-18, DT was prolonged to 32.3±0.5 h (p < 0.01). Similarly, proliferation was significantly delayed with other cells (YCC-1: 25.4±0.2 vs. 33.9±0.4 h; MKN-28: 27.4±0.1 vs. 36.3±0.5 h; MKN-74: 27.7±0.2 vs. 39.3±0.8 h, p < 0.05). However, RAV18 did not affect proliferation in cells with low ADAM9 expression (Fig. 3B).
We investigated the effect of RAV-18 on cell adhesion. At a dose of 20 µg/mL RAV-18, adhesion to Matrigel was unchanged in any of cells tested (Fig. S3). However, high ADAM9 expressing cells demonstrated reduced invasion by 60% in SNU-638, 47% in YCC-1, 31% in MKN-74, and 51% in MKN-28. (Fig. 3C, p < 0.05), however there was no effect of RAV-18 in the low ADAM9 expressing cells. These findings can transfer that ADAM9 regulates cancer proliferation and invasion, which is dependent on its protease activity.
Phospho-EGFR and phospho-ERK were decreased in cells that responded to RAV18, which accords to the results of siADAM9 transfection. There was no effect of RAV-18 on phospho-EGFR and phospho-ERK signaling in the low-ADAM9 cells that did not respond to RAV-18 (Fig. 3D).
Effect of hypoxia on ADAM9 protease activity and cancer invasion
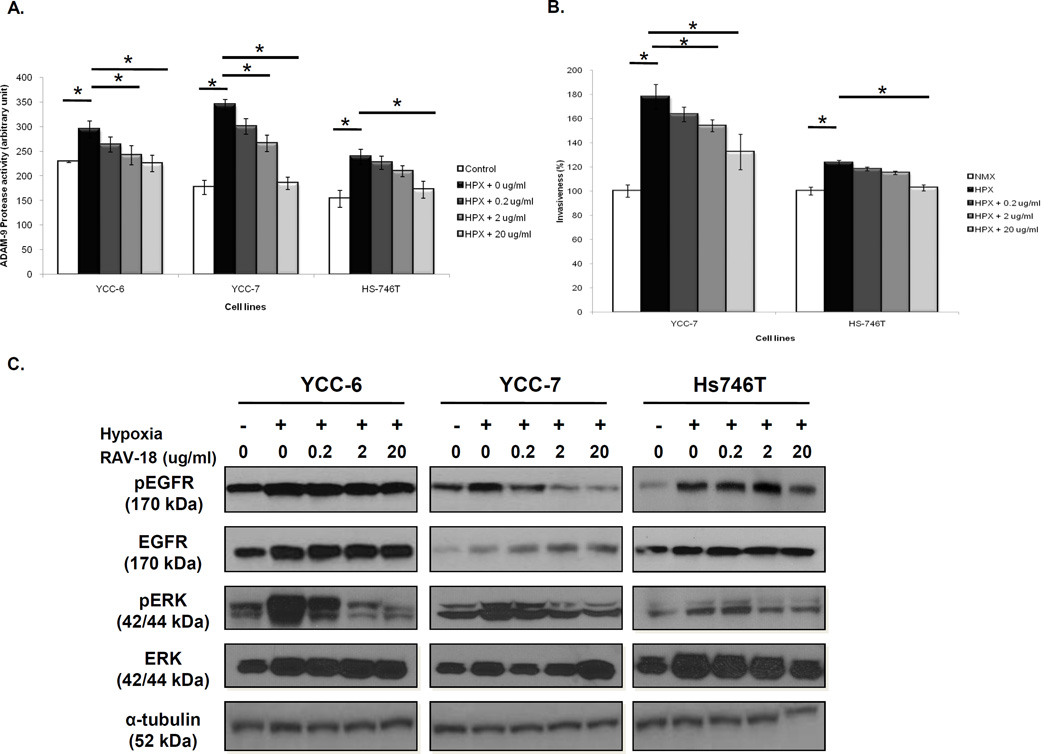
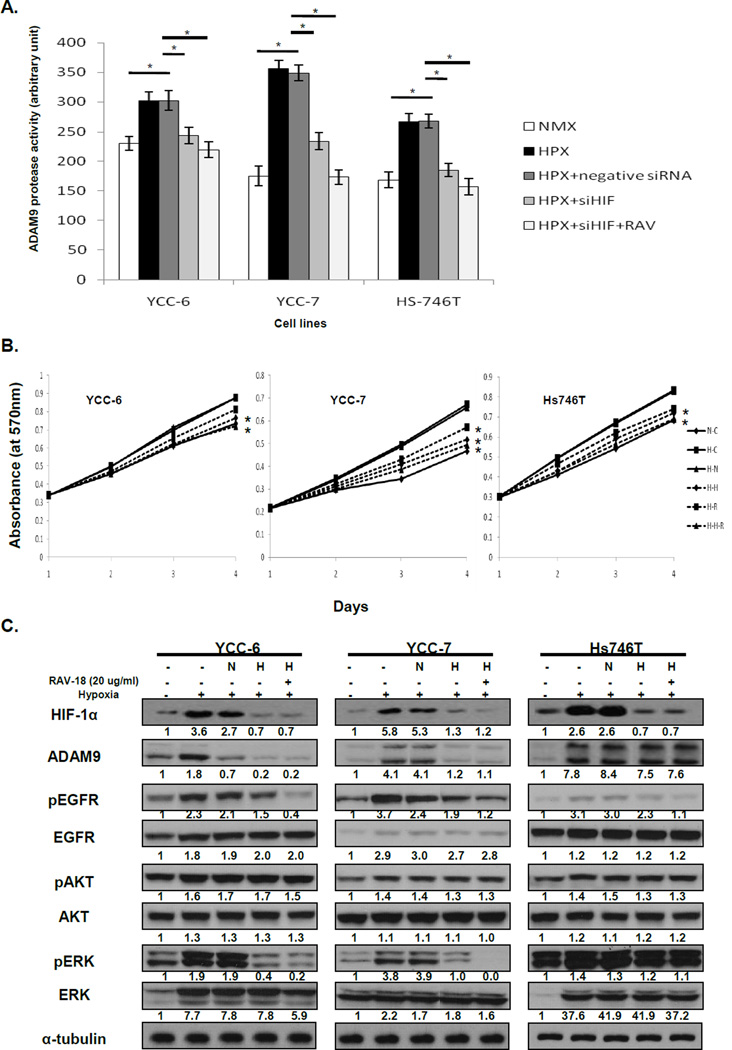
Our hypothesis is that the effect of RAV-18 depends on its inhibition of ADAM9 proteolytic activity and if so, cells with low ADAM9 should respond to RAV-18 if ADAM9 is induced. We investigated the influence of hypoxia on the constitutively low ADAM9 expressing cells and found that exposure to 1% O2 for 24 h increased ADAM9 protease activity in YCC-6, YCC-7 and Hs746T cells, and this was returned to non-hypoxia levels by RAV-18 treatment in a dose dependent manner (Fig. 4A).
Figure 4. Modulation of ADAM9 protease activity, invasion, and downward signaling in GC cells under hypoxic condition.
(A) ADAM9 protease activities were increased in hypoxic conditions. When treated with RAV-18, ADAM9 the protease activities were dose-dependently decreased. (B) Invasiveness also increased in hypoxia and was decreased following RAV-18 treatment (*; p < 0.05). (C) After exposure to hypoxia, EGFR and ERK expression was increased in all low ADAM9 cell lines. Additionally, phospho-EGFR and phospho-ERK expressions were dose-dependently decreased when cells received RAV-18 treatment.
We also found that the invasiveness of cells with low ADAM9 cells was increased under hypoxic conditions (by 24% in YCC-7 and 13% in Hs746T, p < 0.05). RAV-18 treatment significantly reduced the invasiveness in hypoxia (by 30% in YCC-7 and 14% in Hs746T at 20 µg/mL RAV-18, p < 0.05) (Fig. 4B).
When these cells were incubated under hypoxic conditions, phospho- and total EGFR and ERK expressions were, as expected, induced. Additionally, treatment of RAV-18 abrogated the EGFR and ERK phosphorylation (Fig. 4C). Taken together, these findings suggest that hypoxia, a feature present in many solid tumors including GC, increases ADAM9 expression and activity, together with an increased invasion, that may contribute to the growth and spread of GC.
Relationship with ADAM9 and hypoxia
In order to obtain further information on the relationship between ADAM9 and hypoxia, we treated low ADAM9 cells with HIF-1α siRNA under hypoxic condition. ADAM9 protease activity was significantly decreased when HIF-1α was suppressed, and on double siRNA inhibition of HIF-1α and ADAM9, there was an additional effect in suppressing ADAM9 activity to normoxic levels (Fig. 5A, p < 0.05). Under hypoxic conditions, cell proliferation was also increased and this effect was prevented with the treatment RAV-18 and HIF-1α siRNA (Fig. 5B).
Figure 5. HIF-1α regulates the ADAM9 expression under hypoxic condition.
(A) Hypoxia dependent ADAM9 protease activity was decreased when cells were treated with HIF-1α siRNA under hypoxic condition. HIF-1α siRNA and RAV-18 combined to inhibited ADAM9 protease activity more than HIF-1α siRNA treatment alone (*; p < 0.05). NMX; Normoxia, HPX; Hypoxia, siHIF; siRNA against HIF1α and RAV; RAV-18 (B) Cell proliferation was increased in low ADAM9 cells under hypoxic condition and decreased by HIF-1α siRNA transfection of cells, (*; p < 0.05). N-C; Control on Normoxia, H-C; control on Hypoxia, H-N; negative siRNA on Hypoxia, H-H; siRNA against HIF1α on hypoxia, H-R; RAV-18 on hypoxia and H-H-R; combination of siHIF1α and RAV-18 on hypoxia (C) Increased expression of HIF-1α as well as ADAM9 and its down-stream signaling was inhibited by HIF-1α siRNA transfection. When HIF-1α siRNA and RAV-18 were used in combination, HIF-1α expression was not changed comparing to HIF-1α siRNA treatment alone. However, phospho-EGFR and phosph-ERK expression were decreased more than HIF-1α siRNA treatment alone. Phospho-AKT expression was not changed when HIF-1α siRNA, with or without RAV-18 was used. N; negative siRNA and H; siRNA against HIF1α
In immunoblots, we detected that ADAM9 and HIF-1α expression in the cells were increased under hypoxic condition. When HIF-1α siRNA were transfected, ADAM9 expression and EGFR and ERK activity was decreased, while AKT activity was unchanged. On double inhibition of HIF-1α and ADAM9 these effects were pronounced, while HIF-1α expression was not different compared to HIF-1α siRNA alone (Fig. 5C). Thus, hypoxia and HIF-1α induction may be a factor that regulates the ADAM9 expression and its down-stream signaling which lead to cancer progression.
In vivo experiment of RAV-18 for therapeutic effect
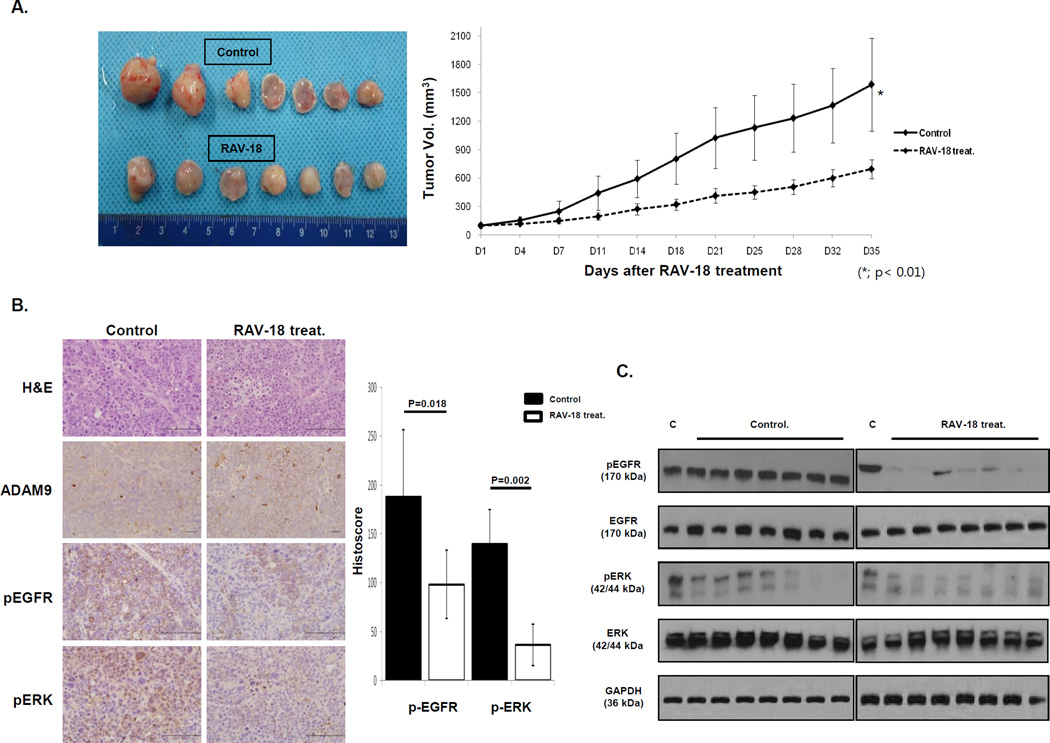
To investigate whether ADAM9 protease activity was related to cancer progression in vivo, we conducted a xenograft study using BALB/c nu/nu mice. Mice were inoculated subcutaneously with MKN-28 (high ADAM9 protease activity) cells. At three weeks after inoculating the cells, when the tumor volumes reached around 100 mm3, mice were split into two groups and treated with either saline or 50 mg/Kg RAV-18 intraperitoneally for two weeks. The RAV-18-treated group showed suppressed tumor growth compared to the saline treated group (mean tumor volume: 692±219 vs. 1,568±557 mm3, p < 0.01) (Fig. 6A).
Figure 6. In vivo xenograft model of ADAM9.
(A) Photographs of tumors from control and RAV-18 treated groups. Tumor volumes increased in both control and RAV-18 treated groups, however, the tumor volumes of the control group were larger than in the RAV-18 treated group (p < 0.01). (B) Representative phospho-EGFR and phospho-ERK immunostaining (original images were captured at 400× magnification) and H&E staining. Average of phospho-EGFR histoscores in the control group was approximately two times higher than in the RAV-18 treated group (p = 0.018). Furthermore, the average phospho-ERK histoscores in controls was approximately three times higher than in RAV-18 treated groups (p = 0.002). (C) Western blot analysis showed that phospho-EGFR and phospho-ERK expressions were lower in the RAV-18 treated group. C: MKN-28 cell whole lysate.
Subsequently, MKN-28 cells were inoculated intraperitoneally into mice. After three weeks, saline and RAV-18 were administered as above. At five weeks of treatment, tumor nodules were significantly smaller in the RAV-18 group compared to the control group; the average tumor nodules in RAV-18-treated group were 58.3±17.8, while those of the control were 96.3±19.6, indicating a 40% reduction of tumor formation (Fig. S4, p = 0.0001). Immunoblot showed a decrease of phospho-EGFR and phospho-ERK expression following RAV-18 treatment as had been seen in vitro studies. The average p-EGFR histoscore of the control group was 188.1±68.4, while that of the RAV-18 group was 98.6±34.9 (p = 0.018). The average p-ERK histoscore was 139.3±35.7 in the control group and 63.4±21.2 in the RAV-18 group (p = 0.002) (Fig. 6B and C). These findings suggest that ADAM9 protease activity regulates GC tumorigenesis, including in a model for intraperitoneal tumor spread, and may be a therapeutic target in GC.
DISCUSSION
In this study, we have demonstrated that ADAM9 is expressed in GC cells, which related to cancer proliferation and invasion. Additionally, using the protease domain-specific blocking antibody RAV-18 we showed that this effect depends on the protease activity of ADAM9. The level of the ADAM9 enzymatic activity in cells with low ADAM9 expression is induced by hypoxia, and inhibited by RAV-18. We further showed that ADAM9 protease activity regulates GC tumorigenesis, including intraperitoneal metastatic spread. Taken together, these findings suggest that inhibition of ADAM9 activity is a rational therapeutic approach to suppress GC progression and metastasis.
There has been an earlier report demonstrating that ADAM9 transcription is increased in GC(12). Although little is known about the regulatory mechanism of ADAM9, some recent evidences suggest a role for post-translational modification is more important. In one prostate cancer model, induction of ADAM9 is regulated by stress through the accumulation of reactive oxygen species (ROS) as a common mediator, and removing ROS, or the use of antioxidants resulted in a marked reduction of ADAM9(11). These findings suggest a link between stress-induced signaling and ADAM9, with ROS serving as a common mediator(15). We have shown that another stress factor, hypoxia, a common feature found in many solid tumors, leads to an increase in the expression of ADAM9 protein alongside enzymatic activity in HIF-1 dependent way. We found that post-translational modification and processing to a “mature” cleaved (84 kDa) form, is correlated with its enzymatic activity. Although all ADAMs possess the MMP-like domain adjacent to the pro-domain, only 50% exhibit protease activity. Currently, protease activity is the best-defined function of ADAMs, with most putative substrates currently identified as being trans-membrane proteins(16).
One of the limitations of our study is the substrate we used for ADAM9 protease activity is not only specific to ADAM9, also reactive to other ADAMs - especially ADAM10 and ADAM17(17–21). As in Fig. S5, [1] changes of protease activity after siRNA knock-down is dependent on basal expression level of each ADAM, and [2] when co-treated with RAV-18, additive effect is prominent only in high ADAM9-expressing cells, implying that RAV-18 works specifically to ADAM9.
Several explanations are possible about how ADAM9 plays a role in cancer progression. There is evidence that ADAM9 mediates the release of growth factor ligands, which regulate EGFR signaling. In the stomach, trans-activation of the EGFR by peptides such as gastrin, angiotensin II, bradykinin, bombesin, or substance P, is mediated by shedding of EGFR-ligands by ADAMs(22, 23). ADAM9 also releases some trans-membrane protein-derived ligands, such as heparin-binding-EGF, amphiregulin, TGFα, and TNFα(24, 25). We herein found that ADAM9 knock-down blocks EGFR phosphorylation and down-stream ERK activation, and that RAV-18 inhibited tumor growth, in line with EGFR and ERK phosphorylation in vivo. Thus, inhibition of EGFR ligand shedding by ADAM9 with subsequent deceased EGFR activation and down-stream signaling could be a mechanism for the antitumor activity of RAV-18.
It is intriguing that hypoxia induces ADAM9 activity, relating to cancer cell proliferation and invasion, which is reversed by RAV-18 although not beyond at the levels in normoxia. This does not appear to be related to the processing of ADAM9 to the cleaved form as this was detected in both low and high ADAM9 expressing cell lines. We speculate that there are two forms of ADAM9, one not responsive to inhibition by RAV-18, which is perhaps intracellular located or masked, and another induced by hypoxia that is accessible to inhibition by RAV-18, that is responsible for the growth stimulatory and invasive effects of ADAM9. Another point is that ADAM9 was induced by hypoxia in Hs746T cells but its downstream signaling was not altered by HIF-1α knock-down or RAV-18 treatment. It is possible that Hs746T is more dependent on other ADAM proteins – such as ADAM10 and ADMA17 – rather than ADAM9 for its biological activity, as shown in Fig. S2.
We showed that proliferation and invasion is affected by RAV18, but not adhesion. The disintegrin domain is found in all ADAMs and is located downstream of the metalloproteinase domain. We believe that adhesion proteins are important for cellular adhesion, migration, and signal transduction(26). However most information on the binding of disintegrin to integrin comes from in vitro studies using recombinant disintegrin domains(16). Thus, the biological relevance of our finding that RAV-18 does not alter adhesion is unclear at this time.
Our in vivo studies demonstrate that ADAM9-targeting antibody is a reasonable option for treatment of GC. One limitation is a still lack of information on the actual incidence of ADAM9 expression in large-scale clinical samples. Our results suggest that only high level ADAM9 expressing tumors, and those where ADAM9 is induced in response to hypoxia will be responsive to ADAM9 antibody. The subcellular location of ADAM9 could also play a role in the response to antibody treatment.
Collectively, our work shows that ADAM9 plays an important role in GC proliferation and invasion, and that while expressed at high levels in some cancer cells that are responsive to functional inhibition and antitumor activity of a catalytic site directed antibody, other GC cells have low levels of expression and only when exposed to hypoxia do ADAM9 levels increase and the cells become responsive to anti-ADAM9 treatment. These finding suggest that ADAM9 could be an effective therapeutic target for therapy of metastatic GC.
Supplementary Material
Acknowledgments
We thank Macrogenics Incorporation for generously supporting the ADAM9 inhibitor RAV-18.
Funding
For this study, H.C. Jeung received a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1420060). G. Powis was supported by grants CA163541 and CA172670 from the National Cancer Institute. J.M. Kim, H.C. Jeung, S.Y. Rha, T.S. Kim, K.H. Park, and H.C. Chung received the Public Welfare and Safety Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0020841).
Footnotes
Disclosure of conflicts of interest
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Albelda SM. Role of integrins and other cell adhesion molecules in tumor progression and metastasis. Laboratory investigation. 1993;68:4–17. [PubMed] [Google Scholar]
- 2.Nicolson GL. Tumor and host molecules important in the organ preference of metastasis. Seminars in cancer biology. 1991;2:143–154. [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Becherer JDBC. Biochemical properties and functions of membrane-anchored metalloprotease-disintegrin proteins (ADAMs) Current topics in developmental biology. 2003;54:101–123. doi: 10.1016/s0070-2153(03)54006-6. [DOI] [PubMed] [Google Scholar]
- 5.Grützmann R, Lüttges J, Sipos B, Ammerpohl O, Dobrowolski F, Alldinger I, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. British Journal of Cancer. 2004;90:1053–1058. doi: 10.1038/sj.bjc.6601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirao T, Nanba D, Tanaka M, Ishiguro H, Kinugasa Y, Doki Y, et al. Overexpression of ADAM9 enhances growth factor-mediated recycling of E-cadherin in human colon cancer cell line HT29 cells. Experimental cell research. 2006;312:331–339. doi: 10.1016/j.yexcr.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 7.Lendeckel U, Kohl J, Arndt M, Carl McGrath S, Donat H, Röcken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. Journal of cancer research and clinical oncology. 2005;131:41–48. doi: 10.1007/s00432-004-0619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada D, Ohuchida K, Mizumoto K, Ohhashi S, Yu J, Egami T, et al. Increased expression of ADAM 9 and ADAM 15 mRNA in pancreatic cancer. Anticancer research. 2007;27:793–799. [PubMed] [Google Scholar]
- 9.Edwards DR, Handsley M, Pennington CJ. The ADAM metalloproteinases. Molecular aspects of medicine. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer OM, Hart S, Gschwind A, Prenzel N, Ullrich A. Oxidative and osmotic stress signaling in tumor cells is mediated by ADAM proteases and heparin-binding epidermal growth factor. Molecular and cellular biology. 2004;24:5172–5183. doi: 10.1128/MCB.24.12.5172-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung SY, Kubo H, Shigemura K, Arnold RS, Logani S, Wang R, et al. Oxidative stress induces ADAM9 protein expression in human prostate cancer cells. Cancer research. 2006;66:9519–9526. doi: 10.1158/0008-5472.CAN-05-4375. [DOI] [PubMed] [Google Scholar]
- 12.Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rcken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. International journal of oncology. 2005;26:17–24. [PubMed] [Google Scholar]
- 13.Ciss MA, Gandreuil C, Hernandez J, Martinez J, Checler F, Vincent B. Design and characterization of a novel cellular prion-derived quenched fluorimetric substrate of alpha-secretase. Biochemical and biophysical research communications. 2006;347:254–260. doi: 10.1016/j.bbrc.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 14.Roghani M, Becherer JD, Moss ML, Atherton RE, Erdjument-Bromage H, Arribas J, et al. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. The Journal of biological chemistry. 1999;274:3531–3540. doi: 10.1074/jbc.274.6.3531. [DOI] [PubMed] [Google Scholar]
- 15.Shigemura K, Sung SY, Kubo H, Arnold RS, Fujisawa M, Gotoh A, et al. Reactive oxygen species mediate androgen receptor- and serum starvation-elicited downstream signaling of ADAM9 expression in human prostate cancer cells. The Prostate. 2007;67:722–731. doi: 10.1002/pros.20565. [DOI] [PubMed] [Google Scholar]
- 16.Duffy MJ, Mullooly M, O'Donovan N, Sukor S, Crown J, Pierce A, et al. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clinical Proteomics. 2011;8:9. doi: 10.1186/1559-0275-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canault M, Leroyer AS, Peiretti F, Lesèche G, Tedgui A, Bonardo B, et al. Microparticles of human atherosclerotic plaques enhance the shedding of the tumor necrosis factor-alpha converting enzyme/ADAM17 substrates, tumor necrosis factor and tumor necrosis factor receptor-1. The American journal of pathology. 2007;171:1713–1723. doi: 10.2353/ajpath.2007.070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiss K, Cornelsen I, Husmann M, Gimpl G, Bhakdi S. Unsaturated fatty acids drive disintegrin and metalloproteinase (ADAM)-dependent cell adhesion, proliferation, and migration by modulating membrane fluidity. Journal of biological chemistry. 2011;286:26931–26942. doi: 10.1074/jbc.M111.243485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barsoum IB, Hamilton TK, Li X, Cotechini T, Miles EA, Siemens DR, et al. Hypoxia induces escape from innate immunity in cancer cells via increased expression of ADAM10: role of nitric oxide. Cancer research. 2011;71:7433–7441. doi: 10.1158/0008-5472.CAN-11-2104. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, He L, Yuan P, Wang P, Lu Y, Tong F, et al. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncology Reports. 2012;28:1709–1718. doi: 10.3892/or.2012.2003. [DOI] [PubMed] [Google Scholar]
- 21.Shou Z, Jin X, Zhao Z. Upregulated expression of ADAM17 is a prognostic marker for patients with gastric cancer. Annals of surgery. 2012;256:1014–1022. doi: 10.1097/SLA.0b013e3182592f56. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki Y, Shinomura Y, Tsutsui S, Zushi S, Higashimoto Y, Kanayama S, et al. Gastrin induces heparin-binding epidermal growth factor-like growth factor in rat gastric epithelial cells transfected with gastrin receptor. Gastroenterology. 1999;116:78–89. doi: 10.1016/s0016-5085(99)70231-3. [DOI] [PubMed] [Google Scholar]
- 23.Park JW, Baek NS, Lee SC, Oh SJ, Jang SH, Kim IH, et al. Preclinical efficacy testing for stomach and liver cancers. Cancer Research and Treatment. 2014;46:186–193. doi: 10.4143/crt.2014.46.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi Y, Hirata M, Hasuwa H, Iwamoto R, Umata T, Miyado K, et al. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. The EMBO journal. 1998;17:7260–7272. doi: 10.1093/emboj/17.24.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schfer B, Gschwind A, Ullrich A. Multiple G-protein-coupled receptor signals converge on the epidermal growth factor receptor to promote migration and invasion. Oncogene. 2004;23:991–999. doi: 10.1038/sj.onc.1207278. [DOI] [PubMed] [Google Scholar]
- 26.Stupack DG. The biology of integrins. Oncology. 2007;21:6–12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.